Biotin Auxotrophy and Biotin Enhanced Germ Tube Formation in Candida albicans
Abstract
:1. Introduction
2. Materials and Methods
2.1. C. albicans Strains and Growth Conditions
2.2. Analysis of Germ Tube Formation
2.3. Analysis of Biotin Auxotrophy
2.4. Effects of CO2 on Biotin Auxotrophy
2.5. Gene Expression Analysis
2.6. Mitochondrial Targeting Peptides (mTPs) Predictions
3. Results
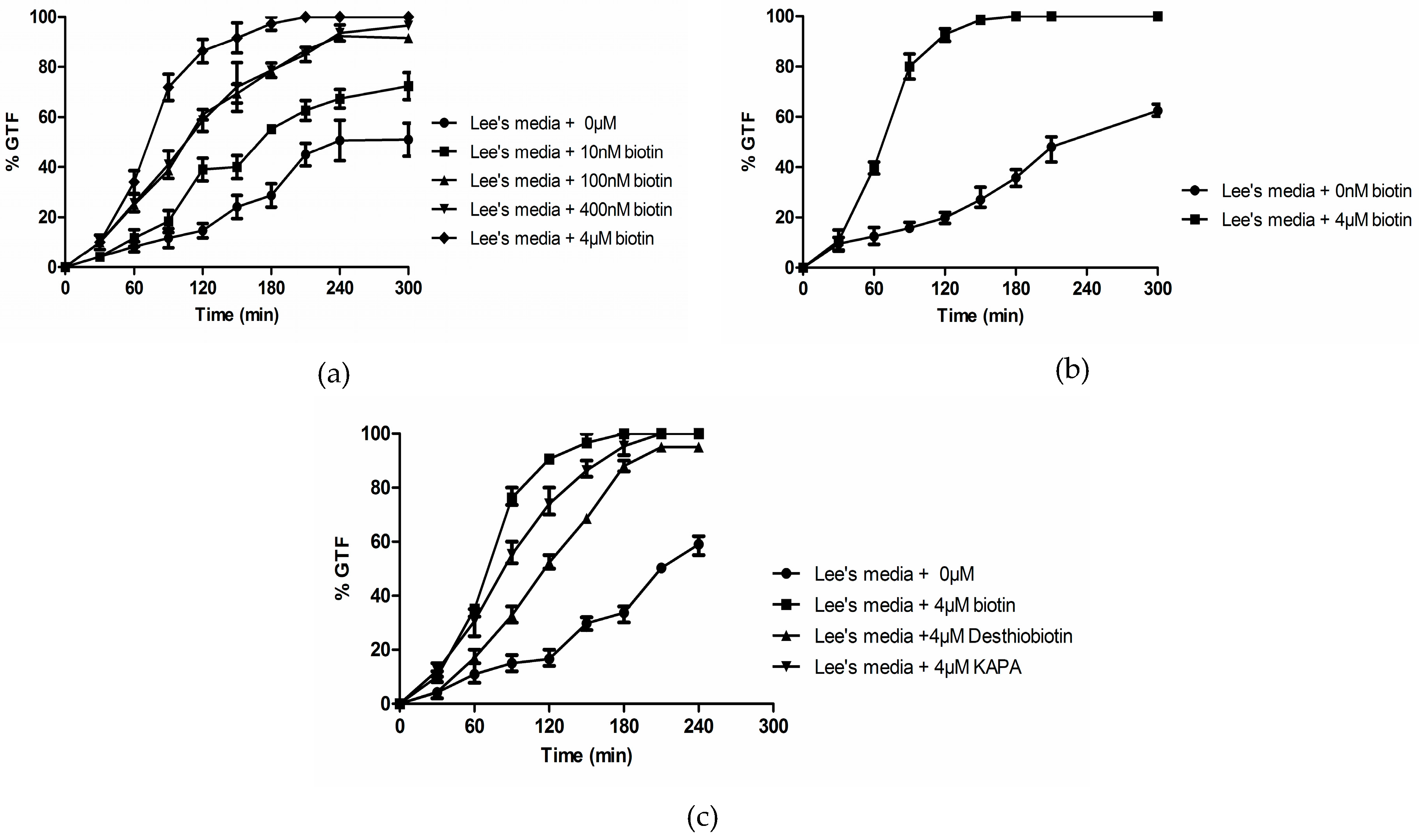
3.1. Biotin Senhances Germ Tube Formation in both GPP and Lee’s Medium
3.2. Biotin Enhances Germ Tube Formation in Serum
3.3. Biotin Does Not Enhance Germ Tube Formation in N-Acetylglucosamine
3.4. Effects of Biotin on ∆efg1, ∆cph1, and ∆cdc35 Deletion Mutants
3.5. Biotin Starvation Upregulates BIO2, BIO3, BIO4, and Biotin Protein Ligase BPL1
3.6. Biotin Auxotrophy and Carbon Dioxide
3.7. C. albicans 6713
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Odds, F.C. Candida and candidosis: A review and bibliography. J. Basic Microb. 1988, 30, 382–383. [Google Scholar]
- Lee, K.L.; Buckley, H.R.; Campbell, C.C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 1975, 13, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Mattia, E.; Carruba, G.; Angiolella, L.; Cassone, A. Induction of germ tube formation by N-acetyl-d-glucosamine in Candida albicans: Uptake of inducer and germinative response. J. Bacteriol. 1982, 152, 555–562. [Google Scholar] [PubMed]
- Simonetti, N.; Strippoli, V.; Cassone, A. Yeast-mycelial conversion induced by N-acetyl-d-glucosamine in Candida albicans. Nature 1974, 250, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Land, G.A.; McDonald, W.C.; Stjernholm, R.L.; Friedman, T.L. Factors affecting filamentation in Candida albicans: Relationship of the uptake and distribution of proline to morphogenesis. Infec. Immun. 1975, 11, 1014–1023. [Google Scholar]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microb. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, R.; Hornby, J.M.; Nickerson, K.W. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Ch. 2004, 48, 2350–2354. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.A.; Sciascia, Q.L.; Sanders, R.J.; Norris, G.E.; Edwards, P.J.; Sullivan, P.A.; Farley, P.C. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 2004, 150, 3041–3049. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Lee, R.T.; Fang, H.M.; Wang, Y.M.; Li, R.; Zou, H.; Zhu, Y.; Wang, Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase cyr1p. Cell. Host Microbe 2008, 4, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Hasim, S.; Tati, S.; Madayiputhiya, N.; Nandakumar, R.; Nickerson, K.W. Histone biotinylation in Candida albicans. FEMS Yeast Res. 2013, 13, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Kogl, F.; Tonnis, B. Über das bios-problem. Darstellung von krystallisiertem biotin aus eigelb. Hoppe Seyler’s Z. Physiol. Chem. 1936, 242, 43–73. (In German) [Google Scholar] [CrossRef]
- Suomalainen, H.; Oura, E. Yeast nutrition and solute uptake. In The Yeasts; Rose, A.H., Harrison, J.S., Eds.; Academic Press London: New York, NY, USA, 1971; pp. 3–74. [Google Scholar]
