Inclusion Body Bead Size in E. coli Controlled by Physiological Feeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioreactor Cultivations
2.2. Cultivation Scheme and qs Adaption
2.3. Analytics
2.3.1. Process Analytics
2.3.2. Product Analytics
IB Preparation:
IB Bead Size:
IB Titer:
3. Results
3.1. Static qs,C Feed-Forwards Feeding for Size Determination
3.2. Model-Based Approach for Prediction of IB Bead Size
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| CDW | cell dry weight [g/L] |
| CQA | critical quality attribute |
| dSn | cumulative sugar uptake rate [g/g] |
| DSP | downstream processing |
| Fin | Feed into the reactor [mL/h] |
| HPLC | high pressure liquid chromatography |
| IB | inclusion body |
| qs,C | specific substrate uptake rate [g/g/h] |
| SEM | scanning electron microscopy |
| T | temperature [°C] |
References
- Gupta, S.K.; Shukla, P. Microbial platform technology for recombinant antibody fragment production: A review. Crit. Rev. Microbiol. 2017, 43, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G. Biopharmaceutical benchmarks 2010. Nat. Biotechnol. 2010, 28, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Wurm, D.J.; Veiter, L.; Ulonska, S.; Eggenreich, B.; Herwig, C.; Spadiut, O. The E. coli pET expression system revisited-mechanistic correlation between glucose and lactose uptake. Appl. Microbiol. Biotechnol. 2016, 100, 8721–8729. [Google Scholar] [CrossRef] [PubMed]
- Meuris, L.; Santens, F.; Elson, G.; Festjens, N.; Boone, M.; Dos Santos, A.; Devos, S.; Rousseau, F.; Plets, E.; Houthuys, E.; et al. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat. Biotechnol. 2014, 32, 485–489. [Google Scholar] [CrossRef] [PubMed]
- DeLisa, M.P.; Li, J.; Rao, G.; Weigand, W.A.; Bentley, W.E. Monitoring GFP-operon fusion protein expression during high cell density cultivation of Escherichia coli using an on-line optical sensor. Biotechnol. Bioeng. 1999, 65, 54–64. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.; Ramadan, H.A.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of Biopharmaceuticals in E. coli: Current Scenario and Future Perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [PubMed]
