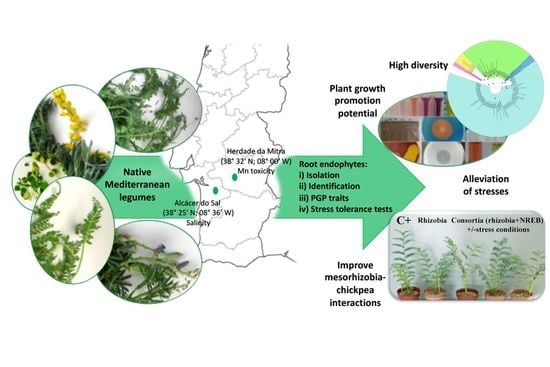
Mediterranean Native Leguminous Plants: A Reservoir of Endophytic Bacteria with Potential to Enhance Chickpea Growth under Stress Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Isolation of Culturable Endophytic Bacteria from Legume Roots
2.3. Screening for Plant Growth-Promoting Characteristics in vitro
2.4. Tolerance to Salt, Aluminium, and Manganese
2.5. Gnotobiotic Root Elongation Assay
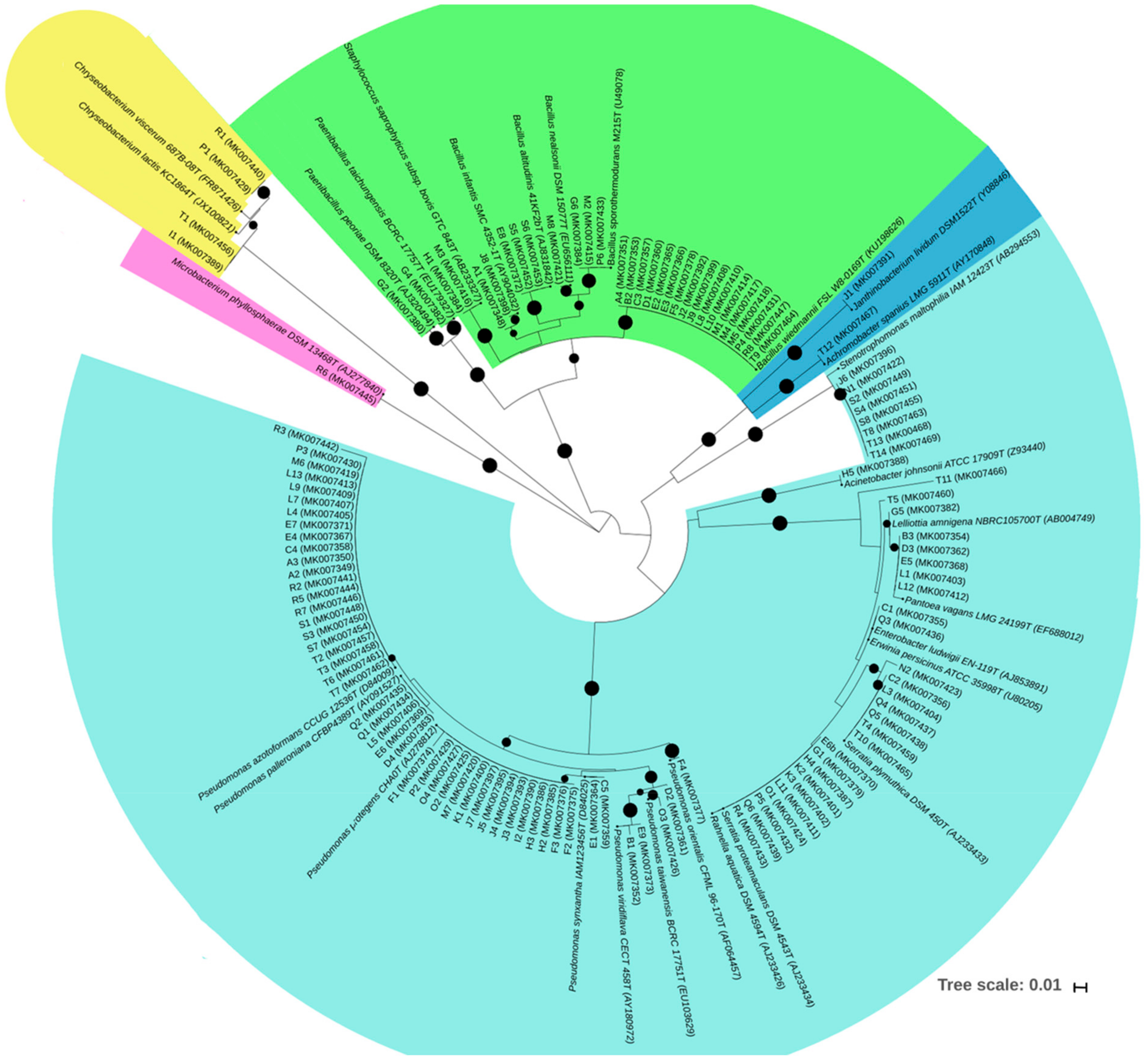
2.6. 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.7. Plant Growth Promotion Assays under Control and Stress Conditions
2.8. Statistical Analysis
3. Results
3.1. Native Legumes Surveyed
3.2. Bacterial Endophytes Diversity and Distribution Among Legume Host Species and Sites
3.3. Potential of Bacterial Endophytes for Plant Growth Promotion and Cellulase Production
3.4. Salt and Metals Tolerance and Promotion of Canola Root Length
3.5. Effect of Non-Rhizobial Endophytes on Symbiotic Performance of Mesorhizobium—Chickpea under Control and Stress Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orozco-Mosqueda, M.D.; Rocha-Granados, M.D.; Glick, B.R.; Santoyo, G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018, 208, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Rho, H.; Hsieh, M.; Kandel, S.L.; Cantillo, J.; Doty, S.L.; Kim, S.H. Do endophytes promote growth of host plants under stress? A meta-analysis on plant stress mitigation by endophytes. Microb. Ecol. 2018, 75, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.L.; Van Trappen, S.; Thompson, F.L.; Rocha, R.C.S.; Barbosa, H.R.; De Vos, P.; Moreira, C.A. Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.K.; Samanta, R.; Yadav, R.N.S. Inside the plant: Addressing bacterial endophytes in biotic stress alleviation. Arch. Microbiol. 2019, 201, 415–429. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Lata, R.; Chowdhury, S.; Gond, S.K.; White, J.F. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 2018, 66, 268–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, F.T.; Wallenstein, M.D. Below-ground connections underlying above-ground food production: A framework for optimising ecological connections in the rhizosphere. J. Ecol. 2017, 105, 913–920. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Ralli, P.; Tsaftaris, A. Barcoding the major Mediterranean leguminous crops by combining universal chloroplast and nuclear DNA sequence targets. Genet. Mol. Res. 2012, 11, 2548–2558. [Google Scholar] [CrossRef]
- Davis, P.H.; Lathyrus, L. Flora of Turkey and the East Aegean Islands; Davis, P.H., Ed.; Edinburgh University Press: Edinburgh, UK, 1970; pp. 328–389. [Google Scholar]
- Maxted, N.; Bennett, S.J. Legume diversity in the Mediterranean region. In Plant Genetic Resources of Legumes in the Mediterranean; Maxted, N., Bennett, S.J., Eds.; Klumer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 51–75. [Google Scholar]