- Hall, C.; Dietrich, F.S. The reacquisition of biotin prototrophy in saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 2007, 177, 2293–2307. [Google Scholar] [CrossRef] [PubMed]
- Phalip, V.; Kuhn, I.; Lemoine, Y.; Jeltsch, J.M. Characterization of the biotin biosynthesis pathway in saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene 1999, 232, 43–51. [Google Scholar] [CrossRef]
- McVeigh, I.; Bell, E. The amino acid and vitamin requirements of Candida albicans Y-475 and Mycoderma vini Y-939. Bull. Torrey Botan. Club 1951, 134–144. [Google Scholar] [CrossRef]
- Firestone, B.Y.; Koser, S.A. Growth promoting effect of some biotin analogues for Candida albicans. J. Bacteriol. 1960, 79, 674–676. [Google Scholar] [PubMed]
- Inglis, D.O.; Arnaud, M.B.; Binkley, J.; Shah, P.; Skrzypek, M.S.; Wymore, F.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida genome database incorporates multiple Candida species: Multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucl. Acids Res. 2012, 40, D667–D674. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.K.; Nickerson, K.W. Nutritional control of dimorphism inceratocystis ulmi. Exp. Mycol. 1981, 5, 148–154. [Google Scholar] [CrossRef]
- Yamaguchi, H. Mycelial development and chemical alteration of Candida albicans from biotin insufficiency. Sabouraudia 1974, 12, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Gillum, A.M.; Tsay, E.Y.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984, 198, 179–182. [Google Scholar]
- Lo, H.J.; Kohler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef]
- Rocha, C.R.; Schroppel, K.; Harcus, D.; Marcil, A.; Dignard, D.; Taylor, B.N.; Thomas, D.Y.; Whiteway, M.; Leberer, E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell. 2001, 12, 3631–3643. [Google Scholar] [CrossRef] [PubMed]
- Kohrer, K.; Domdey, H. Preparation of high molecular weight RNA. Methods Enzymol. 1991, 194, 398–405. [Google Scholar] [PubMed]
- Navarathna, D.H.; Roberts, D.D. Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radic. Biol. Med. 2010, 49, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
- Pendrak, M.L.; Chao, M.P.; Yan, S.S.; Roberts, D.D. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J. Biol. Chem. 2004, 279, 3426–3433. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Horton, P. Psort: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–36. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Casadio, R. TPpred2: Improving the prediction of mitochondrial targeting peptide cleavage sites by exploiting sequence motifs. Bioinformatics 2014, 30, 2973–2974. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Method. Enzymol. 2002, 350, 3–41. [Google Scholar]
- Mosel, D.D.; Dumitru, R.; Hornby, J.M.; Atkin, A.L.; Nickerson, K.W. Farnesol concentrations required to block germ tube formation in Candida albicans in the presence and absence of serum. Appl. Environ. Microb. 2005, 71, 4938–4940. [Google Scholar] [CrossRef] [PubMed]
- Mock, D.M.; Lankford, G.L.; Mock, N.I. Biotin accounts for only half of the total avidin-binding substances in human serum. J. Nutr. 1995, 125, 941–946. [Google Scholar] [PubMed]
