Transmission of Bacterial Endophytes
Abstract
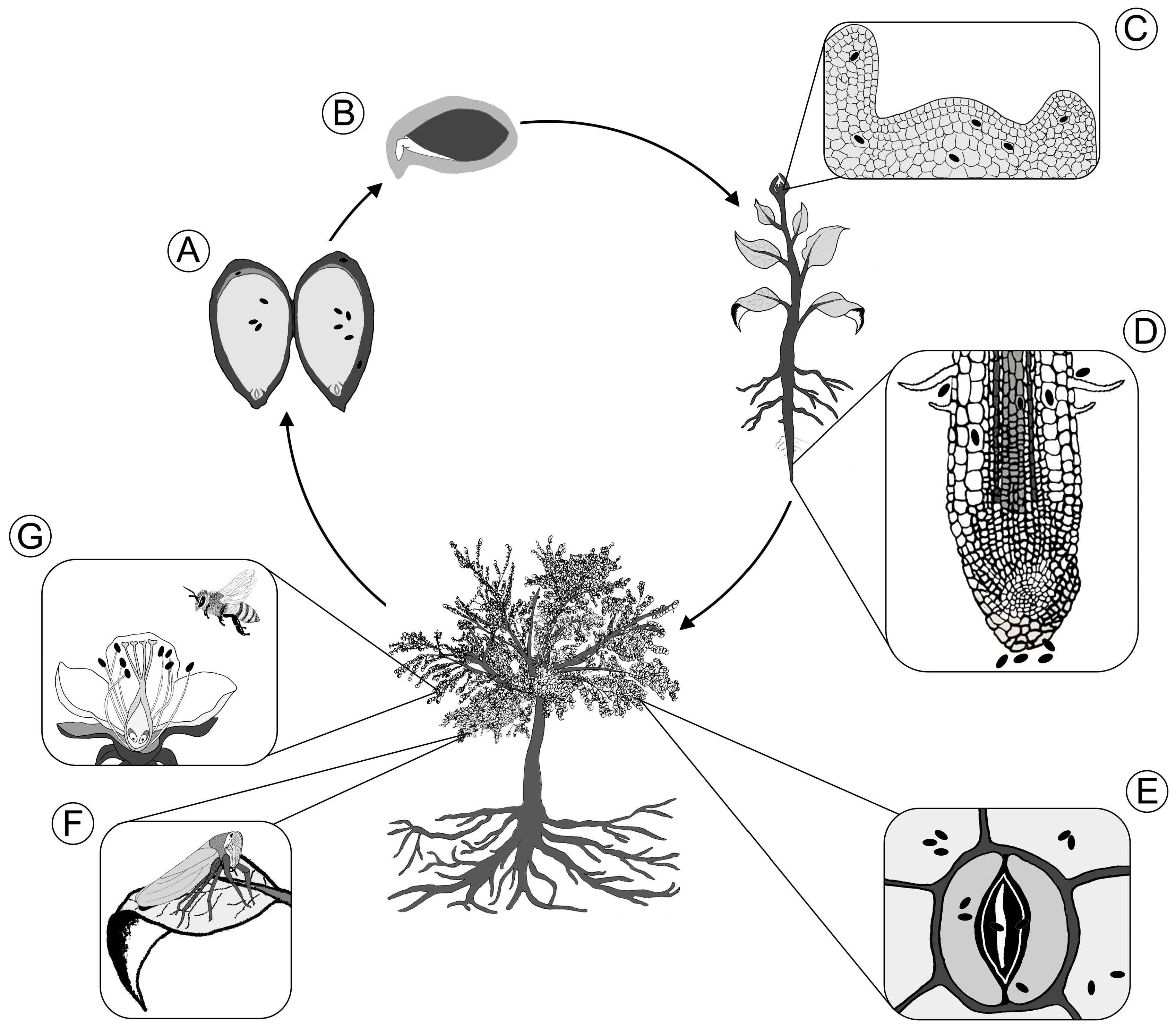
:1. Introduction
2. Vertical Transmission
2.1. Vertical Transfer via Seeds
2.2. Vertical Transfer via Pollen
3. Horizontal Transmission
3.1. Colonization of Seed and Root via Soil
3.1.1. Endophytic Colonization of the Spermosphere
3.1.2. Colonization of the Root Endosphere via the Rhizosphere
3.2. Entry into Aerial Tissues
3.2.1. Aerial Dispersal of the Plant Microbiome
3.2.2. Endophytic Leaf Colonization via Stomata
3.2.3. Floral Transmission of Bacterial Endophytes
3.2.4. Endophyte Transmission by Plant-Feeding Insects
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hallman, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants: Bacterial seed endophytes. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Nair, D.N.; Padmavathy, S. Impact of Endophytic Microorganisms on Plants, Environment and Humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Rajkumar, M.; Luo, Y.; Freitas, H. Inoculation of endophytic bacteria on host and non-host plants—Effects on plant growth and Ni uptake. J. Hazard. Mater. 2011, 195, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Arshad, M.; Hashmi, I.; Karthikeyan, R.; Gentry, T.J.; Schwab, A.P. Biodegradation of phenol and benzene by endophytic bacterial strains isolated from refinery wastewater-fed Cannabis sativa. Environ. Technol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Blain, N.P.; Helgason, B.L.; Germida, J.J. Endophytic root bacteria associated with the natural vegetation growing at the hydrocarbon-contaminated Bitumount Provincial Historic site. Can. J. Microbiol. 2017, 63, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Moyes, A.B.; Kueppers, L.M.; Pett-Ridge, J.; Carper, D.L.; Vandehey, N.; O’Neil, J.; Frank, A.C. Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 2016, 210, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Doty, S.L.; Sher, A.W.; Fleck, N.D.; Khorasani, M.; Bumgarner, R.E.; Khan, Z.; Ko, A.W.K.; Kim, S.-H.; DeLuca, T.H. Variable Nitrogen Fixation in Wild Populus. PLoS ONE 2016, 11, e0155979. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.S.; Six, D.L.; Adams, S.M.; Holben, W.E. In vitro interactions between yeasts and bacteria and the fungal symbionts of the mountain pine beetle (Dendroctonus ponderosae). Microb. Ecol. 2008, 56, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Elbeltagy, A.; Nishioka, K.; Sato, T.; Suzuki, H.; Ye, B.; Hamada, T.; Isawa, T.; Mitsui, H.; Minamisawa, K. Endophytic Colonization and in Planta Nitrogen Fixation by a Herbaspirillum sp. Isolated from Wild Rice Species. Appl. Environ. Microbiol. 2001, 67, 5285–5293. [Google Scholar] [CrossRef] [PubMed]
- Bright, M.; Bulgheresi, S. A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 2010, 8, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Herre, E.A.; Knowlton, N.; Mueller, U.G.; Rehner, S.A. The evolution of mutualisms: Exploring the paths between conflict and cooperation. Trends Ecol. Evol. 1999, 14, 49–53. [Google Scholar] [CrossRef]
- Moran, N.A. Symbiosis. Curr. Biol. 2006, 16, R866–R871. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L. Epichloë festucae and related mutualistic symbionts of grasses. Fungal Genet. Biol. 2001, 33, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.M.; Sherratt, T.N. Horizontally acquired mutualisms, an unsolved problem in ecology? Oikos 2001, 92, 377–384. [Google Scholar] [CrossRef]
- Foster, K.R.; Wenseleers, T. A general model for the evolution of mutualisms. J. Evol. Biol. 2006, 19, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Hardoim, C.C.; Van Overbeek, L.S.; Van Elsas, J.D. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Reiter, B.; Sessitsch, A.; Nowak, J.; Clément, C.; Ait Barka, E. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 2005, 71, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Guelich, G.; Phan, H.; Redman, R.; Doty, S. Bacterial and Yeast Endophytes from Poplar and Willow Promote Growth in Crop Plants and Grasses. ISRN Agron. 2012, 2012, 890280. [Google Scholar] [CrossRef]
- Carroll, G. Fungal endophytes in stems and leaves: From latent pathogen to mutualistic symbiont. Ecology 1988, 69, 2–9. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant Microbe Interact. 2015, 212, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Dennis, P.G.; Paungfoo-Lonhienne, C.; Weber, L.; Brackin, R.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; von Maltzahn, G. A New Approach to Modify Plant Microbiomes and Traits by Introducing Beneficial Bacteria at Flowering into Progeny Seeds. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.-X.; Chi, F.; Yang, M.-F.; Shen, S.-H.; Jing, Y.-X.; Dazzo, F.B.; Cheng, H.-P. Movement of rhizobia inside tobacco and lifestyle alternation from endophytes to free-living rhizobia on leaves. J. Microbiol. Biotechnol. 2010, 20, 238–244. [Google Scholar] [PubMed]