- Spadiut, O.; Capone, S.; Krainer, F.; Glieder, A.; Herwig, C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014, 32, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cass, B.; Pham, P.L.; Kamen, A.; Durocher, Y. Purification of recombinant proteins from mammalian cell culture using a generic double-affinity chromatography scheme. Protein Expr. Purif. 2005, 40, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Steen, R.; Dahlberg, A.E.; Lade, B.N.; Studier, F.W.; Dunn, J.J. T7 RNA polymerase directed expression of the Escherichia coli rrnB operon. EMBO J. 1986, 5, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Rosenberg, A.H.; Dunn, J.J.; Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzym. 1990, 185, 60–89. [Google Scholar]
- Dubendorff, J.W.; Studier, F.W. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 1991, 219, 45–59. [Google Scholar] [CrossRef]
- Neubauer, P.; Hofmann, K. Efficient use of lactose for the lac promoter-controlled overexpression of the main antigenic protein of the foot and mouth disease virus in Escherichia coli under fed-batch fermentation conditions. FEMS Microbiol. Rev. 1994, 14, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Lyakhov, D.L.; He, B.; Zhang, X.; Studier, F.W.; Dunn, J.J.; McAllister, W.T. Pausing and termination by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1998, 280, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21 (DE3) carrying a synthetic metabolic pathway. Microb. Cell Factories 2015, 14, 201. [Google Scholar] [CrossRef] [PubMed]
- Marbach, A.; Bettenbrock, K. LAC operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef] [PubMed]
- García-Fruitós, E.; Vázquez, E.; Díez-Gil, C.; Corchero, J.L.; Seras-Franzoso, J.; Ratera, I.; Veciana, J.; Villaverde, A. Bacterial inclusion bodies: Making gold from waste. Trends Biotechnol. 2012, 30, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ramón, A.; Señorale-Pose, M.; Marín, M. Inclusion bodies: Not that bad…. Front. Microbiol 2014, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Palmer, I.; Wingfield, P.T. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr. Protoc. Protein Sci. 2012. [Google Scholar] [CrossRef]
- Villaverde, A.; Corchero, J.L.; Seras-Franzoso, J.; Garcia-Fruitós, E. Functional protein aggregates: Just the tip of the iceberg. Nanomedicine 2015, 10, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Strukelj, B. Current state and recent advances in biopharmaceutical production in Escherichia coli, yeasts and mammalian cells. J. Ind. Microbiol. Biotechnol. 