- Porqueddu, C.; Ates, S.; Louhaichi, M.; Kyriazopoulos, A.P.; Moreno, G.; del Pozo, A.; Ovalle, C.; Ewing, M.A.; Nichols, P.G.H. Grasslands in “Old World’ and “New World’ Mediterranean-climate zones: Past trends, current status and future research priorities. Grass Forage Sci. 2016, 71, 1–35. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Minamisawa, K. Plant-Microbe Communications for Symbiosis. Plant. Cell Physiol. 2010, 51, 1377–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partida-Martinez, L.P.; Heil, M. The microbe-free plant: Fact or artifact? Front. Plant. Sci. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Alho, L.; Carvalho, M.; Brito, I.; Goss, M.J. The effect of arbuscular mycorrhiza fungal propagules on the growth of subterranean clover (Trifolium subterraneum L.) under Mn toxicity in ex situ experiments. Soil Use Manag. 2015, 31, 337–344. [Google Scholar] [CrossRef]
- Brígido, C.; van Tuinen, D.; Brito, I.; Alho, L.; Goss, M.J.; Carvalho, M. Management of the biological diversity of AM fungi by combination of host plant succession and integrity of extraradical mycelium. Soil Biol. Biochem. 2017, 112, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Monteiro-Henriques, T.; Martins, M.J.; Cerdeira, J.O.; Silva, P.; Arsenio, P.; Silva, A.; Bellu, A.; Costa, J.C. Bioclimatological mapping tackling uncertainty propagation: Application to mainland Portugal. Int. J. Climatol. 2016, 36, 400–411. [Google Scholar] [CrossRef]
- Talavera, S.; Aedo, C.; Castroviejo, S.; Herrero, A.; Romero Zarco, C.; Salgueiro, E.J.; Velayos, M. Flora iberica Real Jardín Botánico: Consejo Superior de Investigaciones Científicas; Castroviejo, S., Talavera, S., Eds.; CSIC: Madrid, Spain, 2000; Volume VII(II). [Google Scholar]
- Talavera, S.; Aedo, C.; Castroviejo, S.; Romero Zarco, C.; Sáez, L.; Salgueiro, E.J.; Velayos, M. Flora iberica Real Jardín Botánico: Consejo Superior de Investigaciones Científicas; CSIC: Madrid, Spain, 1999; Volume VII (I). [Google Scholar]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Félix, M.d.R.; Oliveira, S.; Carvalho, M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef]
- Gupta, R.; Singal, R.; Skankar, A.; Kuhad, R.C.; Saxena, R.K. A modified plate assay for screening phosphate solubilizing microorganisms. J. Gen. Appl. Microbiol. 1994, 40, 255–260. [Google Scholar] [CrossRef]
- Brígido, C.; Duan, J.; Glick, B.R. Methods to study 1-aminocyclopropane-1-carboxylate (ACC) deaminase in plant growth-promoting bacteria. In Handbook for Azospirillum: Technical Issues and Protocols; Cassán, F.D., Okon, Y., Creus, C.M., Eds.; Springer: Cham, Switzerland, 2015; pp. 287–305. [Google Scholar]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [PubMed]
- Nascimento, F.; Brígido, C.; Alho, L.; Glick, B.R.; Oliveira, S. Enhanced chickpea growth-promotion ability of a Mesorhizobium strain expressing an exogenous ACC deaminase gene. Plant. Soil 2012, 353, 221–230. [Google Scholar] [CrossRef]