- James, E.K.; Gyaneshwar, P.; Mathan, N.; Barraquio, W.L.; Reddy, P.M.; Iannetta, P.P.; Olivares, F.L.; Ladha, J.K. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant. Microbe Interact. 2002, 15, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jacques, M.-A.; Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 2017, 37, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.B. The seed microbiome: Origins, interactions, and impacts. Plant Soil 2017. [Google Scholar] [CrossRef]
- Rodríguez, C.E.; Mitter, B.; Barret, M.; Sessitsch, A.; Compant, S. Commentary: Seed bacterial inhabitants and their routes of colonization. Plant Soil 2017. [Google Scholar] [CrossRef]
- Charkowski, A.O.; Sarreal, C.Z.; Mandrell, R.E. Wrinkled alfalfa seeds harbor more aerobic bacteria and are more difficult to sanitize than smooth seeds. J. Food Prot. 2001, 64, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Cottyn, B.; Regalado, E.; Lanoot, B.; De Cleene, M.; Mew, T.; Swings, J. Bacterial populations associated with rice seed in the tropical environment. Phytopathology 2001, 91, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; del Valle, M.V.; Pérez, A.; Zepeda, A.; Zenteno, E. Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol. Biochem. 2001, 33, 167–172. [Google Scholar] [CrossRef]
- Kaga, H.; Mano, H.; Tanaka, F.; Watanabe, A.; Kaneko, S.; Morisaki, H. Rice seeds as sources of endophytic bacteria. Microbes Environ. 2009, 24, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Okunishi, S.; Sako, K.; Mano, H.; Imamura, A.; Morisaki, H. Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes Environ. 2005, 20, 168–177. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Irizarry, I.; Bergen, M.; Kharwar, R.N.; White, J.F. Seed vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 2017, 122, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zuo, S.; Zou, Y.; Wang, J.; Song, W. Investigation on diversity and population succession dynamics of endophytic bacteria from seeds of maize (Zea mays L., Nongda108) at different growth stages. Ann. Microbiol. 2013, 63, 71–79. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed]
- Mastretta, C.; Taghavi, S.; van Der Lelie, D.; Mengoni, A.; Galardi, F.; Gonnelli, C.; Barac, T.; Boulet, J.; Weyens, N.; Vangronsveld, J. Endophytic bacteria from seeds of Nicotiana tabacum can reduce cadmium phytotoxicity. Int. J. Phytoremediat. 2009, 11, 251–267. [Google Scholar] [CrossRef]
- Vega, F.E.; Pava-Ripoll, M.; Posada, F.; Buyer, J.S. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 2005, 45, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Developmental Peculiarities and Seed-Borne Endophytes in Quinoa: Omnipresent, Robust Bacilli Contribute to Plant Fitness. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Rogel, M.A.; Ormeno-Orrillo, E.; Martínez-Romero, J.; Martínez-Romero, E. Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 2010, 33, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of Grapevine Flowers, Berries, and Seeds: Identification of Cultivable Bacteria, Comparison with Other Plant Parts, and Visualization of Niches of Colonization. Microb. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Zawoznik, M.S.; Vázquez, S.C.; Díaz Herrera, S.M.; Groppa, M.D. Search for endophytic diazotrophs in barley seeds. Braz. J. Microbiol. 2014, 45, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Fürnkranz, M.; Lukesch, B.; Müller, H.; Huss, H.; Grube, M.; Berg, G. Microbial Diversity Inside Pumpkins: Microhabitat-Specific Communities Display a High Antagonistic Potential against Phytopathogens. Microb. Ecol. 2012, 63, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Puente, M.E.; Li, C.Y.; Bashan, Y. Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 2009, 66, 402–408. [Google Scholar] [CrossRef]
- Goggin, D.E.; Emery, R.J.N.; Kurepin, L.V.; Powles, S.B. A potential role for endogenous microflora in dormancy release, cytokinin metabolism and the response to fluridone in Lolium rigidum seeds. Ann. Bot. 2015, 115, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araújo, W.L. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2008, 287, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Cankar, K.; Kraigher, H.; Ravnikar, M.; Rupnik, M. Bacterial endophytes from seeds of Norway spruce (Picea abies L. Karst). FEMS Microbiol. Lett. 2005, 244, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Alibrandi, P.; Cardinale, M.; Rahman, M.M.; Strati, F.; Ciná, P.; de Viana, M.L.; Giamminola, E.M.; Gallo, G.; Schnell, S.; De Filippo, C. The seed endosphere of Anadenanthera colubrina is inhabited by a complex microbiota, including Methylobacterium spp. and Staphylococcus spp. with potential plant-growth promoting activities. Plant Soil 2017, 1–19. [Google Scholar] [CrossRef]