2013, 40, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Slouka, C.; Kopp, J.; Hutwimmer, S.; Strahammer, M.; Strohmer, D.; Eitenberger, E.; Schwaighofer, A.; Herwig, C. Custom Made Inclusion Bodies: Impact of classical process parameters and physiological parameters on Inclusion Body quality attributes. Microb. Cell Factories 2018, 17, 148. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, P.T. Preparation of Soluble Proteins from Escherichia coli. Curr. Protoc. Protein Sci. 2014. [Google Scholar] [CrossRef]
- Hrabárová, E.; Achbergerová, L.; Nahálka, J. Insoluble protein applications: The use of bacterial inclusion bodies as biocatalysts. In Insoluble Proteins; Springer: New York, NY, USA, 2015; pp. 411–422. [Google Scholar]
- Peternel, Š.; Komel, R. Isolation of biologically active nanomaterial (inclusion bodies) from bacterial cells. Microb. Cell Factories 2010, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jevševar, S.; Gaberc-Porekar, V.; Fonda, I.; Podobnik, B.; Grdadolnik, J.; Menart, V. Production of nonclassical inclusion bodies from which correctly folded protein can be extracted. Biotechnol. Prog. 2005, 21, 632–639. [Google Scholar] [CrossRef] [PubMed]
- García-Fruitós, E.; González-Montalbán, N.; Morell, M.; Vera, A.; Ferraz, R.M.; Arís, A.; Ventura, S.; Villaverde, A. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Factories 2005, 4, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Fruitós, E.; Rodríguez-Carmona, E.; Díez-Gil, C.; Ferraz, R.M.; Vázquez, E.; Corchero, J.L.; Cano-Sarabia, M.; Ratera, I.; Ventosa, N.; Veciana, J. Surface cell growth engineering assisted by a novel bacterial nanomaterial. Adv. Mater. 2009, 21, 4249–4253. [Google Scholar] [CrossRef]
- García-Fruitós, E.; Arís, A.; Villaverde, A. Localization of functional polypeptides in bacterial inclusion bodies. Appl. Environ. Microbiol. 2007, 73, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Díez-Gil, C.; Krabbenborg, S.; García-Fruitós, E.; Vazquez, E.; Rodríguez-Carmona, E.; Ratera, I.; Ventosa, N.; Seras-Franzoso, J.; Cano-Garrido, O.; Ferrer-Miralles, N. The nanoscale properties of bacterial inclusion bodies and their effect on mammalian cell proliferation. Biomaterials 2010, 31, 5805–5812. [Google Scholar] [CrossRef] [PubMed]