- Nyalala, J.O.; Livaniou, E.; Leondiadis, L.; Evangelatos, G.P.; Ithakissios, D.S. Indirect enzyme-linked method for determining biotin in human serum. J. Immunoassay 1997, 18, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Navarathna, D.H.; Hornby, J.M.; Hoerrmann, N.; Parkhurst, A.M.; Duhamel, G.E.; Nickerson, K.W. Enhanced pathogenicity of Candida albicans pre-treated with subinhibitory concentrations of fluconazole in a mouse model of disseminated candidiasis. J. Antimicrob. Chemot. 2005, 56, 1156–1159. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kohler, J.; Fink, G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef] [PubMed]
- Doedt, T.; Krishnamurthy, S.; Bockmuhl, D.P.; Tebarth, B.; Stempel, C.; Russell, C.L.; Brown, A.J.; Ernst, J.F. Apses proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell. 2004, 15, 3167–3180. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.A.; O’Gaora, P.; Byrne, K.P.; Butler, G. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Lichstein, H.C. Microbial nutrition. Annu. Rev. Microbiol. 1960, 14, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Samols, D.; Thornton, C.G.; Murtif, V.L.; Kumar, G.K.; Haase, F.C.; Wood, H.C. Evolutionary conservation among biotin enzymes. J. Bol. Chem. 1988, 263, 6461–6464. [Google Scholar]
- Bahn, Y.S.; Muhlschlegel, F.A. CO2 sensing in fungi and beyond. Curr. Opin. Microbiol. 2006, 9, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Sims, W. Effect of carbon dioxide on the growth and form of Candida albicans. J. Med. Microbiol. 1986, 22, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H. Dimorphism in Candida albicans. I. Morphology-dependent changes in cellular content of macromolecules and respiratory activity. J. Gen. Appl. Microbiol. 1974, 20, 87–99. [Google Scholar] [CrossRef]
- Yamaguchi, H. Effect of biotin insufficiency on composition and structure of cell wall of Candida albicans in relation to its mycelial morphogenesis. J. Gen. Appl. Microbiol. 1974, 20, 271–228. [Google Scholar] [CrossRef]
- Shapiro, R.S.; Uppuluri, P.; Zaas, A.K.; Collins, C.; Senn, H.; Perfect, J.R.; Heitman, J.; Cowen, L.E. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 2009, 19, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Stokes, C.; Moran, G.P.; Spiering, M.J.; Cole, G.T.; Coleman, D.C.; Sullivan, D.J. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet. Biol. 2007, 44, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, V.; Pugliese, A.; Gioannini, P. Growth of Candida albicans in a minimal synthetic medium without biotin. Mycopathologia 1987, 100, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Zakikhany, K.; Naglik, J.R.; Schmidt-Westhausen, A.; Holland, G.; Schaller, M.; Hube, B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell. Microbiol. 2007, 9, 2938–2954. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.Y.; Rodarte, G.; Murillo, L.A.; Jones, T.; Davis, R.W.; Dungan, J.; Newport, G.; Agabian, N. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 2004, 53, 1451–1469. [Google Scholar] [CrossRef] [PubMed]