- Glassner, H.; Zchori-Fein, E.; Yaron, S.; Sessitsch, A.; Sauer, U.; Compant, S. Bacterial niches inside seeds of Cucumis melo L. Plant Soil 2017, 1–13. [Google Scholar] [CrossRef]
- Rout, M.E.; Chrzanowski, T.H.; Westlie, T.K.; Deluca, T.H.; Callaway, R.M.; Holben, W.E. Bacterial endophytes enhance competition by invasive plants. Am. J. Bot. 2013, 100, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Herrera, S.D.; Grossi, C.; Zawoznik, M.; Groppa, M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 2016, 186, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, K.; Garrison, N.K.; Hinton, D.M.; Bacon, C.W.; Khush, G.S.; Peck, H.D.; Datta, N. Identification and characterization of bacterial endophytes of rice. Mycopathologia 1996, 134, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Saikkonen, K.; Saari, S.; Helander, M. Defensive mutualism between plants and endophytic fungi? Fungal Divers. 2010, 41, 101–113. [Google Scholar] [CrossRef]
- Hodgson, S.; de Cates, C.; Hodgson, J.; Morley, N.J.; Sutton, B.C.; Gange, A.C. Vertical transmission of fungal endophytes is widespread in forbs. Ecol. Evol. 2014, 4, 1199–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, I.M. Bacterial Leaf Nodule Symbiosis. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 1990; Volume 17, pp. 163–234. ISBN 978-0-12-005917-1. [Google Scholar]
- Lemaire, B.; Janssens, S.; Smets, E.; Dessein, S. Endosymbiont Transmission Mode in Bacterial Leaf Nodulation as Revealed by a Population Genetic Study of Psychotria leptophylla. Appl. Environ. Microbiol. 2012, 78, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Gagne-Bourgue, F.; Aliferis, K.A.; Seguin, P.; Rani, M.; Samson, R.; Jabaji, S. Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. J. Appl. Microbiol. 2013, 114, 836–853. [Google Scholar] [CrossRef] [PubMed]
- Ringelberg, D.; Foley, K.; Reynolds, C.M. Bacterial endophyte communities of two wheatgrass varieties following propagation in different growing media. Can. J. Microbiol. 2012, 58, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zuo, S.; Xu, L.; Zou, Y.; Song, W. Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch. Microbiol. 2012, 194, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence Shapes the Structure of the Seed Microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Beckers, B.; Thijs, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. The effects of the growth substrate on cultivable and total endophytic assemblages of Arabidopsis thaliana. Plant Soil 2016, 405, 325–336. [Google Scholar] [CrossRef]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Changes in the population of seed bacteria of transgenerationally Cd-exposed Arabidopsis thaliana. Plant Biol. 2013, 15, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.K.; Sinclair, J.B. Principles of Seed Pathology, 2nd ed.; Lewis Publishers: Boca Raton, FL, USA, 1997; ISBN 978-0-87371-670-3. [Google Scholar]
- Puente, M.E.; Li, C.Y.; Bashan, Y. Rock-degrading endophytic bacteria in cacti. Environ. Exp. Bot. 2009, 66, 389–401. [Google Scholar] [CrossRef]
- Clark, S.E. Organ formation at the vegetative shoot meristem. Plant Cell 1997, 9, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Pirttilä, A.M. Endophytic Bacteria in Tree Shoot Tissues and Their Effects on Host. In Endophytes of Forest Trees: Biology and Applications; Pirttila, A.M., Frank, A.C., Eds.; Springer: New York, NY, USA, 2011; Volume 80, pp. 139–149. [Google Scholar]
- Van Aken, B.; Peres, C.M.; Doty, S.L.; Yoon, J.M.; Schnoor, J.L. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoids × nigra DN34). Int. J. Syst. Evol. Microbiol. 2004, 54, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, K.; Ulrich, A.; Ewald, D. Paenibacillus—A predominant endophytic bacterium colonizing tissue cultures of woody plants. Plant Cell Tissue Organ Cult. 2008, 93, 347–351. [Google Scholar] [CrossRef]
- Pirttilä, A.M.; Laukkanen, H.; Pospiech, H.; Myllyla, R.; Hohtola, A. Detection of intracellular bacteria in the buds of Scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl. Environ. Microbiol. 2000, 66, 3073–3077. [Google Scholar] [CrossRef] [PubMed]