- Spadiut, O.; Zalai, D.; Dietzsch, C.; Herwig, C. Quantitative comparison of dynamic physiological feeding profiles for recombinant protein production with Pichia pastoris. Bioprocess Biosyst. Eng. 2014, 37, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.; Gunderson, C.W.; Mateescu, E.M.; Zhang, Z.; Hwa, T. Interdependence of cell growth and gene expression: Origins and consequences. Science 2010, 330, 1099–1102. [Google Scholar] [CrossRef] [PubMed]
- Rozkov, A.; Avignone-Rossa, C.; Ertl, P.; Jones, P.; O’Kennedy, R.; Smith, J.; Dale, J.; Bushell, M. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol. Bioeng. 2004, 88, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, W.N.; Thurrold, P.; Brillmann, M.; Kager, J.; Fricke, J.; Herwig, C. Generic biomass estimation methods targeting physiologic process control in induced bacterial cultures. Eng. Life Sci. 2016, 16, 720–730. [Google Scholar] [CrossRef]
- Reichelt, W.N.; Brillmann, M.; Thurrold, P.; Keil, P.; Fricke, J.; Herwig, C. Physiological capacities decline during induced bioprocesses leading to substrate accumulation. Biotechnol. J. 2017, 12, 1600547. [Google Scholar] [CrossRef] [PubMed]
- Achmüller, C.; Kaar, W.; Ahrer, K.; Wechner, P.; Hahn, R.; Werther, F.; Schmidinger, H.; Cserjan-Puschmann, M.; Clementschitsch, F.; Striedner, G.; et al. N(pro) fusion technology to produce proteins with authentic N termini in E. Coli. Nat. Methods 2007, 4, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gómez, K.; Flores, N.; Castañeda, H.M.; Martínez-Batallar, G.; Hernández-Chávez, G.; Ramírez, O.T.; Gosset, G.; Encarnación, S.; Bolivar, F. New insights into Escherichia coli metabolism: Carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb. Cell Factories 2012, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.; Slouka, C.; Ulonska, S.; Kager, J.; Fricke, J.; Spadiut, O.; Herwig, C. Impact of Glycerol as Carbon Source onto Specific Sugar and Inducer Uptake Rates and Inclusion Body Productivity in E. coli BL21 (DE3). Bioengineering 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Wurm, D.J.; Herwig, C.; Spadiut, O. How to Determine Interdependencies of Glucose and Lactose Uptake Rates for Heterologous Protein Production with E. coli. Methods Mol. Biol. 2017, 1586, 397–408. [Google Scholar] [PubMed]
- Slouka, C.; Wurm, D.J.; Brunauer, G.; Welzl-Wachter, A.; Spadiut, O.; Fleig, J.; Herwig, C. A Novel Application for Low Frequency Electrochemical Impedance Spectroscopy as an Online Process Monitoring Tool for Viable Cell Concentrations. Sensors 2016, 16, 1900. [Google Scholar] [CrossRef] [PubMed]