- Arnal, N.; Alban, C.; Quadrado, M.; Grandjean, O.; Mireau, H. The arabidopsis BIO2 protein requires mitochondrial targeting for activity. Plant. Mol. Biol. 2006, 62, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ito, K.; Shimoi, H. Identification and characterization of a novel biotin biosynthesis gene in saccharomyces cerevisiae. Appl. Environ. Microb. 2005, 71, 6845–6855. [Google Scholar] [CrossRef] [PubMed]








| C. albicans Strain | Genotype | Sources |
|---|---|---|
| SC5314 | Wild-type | [21] |
| A72 | Wild-type | ATCC * MYA-2430 |
| HLC52 | ura3::λimm434/ura3::λimm434 | [22] |
| efg1::hisG/efg1::hisG-URA3-hisG | ||
| JCK19 | ura3::λimm434/ura3::λimm434 | [22] |
| cph1::hisG/cph1::hisG-URA3-hisG | ||
| HLC54 | ura3::imm434/ura3::imm434 | [22] |
| cph1::hisG/cph1::hisG efg1::hisG/efg1::hisG URA3::hisG | ||
| CR216 | ura3::λimm434/ura3::λimm434 | [23] |
| cdc35::hisG-URA3-hisG/cdc35::hisG |
| Gene Abbreviation | Primer Sequences (Forward -F and Reverse -R) | Source |
|---|---|---|
| BIO2 | F: 5′-GCACCCAGAATCATTGCCAA-3′ | This study |
| orf19.2593 | R: 5′-ACTGCTCGTGTTCCTTCATG-3′ | |
| BIO4 | F: 5′-AGTAGCTCGGAGTGGATTGG-3′ | This study |
| orf19.2590 | R: 5′-TTAGAATGAGGGATGTTCGCA-3′ | |
| BIO32 | F: 5′-GTGGACGAGGATTATTTTGGGGAA-3′ | This study |
| orf19.3567 | R: 5′-TCCGTCTATTGTTCCCTTTCCA-3′ | |
| BIO3 | F: 5′-AAACTGGAGCCTGGGAAACT-3′ | This study |
| orf19.2591 | R: 5′-GGCGAACCCAAACACCTAAA-3′ | |
| BPL1 | F: 5′-GTTGAATGAGATCAGACGTGGA-3′ | This study |
| orf19.7645 | R: 5′-GCCATTGTCAACGTCCACTT-3′ | |
| CDC36 | F: 5′-GACCGTCCAGTATAAATCCACCAC-3′ | Pendrak et al. [26] |
| R: 5′-TCAAGACGGGCTCCACATTACTAT-3′ |
| Percent Germ Tube Formation 1 | ||||
|---|---|---|---|---|
| # Cell Washes | 0.01 | 0.1 | 1.0 | 5.0% Serum |
| 0 | 3 | 0 | 52 | 90 |
| 3 | 0 | 14 | 71 | 80 |
| 6 | 7 | 44 | 93 | 98 |
| 8 | 0 | 80 | 95 | 98 |
| Glucose Salts with Biotin (nM) | |||
|---|---|---|---|
| Incubation | 0 | 5, 50, and 4000 | YPD 2 |
| 30 °C | 0.5 3 | 4 ± 0.07 1 (0.9) | 6 ± 0.51 (1.4) |
| 37 °C | 0.5 3 | 3.5 ± 0.15 (0.9) | 6 ± 1.02 (1.4) |
| 30 °C + CO2 | 1.0 4 | 3.5 ± 0.05 (0.8) | 5.5 ± 0.45 (1.4) |
| 37 °C + CO2 | 0.5 3 | 7 ± 1.55 (0.8) | 6 ± 0.88 (1.4) |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad Hussin, N.; Pathirana, R.U.; Hasim, S.; Tati, S.; Scheib-Owens, J.A.; Nickerson, K.W. Biotin Auxotrophy and Biotin Enhanced Germ Tube Formation in Candida albicans. Microorganisms 2016, 4, 37. https://doi.org/10.3390/microorganisms4030037
Ahmad Hussin N, Pathirana RU, Hasim S, Tati S, Scheib-Owens JA, Nickerson KW. Biotin Auxotrophy and Biotin Enhanced Germ Tube Formation in Candida albicans. Microorganisms. 2016; 4(3):37. https://doi.org/10.3390/microorganisms4030037
Chicago/Turabian StyleAhmad Hussin, Nur, Ruvini U. Pathirana, Sahar Hasim, Swetha Tati, Jessica A. Scheib-Owens, and Kenneth W. Nickerson. 2016. "Biotin Auxotrophy and Biotin Enhanced Germ Tube Formation in Candida albicans" Microorganisms 4, no. 3: 37. https://doi.org/10.3390/microorganisms4030037