- Laukkanen, H.; Soini, H.; Kontunen-Soppela, S.; Hohtola, A.; Viljanen, M. A mycobacterium isolated from tissue cultures of mature Pinus sylvestris interferes with growth of Scots pine seedlings. Tree Physiol. 2000, 20, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Kumari, S.; Swarna, G.K.; Prakash, D.P.; Dinesh, M.R. Ubiquitous presence of fastidious endophytic bacteria in field shoots and index-negative apparently clean shoot-tip cultures of papaya. Plant Cell Rep. 2007, 26, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Swarna, G.K.; Roy, P.K.; Patil, P. Identification of culturable and originally non-culturable endophytic bacteria isolated from shoot tip cultures of banana cv. Grand Naine. Plant Cell Tissue Organ Cult. 2008, 93, 55. [Google Scholar] [CrossRef]
- Kamoun, R.; Lepoivre, P.; Boxus, P. Evidence for the occurrence of endophytic prokaryotic contaminants in micropropagated plantlets of Prunus cerasus cv. ‘Montmorency’. In Pathogen and Microbial Contamination Management in Micropropagation; Springer: New York, NY, USA, 1997; pp. 145–148. [Google Scholar]
- Esposito-Polesi, N.P.; de Abreu-Tarazi, M.F.; de Almeida, C.V.; Tsai, S.M.; de Almeida, M. Investigation of Endophytic Bacterial Community in Supposedly Axenic Cultures of Pineapple and Orchids with Evidence on Abundant Intracellular Bacteria. Curr. Microbiol. 2017, 74, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Sekhar, A.C. Live cell imaging reveals extensive intracellular cytoplasmic colonization of banana by normally non-cultivable endophytic bacteria. Aob Plants 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Kumari, S.; Swarna, G.K.; Gowda, T.K.S. Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host-endophyte interaction in vitro and in vivo. Can. J. Microbiol. 2007, 53, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Koskimäki, J.J.; Pirttilä, A.M.; Ihantola, E.-L.; Halonen, O.; Frank, A.C. The Intracellular Scots Pine Shoot Symbiont Methylobacterium extorquens DSM13060 Aggregates around the Host Nucleus and Encodes Eukaryote-Like Proteins. mBio 2015, 6, e00039-15. [Google Scholar] [CrossRef] [PubMed]
- Pirttilä, A.M.; Pospiech, H.; Laukkanen, H.; Myllyla, R.; Hohtola, A. Seasonal variations in location and population structure of endophytes in buds of Scots pine. Tree Physiol. 2005, 25, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Ambika Manirajan, B.; Ratering, S.; Rusch, V.; Schwiertz, A.; Geissler-Plaum, R.; Cardinale, M.; Schnell, S. Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 2016, 18, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Madmony, A.; Chernin, L.; Pleban, S.; Peleg, E.; Riov, J. Enterobacter cloacae, an obligatory endophyte of pollen grains of Mediterranean pines. Folia Microbiol. Praha 2005, 50, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Heydenreich, B.; Bellinghausen, I.; König, B.; Becker, W.-M.; Grabbe, S.; Petersen, A.; Saloga, J. Gram-positive bacteria on grass pollen exhibit adjuvant activity inducing inflammatory T cell responses. Clin. Exp. Allergy 2012, 42, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Obersteiner, A.; Gilles, S.; Frank, U.; Beck, I.; Häring, F.; Ernst, D.; Rothballer, M.; Hartmann, A.; Traidl-Hoffmann, C.; Schmid, M. Pollen-Associated Microbiome Correlates with Pollution Parameters and the Allergenicity of Pollen. PLoS ONE 2016, 11, e0149545. [Google Scholar] [CrossRef] [PubMed]
- Jojima, Y.; Mihara, Y.; Suzuki, S.; Yokozeki, K.; Yamanaka, S.; Fudou, R. Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int. J. Syst. Evol. Microbiol. 2004, 54, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Pollen has a microbiome: Implications for plant reproduction, insect pollination and human allergies: Pollen has a microbiome. Environ. Microbiol. 2017, 19, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Galippe, V. Note sur la présence de micro-organismes dans les tissus végétaux. Comptes Rendus Hebdomadaires des Séances et Mémoires de la Société de Biologie et des ses Filiales et Associées 1887, 39, 410–416. [Google Scholar]
- Hiltner, L. Über neue Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Ber- ücksichtigung der Gründüngung und Brache. Arbeiten der Deustchen Landwirtschafts Gesellesschaft 1904, 98, 59–78. [Google Scholar]
- Nelson, E.B. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 2004, 42, 271–309. [Google Scholar] [CrossRef] [PubMed]
- Schiltz, S.; Gaillard, I.; Pawlicki-Jullian, N.; Thiombiano, B.; Mesnard, F.; Gontier, E. A review: What is the spermosphere and how can it be studied? J. Appl. Microbiol. 2015, 119, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.P.; Baker, C.J.; McKenna, L.; Liu, S.; Buyer, J.S.; Kobayashi, D.Y. Influence of host seed on metabolic activity of Enterobacter cloacae in the spermosphere. Soil Biol. Biochem. 2009, 41, 754–761. [Google Scholar] [CrossRef]