- Wurm, D.J.; Hausjell, J.; Ulonska, S.; Herwig, C.; Spadiut, O. Mechanistic platform knowledge of concomitant sugar uptake in Escherichia coli BL21(DE3) strains. Sci. Rep. 2017, 7, 45072. [Google Scholar] [CrossRef] [PubMed]
- Wurm, D.J.; Quehenberger, J.; Mildner, J.; Eggenreich, B.; Slouka, C.; Schwaighofer, A.; Wieland, K.; Lendl, B.; Rajamanickam, V.; Herwig, C. Teaching an old pET new tricks: Tuning of inclusion body formation and properties by a mixed feed system in E. coli. Appl. Microbiol. Biotechnol. 2018, 102, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Kroll, P.; Hofer, A.; Stelzer, I.V.; Herwig, C. Workflow to set up substantial target-oriented mechanistic process models in bioprocess engineering. Process. Biochem. 2017, 62, 24–36. [Google Scholar] [CrossRef]
- Sagmeister, P.; Wechselberger, P.; Jazini, M.; Meitz, A.; Langemann, T.; Herwig, C. Soft sensor assisted dynamic bioprocess control: Efficient tools for bioprocess development. Chem. Eng. Sci. 2013, 96, 190–198. [Google Scholar] [CrossRef]
- Ehgartner, D.; Sagmeister, P.; Herwig, C.; Wechselberger, P. A novel real-time method to estimate volumetric mass biodensity based on the combination of dielectric spectroscopy and soft-sensors. J. Chem. Technol. Biotechnol. 2015, 90, 262–272. [Google Scholar] [CrossRef]

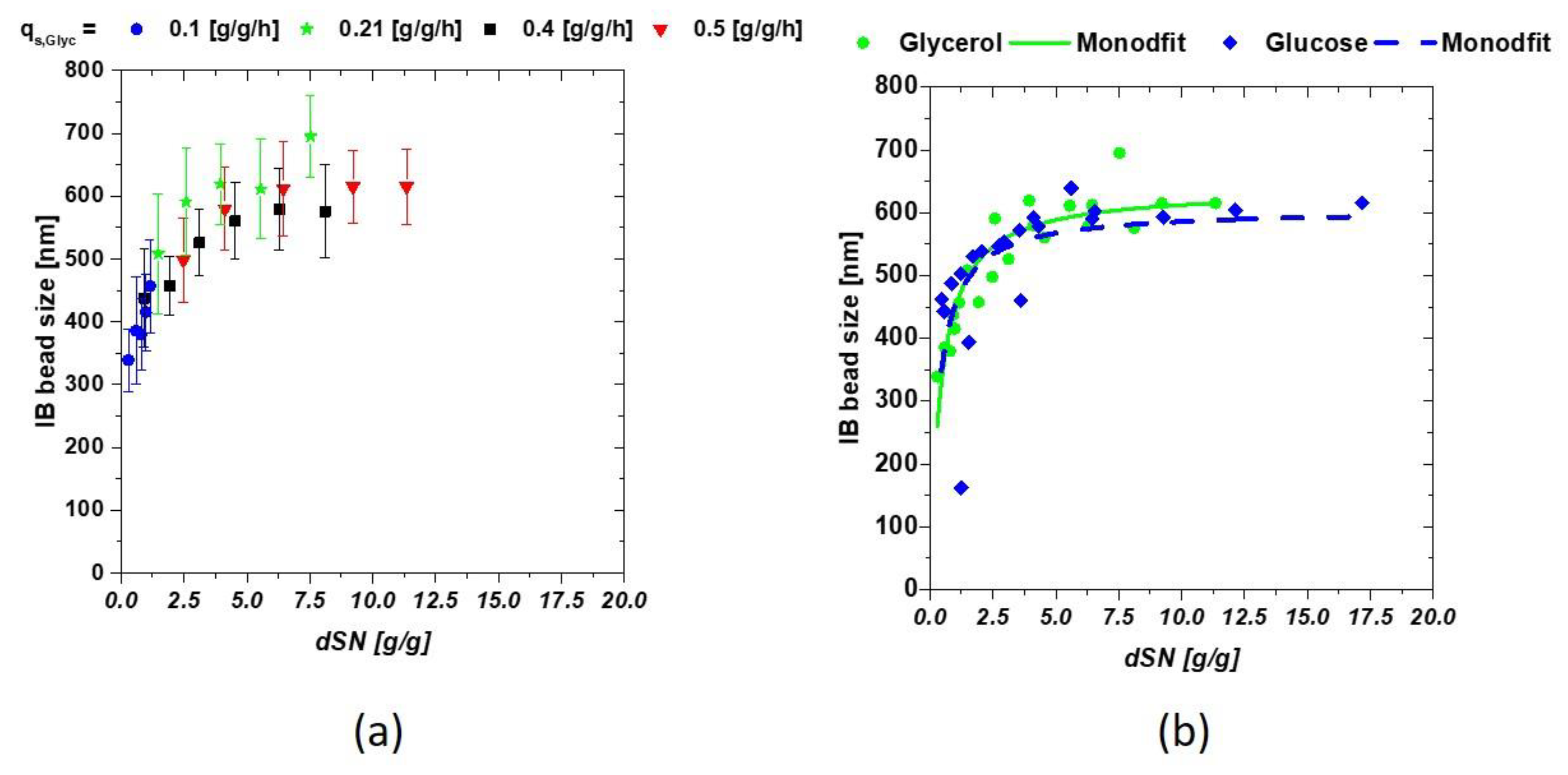
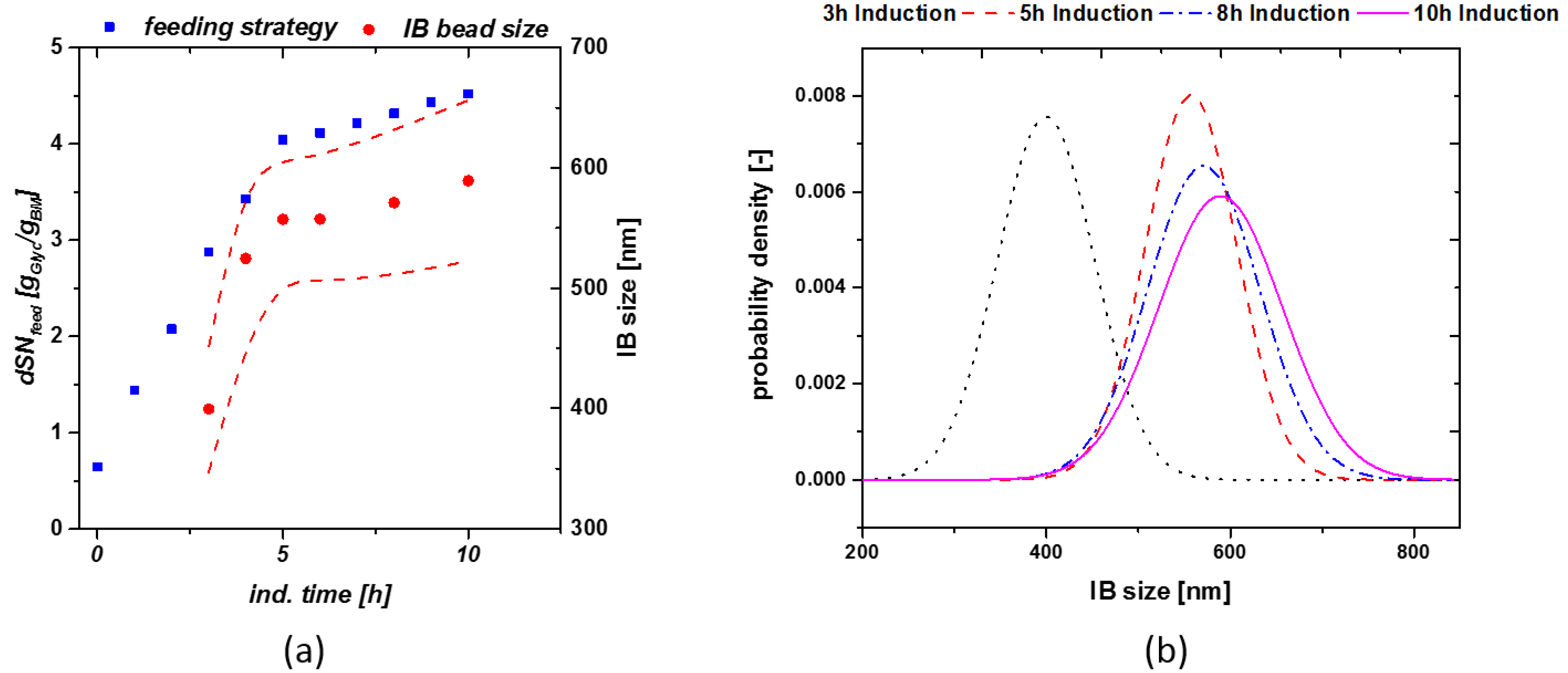

| Phase | Amount of C-source |
|---|---|
| Preculture | 8 g/L |
| Batch-Media | 20 g/L |
| Feed | either 300 g/L or 600 g/L |
| Fit Parameters | Glucose | Glycerol |
|---|---|---|
| Km [g/g] | 0.33 +/− 0.14 | 0.42 +/− 0.06 |
| IBsize,max [nm] | 605.4 +/− 37 | 638 +/− 17 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopp, J.; Slouka, C.; Strohmer, D.; Kager, J.; Spadiut, O.; Herwig, C. Inclusion Body Bead Size in E. coli Controlled by Physiological Feeding. Microorganisms 2018, 6, 116. https://doi.org/10.3390/microorganisms6040116
Kopp J, Slouka C, Strohmer D, Kager J, Spadiut O, Herwig C. Inclusion Body Bead Size in E. coli Controlled by Physiological Feeding. Microorganisms. 2018; 6(4):116. https://doi.org/10.3390/microorganisms6040116
Chicago/Turabian StyleKopp, Julian, Christoph Slouka, Daniel Strohmer, Julian Kager, Oliver Spadiut, and Christoph Herwig. 2018. "Inclusion Body Bead Size in E. coli Controlled by Physiological Feeding" Microorganisms 6, no. 4: 116. https://doi.org/10.3390/microorganisms6040116