- Kageyama, K.; Nelson, E.B. Differential inactivation of seed exudate stimulation of Pythium ultimum sporangium germination by Enterobacter cloacae influences biological control efficacy on different plant species. Appl. Environ. Microbiol. 2003, 69, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.M.; Smith, K.P.; Dodsworth, J.A.; Guenthner, B.; Handelsman, J.; Goodman, R.M. Influence of Tomato Genotype on Growth of Inoculated and Indigenous Bacteria in the Spermosphere. Appl. Environ. Microbiol. 2001, 67, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Vergani, L.; Mapelli, F.; Zanardini, E.; Terzaghi, E.; Di Guardo, A.; Morosini, C.; Raspa, G.; Borin, S. Phyto-rhizoremediation of polychlorinated biphenyl contaminated soils: An outlook on plant-microbe beneficial interactions. Sci. Total Environ. 2017, 575, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; van Breusegem, F.; Eberl, L.; et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, E.E.; Anderson-Furgeson, J.; Estera, K.Y.; Pett-Ridge, J.; de Valpine, P.; Brodie, E.L.; Firestone, M.K. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 2016, 97, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Haichar, F.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clement, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Marschner, H.; Römheld, V.; Horst, W.J.; Martin, P. Root-induced changes in the rhizosphere: Importance for the mineral nutrition of plants. J. Plant Nutr. Soil Sci. 1986, 149, 441–456. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Persello-Cartieaux, F.; Nussaume, L.; Robaglia, C. Tales from the underground: Molecular. Plant Cell Environ. 2003, 26, 189–199. [Google Scholar] [CrossRef]
- Müller, H.; Westendorf, C.; Leitner, E.; Chernin, L.; Riedel, K.; Schmidt, S.; Eberl, L.; Berg, G. Quorum-sensing effects in the antagonistic rhizosphere bacterium Serratia plymuthica HRO-C48. FEMS Microbiol. Ecol. 2009, 67, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Podile, A.R.; Vukanti, R.; Sravani, A.; Kalam, S.; Dutta, S.; Durgeshwar, P.; Rao, V.P. Root colonization and quorum sensing are the driving forces of plant growth promoting rhizobacteria (PGPR) for growth promotion. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2013, 80, 407–413. [Google Scholar] [CrossRef]
- Wei, H.-L.; Zhang, L.-Q. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 2006, 89, 267–280. [Google Scholar] [CrossRef] [PubMed]
- De Weger, L.A.; van der Vlugt, C.I.; Wijfjes, A.H.; Bakker, P.A.; Schippers, B.; Lugtenberg, B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 1987, 169, 2769–2773. [Google Scholar] [CrossRef] [PubMed]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 2016, 90, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; de Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, C.D.; et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Kaplan, H.; Sessitsch, A.; Nowak, J.; Ait Barka, E.; Clément, C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: From the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 2008, 63, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Reinhold-Hurek, B.; van Montagu, M.; Kellenberger, E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 1994, 176, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Hurek, T. Life in grasses: Diazotrophic endophytes. Trends Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.-H.; Cheng, H.-P.; Jing, Y.-X.; Yanni, Y.G.; Dazzo, F.B. Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, inside Rice Plants and Assessment of Benefits to Rice Growth Physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Law, A.D.; Moe, L.A. Nicotiana Roots Recruit Rare Rhizosphere Taxa as Major Root-Inhabiting Microbes. Microb. Ecol. 2016, 71, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Phyllosphere Microbiology; Lindow, S.E.; Hecht-Poinar, E.I.; Elliott, V.J. (Eds.) APS Press/American Phytopathological Society: St. Paul, MN, USA, 2002; ISBN 0-89054-286-4. [Google Scholar]
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef]
- Lighthart, B. Mini-review of the concentration variations found in the alfresco atmospheric bacterial populations. Aerobiologia 2000, 16, 7–16. [Google Scholar] [CrossRef]
- Hill, K.A.; Shepson, P.B.; Galbavy, E.S.; Anastasio, C.; Kourtev, P.S.; Konopka, A.; Stirm, B.H. Processing of atmospheric nitrogen by clouds above a forest environment. J. Geophys. Res. 2007, 112. [Google Scholar] [CrossRef]
- Sattler, B.; Puxbaum, H.; Psenner, R. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 2001, 28, 239–242. [Google Scholar] [CrossRef]
- DeLeon-Rodriguez, N.; Lathem, T.L.; Rodriguez-R, L.M.; Barazesh, J.M.; Anderson, B.E.; Beyersdorf, A.J.; Ziemba, L.D.; Bergin, M.; Nenes, A.; Konstantinidis, K.T. Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. USA 2013, 110, 2575–2580. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Bohannan, B.J.M.; Jaffe, D.A.; Levin, D.A.; Green, J.L. Molecular Evidence for Metabolically Active Bacteria in the Atmosphere. Front. Microbiol. 2016, 7, 772. [Google Scholar] [CrossRef] [PubMed]
- Burrows, S.M.; Butler, T.; Jöckel, P.; Tost, H.; Kerkweg, A.; Pöschl, U.; Lawrence, M.G. Bacteria in the global atmosphere—Part 2: Modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 2009, 9, 9281–9297. [Google Scholar] [CrossRef]
- Burrows, S.M.; Elbert, W.; Lawrence, M.G.; Pöschl, U. Bacteria in the global atmosphere—Part 1: Review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 2009, 9, 9263–9280. [Google Scholar] [CrossRef]
- Womack, A.M.; Bohannan, B.J.M.; Green, J.L. Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Bovallius, A.; Bucht, B.; Roffey, R.; Anäs, P. Long-range air transmission of bacteria. Appl. Environ. Microbiol. 1978, 6, 1231–1232. [Google Scholar]
- Yamaguchi, N.; Ichijo, T.; Sakotani, A.; Baba, T.; Nasu, M. Global dispersion of bacterial cells on Asian dust. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Prospero, J.M.; Blades, E.; Mathison, G.; Naidu, R. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 2005, 21, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Barberán, A.; Henley, J.; Fierer, N.; Casamayor, E.O. Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci. Total Environ. 2014, 487, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Meola, M.; Lazzaro, A.; Zeyer, J. Bacterial Composition and Survival on Sahara Dust Particles Transported to the European Alps. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- McCartney, H.A.; Fitt, B.D.L.; Schmechel, D. Sampling bioaerosols in plant pathology. J. Aerosol Sci. 1997, 28, 349–364. [Google Scholar] [CrossRef]
- Brown, J.K.M. Aerial Dispersal of Pathogens on the Global and Continental Scales and Its Impact on Plant Disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, J.; Constantinidou, H.A.; Barchet, W.R.; Upper, C.D. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl. Environ. Microbiol. 1982, 44, 1059–1063. [Google Scholar] [PubMed]
- Bowers, R.M.; McLetchie, S.; Knight, R.; Fierer, N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011, 5, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Womack, A.M.; Artaxo, P.E.; Ishida, F.Y.; Mueller, R.C.; Saleska, S.R.; Wiedemann, K.T.; Bohannan, B.J.M.; Green, J.L. Characterization of active and total fungal communities in the atmosphere over the Amazon rainforest. Biogeosciences 2015, 12, 6337–6349. [Google Scholar] [CrossRef]
- Joung, Y.S.; Ge, Z.; Buie, C.R. Bioaerosol generation by raindrops on soil. Nat. Commun. 2017, 8, 14668. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, J.; Upper, C.D. Aerial Dispersal of Epiphytic Bacteria over Bean Plants. Appl. Environ. Microbiol. 1985, 50, 1229–1232. [Google Scholar] [PubMed]
- Quinn, C.E.; Sells, I.A.; Graham, D.C. Soft Rot Erwinia Bacteria in the Atmospheric Bacterial Aerosol. J. Appl. Bacteriol. 1980, 49, 175–181. [Google Scholar] [CrossRef]
- Graham, D.C.; Quinn, C.E.; Bradley, L.F. Quantitative Studies on the Generation of Aerosols of Erwinia carotovora var. atroseptica by Simulated Raindrop Impaction on Blackleg-infected Potato Stems. J. Appl. Bacteriol. 1977, 43, 413–424. [Google Scholar] [CrossRef]
- Ercolani, G.L. Epiphytic Survival of Pseudomonas syringae on Hairy Vetch in Relation to Epidemiology of Bacterial Brown Spot of Bean in Wisconsin. Phytopathology 1974, 64, 1330. [Google Scholar] [CrossRef]
- Cafati, C.R. Role of Nonhost Species as Alternate Inoculum Sources of Xanthomonas phaseoli. Plant Dis. 1980, 64, 194. [Google Scholar] [CrossRef]
- Martiny, J.B.H. History Leaves Its Mark on Soil Bacterial Diversity. mBio 2016, 7, e00784-16. [Google Scholar] [CrossRef] [PubMed]
- Andam, C.P.; Doroghazi, J.R.; Campbell, A.N.; Kelly, P.J.; Choudoir, M.J.; Buckley, D.H. A Latitudinal Diversity Gradient in Terrestrial Bacteria of the Genus Streptomyces. mBio 2016, 7, e02200-15. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.J.; Hart, S.C.; Aciego, S.M.; Riebe, C.S.; Blakowski, M.A.; Aronson, E.L. Microbial Community Structure of Subalpine Snow in the Sierra Nevada, California. Arct. Antarct. Alp. Res. 2016, 48, 685–701. [Google Scholar] [CrossRef]
- Carrell, A.A.; Frank, A.C. Pinus flexilis and Piceae engelmannii share a simple and consistent needle endophyte microbiota with a potential role in nitrogen fixation. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Carrell, A.A.; Carper, D.L.; Frank, A.C. Subalpine conifers in different geographical locations host highly similar foliar bacterial endophyte communities. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of Stomata in Plant Innate Immunity and Foliar Bacterial Diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Arnold, E.A. Endophytic fungi: Hidden components of tropical community ecology. In Tropical Forest Community Ecology; Blackwell Scientific, Inc.: Oxford, UK, 2008; pp. 254–271. [Google Scholar]
- Mendgen, K.; Hahn, M.; Deising, H. Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 1996, 34, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.; Chard, J.; Hocart, M.; Lennard, J.; Graham, D. Penetration of potato tuber lenticels by bacteria in relation to biological control of black leg disease. Potato Res. 1996, 39, 333–344. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Barber, C.E.; Daniels, M.J. Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: A system for studying early infection events in bacterial pathogenesis. Mol. Plant-Microbe Interact. 1998, 11, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, D.; Hwang, I. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol. Plant 2015, 8, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, S.Y.; Assmann, S.M. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. Cell Mol. Biol. 2008, 56, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C.; et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef] [PubMed]
- Panchal, S.; Roy, D.; Chitrakar, R.; Price, L.; Breitbach, Z.S.; Armstrong, D.W.; Melotto, M. Coronatine Facilitates Pseudomonas syringae Infection of Arabidopsis Leaves at Night. Front. Plant Sci. 2016, 7, 880. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Hurley, B.; Lee, D.; Mott, A.; Wilton, M.; Liu, J.; Liu, Y.C.; Angers, S.; Coaker, G.; Guttman, D.S.; Desveaux, D. The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS ONE 2014, 9, e114921. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Panchal, S.; Rosa, B.A.; Melotto, M. Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 2013, 103, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Kroupitski, Y.; Golberg, D.; Belausov, E.; Pinto, R.; Swartzberg, D.; Granot, D.; Sela, S. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 2009, 75, 6076–6086. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Zhang, L.; Oblessuc, P.R.; He, S.Y. Stomatal Defense a Decade Later. Plant Physiol. 2017, 174, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Baldotto, L.E.B.; Olivares, F.L.; Bressan-Smith, R. Structural interaction between GFP-labeled diazotrophic endophytic bacterium Herbaspirillum seropedicae RAM10 and pineapple plantlets “Vitória”. Braz. J. Microbiol. 2011, 42, 114–125. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.O.; Pamphile, J.A.; Lúcia, C.; Mello, S.; Da Rocha, C.; Azevedo, J.L. Plant-microbe interactions between maize (Zea mays L.) and endophytic microrganisms observed by Scanning Electron Microscopy. Acta Sci. Biol. Sci. 2004, 26, 357–359. [Google Scholar]
- White, J.F.; Torres, M.S.; Sullivan, R.F.; Jabbour, R.E.; Chen, Q.; Tadych, M.; Irizarry, I.; Bergen, M.S.; Havkin-Frenkel, D.; Belanger, F.C. Occurrence of B acillus amyloliquefaciens as a systemic endophyte of vanilla orchids: B. Amyloliquefaciens as a Systemic Endophyte of Vanilla Orchids. Microsc. Res. Tech. 2014, 77, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Fukami, J.; Nogueira, M.A.; Araujo, R.S.; Hungria, M. Accessing inoculation methods of maize and wheat with Azospirillum brasilense. AMB Express 2016, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Aizenberg-Gershtein, Y.; Izhaki, I.; Halpern, M. Do Honeybees Shape the Bacterial Community Composition in Floral Nectar? PLoS ONE 2013, 8, e67556. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Pérez, S.; Herrera, C.M.; de Vega, C. Zooming-in on floral nectar: A first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 2012, 80, 591–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridman, S.; Izhaki, I.; Gerchman, Y.; Halpern, M. Bacterial communities in floral nectar. Environ. Microbiol. Rep. 2012, 4, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Lenaerts, M.; Brys, R.; Willems, K.; Honnay, O.; Lievens, B. Among-Population Variation in Microbial Community Structure in the Floral Nectar of the Bee-Pollinated Forest Herb Pulmonaria officinalis L. PLoS ONE 2013, 8, e56917. [Google Scholar] [CrossRef] [PubMed]
- Baruzzi, F.; Cefola, M.; Carito, A.; Vanadia, S.; Calabrese, N. Changes in Bacterial Composition of Zucchini Flowers Exposed to Refrigeration Temperatures. Sci. World J. 2012, 2012, 127805. [Google Scholar] [CrossRef] [PubMed]
- Glassner, H.; Zchori-Fein, E.; Compant, S.; Sessitsch, A.; Katzir, N.; Portnoy, V.; Yaron, S. Characterization of endophytic bacteria from cucurbit fruits with potential benefits to agriculture in melons (Cucumis melo L.). FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Aleklett, K.; Hart, M.; Shade, A. The microbial ecology of flowers: An emerging frontier in phyllosphere research. Botany 2014, 92, 253–266. [Google Scholar] [CrossRef]
- Shade, A.; McManus, P.S.; Handelsman, J. Unexpected Diversity during Community Succession in the Apple Flower Microbiome. mBio 2013, 4, e00602-12. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, J. Fire Blight: The Disease and Its Causative Agent, Erwinia Amylovora; Vanneste, J.L., Ed.; CABI Pub: Wallingford, UK; New York, NY, USA, 2000; ISBN 978-0-85199-294-5. [Google Scholar]
- Spinelli, F.; Ciampolini, F.; Cresti, M.; Geider, K.; Costa, G. Influence of Stigmatic Morphology on Flower Colonization by Erwinia amylovora and Pantoea agglomerans. Eur. J. Plant Pathol. 2005, 113, 395–405. [Google Scholar] [CrossRef]
- Wilson, M. Interactions between the Biological Control Agent Pseudomonas fluorescens A506 and Erwinia amylovora in Pear Blossoms. Phytopathology 1993, 83, 117. [Google Scholar] [CrossRef]
- Buban, T.; Orosz-Kovacs, Z.; Farkas, A. The nectary as the primary site of infection by Erwinia amylovora (Burr.) Winslow et al.: A mini review. Plant Syst. Evol. 2003, 238, 183–194. [Google Scholar] [CrossRef]
- Farkas, Á.; Mihalik, E.; Dorgai, L.; Bubán, T. Floral traits affecting fire blight infection and management. Trees 2012, 26, 47–66. [Google Scholar] [CrossRef]
- Louda, S.M. Inflorescence spiders: A cost/benefit analysis for the host plant, Haplopappus venetus Blake (Asteraceae). Oecologia 1982, 55, 185–191. [Google Scholar] [CrossRef] [PubMed]
- McCall, A.C.; Irwin, R.E. Florivory: The intersection of pollination and herbivory. Ecol. Lett. 2006, 9, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Pellmyr, O.; Thien, L.B. Insect Reproduction and Floral Fragrances: Keys to the Evolution of the Angiosperms? Taxon 1986, 35, 76. [Google Scholar] [CrossRef]
- Wardhaugh, C.W.; Stork, N.E.; Edwards, W.; Grimbacher, P.S. The Overlooked Biodiversity of Flower-Visiting Invertebrates. PLoS ONE 2012, 7, e45796. [Google Scholar] [CrossRef] [PubMed]
- Ushio, M.; Yamasaki, E.; Takasu, H.; Nagano, A.J.; Fujinaga, S.; Honjo, M.N.; Ikemoto, M.; Sakai, S.; Kudoh, H. Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- McFrederick, Q.S.; Thomas, J.M.; Neff, J.L.; Vuong, H.Q.; Russell, K.A.; Hale, A.R.; Mueller, U.G. Flowers and Wild Megachilid Bees Share Microbes. Microb. Ecol. 2017, 73, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.T.; Rodríguez, A.M.; Palaciso, I.S.; López, F.G. Pollen production in anemophilous trees. Grana 1996, 35, 38–46. [Google Scholar] [CrossRef]
- Varis, S.; Pakkanen, A.; Galofré, A.; Pulkkinen, P. The extent of south-north pollen transfer in Finnish Scots pine. Silva Fenn. 2009, 43. [Google Scholar] [CrossRef]
- Campbell, I.D.; McDonald, K.; Flannigan, M.D.; Kringayark, J. Long-distance transport of pollen into the Arctic. Nature 1999, 399, 29–30. [Google Scholar] [CrossRef]
- Marques, J.P.R.; Amorim, L.; Spósito, M.B.; Marin, D.; Appezzato-da-Glória, B. Infection of citrus pollen grains by Colletotrichum acutatum. Eur. J. Plant Pathol. 2013, 136, 35–40. [Google Scholar] [CrossRef]
- Nault, L.R. Arthropod Transmission of Plant Viruses: A New Synthesis. Ann. Entomol. Soc. Am. 1997, 90, 521–541. [Google Scholar] [CrossRef]
- Nault, L.R.; Ammar, E.D. Leafhopper and Planthopper Transmission of Plant Viruses. Annu. Rev. Entomol. 1989, 34, 503–529. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect Vectors of Phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-J.; Ahmed, M.Z.; Lv, N.; Shi, P.-Q.; Wang, X.-M.; Huang, J.-L.; Qiu, B.-L. Plantmediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017, 11, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Lòpez-Fernàndez, S.; Mazzoni, V.; Pedrazzoli, F.; Pertot, I.; Campisano, A. A Phloem-Feeding Insect Transfers Bacterial Endophytic Communities between Grapevine Plants. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frank, A.C.; Saldierna Guzmán, J.P.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70. https://doi.org/10.3390/microorganisms5040070
Frank AC, Saldierna Guzmán JP, Shay JE. Transmission of Bacterial Endophytes. Microorganisms. 2017; 5(4):70. https://doi.org/10.3390/microorganisms5040070
Chicago/Turabian StyleFrank, Anna Carolin, Jessica Paola Saldierna Guzmán, and Jackie E. Shay. 2017. "Transmission of Bacterial Endophytes" Microorganisms 5, no. 4: 70. https://doi.org/10.3390/microorganisms5040070