- Duan, J.; Muller, K.M.; Charles, T.C.; Vesely, S.; Glick, B.R. 1-aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern Saskatchewan. Microb. Ecol. 2009, 57, 423–436. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plantarum. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Young, J.P.; Downer, H.L.; Eardly, B.D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J. Bacteriol. 1991, 173, 2271–2277. [Google Scholar] [CrossRef]
- Laranjo, M.; Machado, J.; Young, J.P.W.; Oliveira, S. High diversity of chickpea Mesorhizobium species isolated in a Portuguese agricultural region. FEMS Microbiol. Ecol. 2004, 48, 101–107. [Google Scholar] [CrossRef]
- Juck, D.; Charles, T.; Whyte, L.G.; Greer, C.W. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 2000, 33, 241–249. [Google Scholar] [CrossRef]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative Evaluation of Four Bacteria-Specific Primer Pairs for 16S rRNA Gene Surveys. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL-W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Vonuexkull, H.R.; Mutert, E. Global extent, development and economic-impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Taghavi, S.; Xie, P.; Orbach, M.J.; Alwathnani, H.A.; Rensing, C.; Wei, G. Phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis. Int. J. Phytorem. 2014, 16, 179–202. [Google Scholar] [CrossRef] [PubMed]
- Brígido, C.; Glick, B.R. Phytoremediation using Rhizobia. In Phytoremediation: Management of Environmental Contaminants; Ansari, A.A., Gill, S.S., Gill, G.R., Gill, G.R., Newman, L., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 2, pp. 95–114. [Google Scholar]
- Brígido, C.; Robledo, M.; Menendez, E.; Mateos, P.F.; Oliveira, S. A ClpB chaperone knockout mutant of Mesorhizobium ciceri shows a delay in the root nodulation of chickpea plants. Mol. Plant. Microbe Interact. 2012, 25, 1594–1604. [Google Scholar] [CrossRef]
- Paço, A.; Brígido, C.; Alexandre, A.; Mateos, P.F.; Oliveira, S. The symbiotic performance of chickpea rhizobia can be improved by additional copies of the clpB chaperone gene. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Brígido, C.; Nascimento, F.X.; Duan, J.; Glick, B.R.; Oliveira, S. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Mesorhizobium spp. reduces the negative effects of salt stress in chickpea. FEMS Microbiol. Lett. 2013, 349, 46–53. [Google Scholar]
- Broughton, W.J.; Dilworth, M.J. Control of Leghaemoglobin Synthesis in Snake Beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Gorley, R.N.; Somerfield, P.J.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; Primer-E Ltd.: Plymouth, UK, 2014; Volume 3, 256p. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. Permanova+ for primer: Guide to software and statisticl methods; PRIMER-E Ltd.: Plymouth, UK, 2008; 214p. [Google Scholar]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Mitter, E.K.; de Freitas, J.R.; Germida, J.J. Bacterial root microbiome of plants growing in oil sands reclamation covers. Front. Microbiol. 2017, 8, 849. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; Al-Lawati, A.; Jana, G.A.; Patankar, H.V.; Glick, B.R. Impact of soil salinity on the structure of the bacterial endophytic community identified from the roots of caliph medic (Medicago truncatula). PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Chen, W.; Zong, L.; Yang, J.; Jiao, S.; Lin, Y.; Wang, E.; Wei, G. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol. Ecol. 2017, 26, 1641–1651. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Wang, E.T.; Li, M.; Li, Q.Q.; Zhang, Y.M.; Zhao, S.J.; Jia, X.L.; Zhang, L.H.; Chen, W.F.; Chen, W.X. Effects of rhizobial inoculation, cropping systems and growth stages on endophytic bacterial community of soybean roots. Plant. Soil 2011, 347, 147–161. [Google Scholar] [CrossRef]
- Akinsanya, M.A.; Goh, J.K.; Lim, S.P.; Ting, A.S.Y. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 2015, 6, 159–163. [Google Scholar] [CrossRef]
- Piechulla, B.; Lemfack, M.C.; Kai, M. Effects of discrete bioactive microbial volatiles on plants and fungi. Plant. Cell Environ. 2017, 40, 2042–2067. [Google Scholar] [CrossRef]
- Cardoso, P.; Alves, A.; Silveira, P.; Sa, C.; Fidalgo, C.; Freitas, R.; Figueira, E. Bacteria from nodules of wild legume species: Phylogenetic diversity, plant growth promotion abilities and osmotolerance. Sci. Total Environ. 2018, 645, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.S.; Eichorst, S.A.; Wertz, J.T.; Schmidt, T.M.; Breznak, J.A. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 2004, 70, 4748–4755. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, D.K. Bacteria in Agrobiology: Crop. Ecosystems; Springer Science & Business Media: Berlin, Germany, 2011. [Google Scholar]
- Ardley, J.K.; Parker, M.A.; De Meyer, S.E.; Trengove, R.D.; O’Hara, G.W.; Reeve, W.G.; Yates, R.J.; Dilworth, M.J.; Willems, A.; Howieson, J.G. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov and Microvirga zambiensis sp. nov are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int. J. Syst. Evol. Microbiol. 2012, 62, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Flores-Felix, J.D.; Carro, L.; Velazquez, E.; Valverde, A.; Cerda-Castillo, E.; Garcia-Fraile, P.; Rivas, R. Phyllobacterium endophyticum sp. nov., isolated from nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 2013, 63, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, E.; Carro, L.; Flores-Félix, J.D.; Martínez-Hidalgo, P.; Menéndez, E.; Ramírez-Bahena, M.-H.; Mulas, R.; González-Andrés, F.; Martínez-Molina, E.; Peix, A. The Legume Nodule Microbiome: A Source of Plant Growth-Promoting Bacteria. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer Singapore: Singapore, 2017; pp. 41–70. [Google Scholar] [CrossRef]
- Msaddak, A.; Duran, D.; Rejili, M.; Mars, M.; Ruiz-Argueso, T.; Imperial, J.; Palacios, J.; Rey, L. Diverse bacteria affiliated with the genera Microvirga, Phyllobacterium, and Bradyrhizobium nodulate Lupinus micranthus growing in soils of Northern Tunisia. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Rasche, F.; Velvis, H.; Zachow, C.; Berg, G.; Van Elsas, J.D.; Sessitsch, A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J. Appl. Ecol. 2006, 43, 555–566. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Ding, T.; Palmer, M.W.; Melcher, U. Community terminal restriction fragment length polymorphisms reveal insights into the diversity and dynamics of leaf endophytic bacteria. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G.J. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Schreiter, S.; Ding, G.C.; Heuer, H.; Neumann, G.; Sandmann, M.; Grosch, R.; Kropf, S.; Smalla, K. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, B.; Blaser, M.B.; Klose, M.; Conrad, R. Crop rotation of flooded rice with upland maize impacts the resident and active methanogenic microbial community. Environ. Microbiol. 2016, 18, 2868–2885. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interations with plants and other organisms. Annu. Rev. Plant. Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Chen, H.W.; Chou, J.Y. Variation in indole-3-acetic acid production by wild Saccharomyces cerevisiae and S. paradoxus strains from diverse ecological sources and its effect on growth. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Bianco, C.; Imperlini, E.; Calogero, R.; Senatore, B.; Amoresano, A.; Carpentieri, A.; Pucci, P.; Defez, R. Indole-3-acetic acid improves Escherichia coli’s defences to stress. Arch. Microbiol. 2006, 185, 373–382. [Google Scholar] [CrossRef]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2014, 106, 85–125. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M. Soil microbes and the availability of soil nutrients. Acta Physiol. Plant 2013, 35, 3075–3084. [Google Scholar] [CrossRef]
- Brito, I.; Carvalho, M.; Alho, L.; Goss, M.J. Managing arbuscular mycorrhizal fungi for bioprotection: Mn toxicity. Soil Biol. Biochem. 2014, 68, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.B.; Yuan, L.X.; Zhang, J.L.; Li, H.G.; Bai, Z.H.; Chen, X.P.; Zhang, W.F.; Zhang, F.S. Phosphorus dynamics: from soil to plant. Plant. Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Suero, M.; Khanshour, A.; Martinez, Y.; Rickauer, M. A study on the susceptibility of the model legume plant Medicago truncatula to the soil-borne pathogen Fusarium oxysporum. Eur. J. Plant. Pathol. 2010, 126, 517–530. [Google Scholar] [CrossRef]
- Holtz, M.D.; Chang, K.F.; Hwang, S.F.; Gossen, B.D.; Strelkov, S.E. Characterization of Fusarium spp. associated with lupin in central Alberta, Canada. Can. J. Plant Pathol. 2013, 35, 56–67. [Google Scholar] [CrossRef]
- Nimbalkar, S.B.; Harsulkar, A.M.; Giri, A.P.; Sainani, M.N.; Franceschi, V.; Gupta, V.S. Differentially expressed gene transcripts in roots of resistant and susceptible chickpea plant (Cicer arietinum L.) upon Fusarium oxysporum infection. Physiol. Mol. Plant Pathol. 2006, 68, 176–188. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Brígido, C.; Glick, B.R.; Oliveira, S. Survey of plant growth-promoting mechanisms in native Portuguese chickpea Mesorhizobium isolates. Microb. Ecol. 2017, 73, 900–915. [Google Scholar] [CrossRef]
- Brígido, C.; Oliveira, S. Most acid-tolerant chickpea mesorhizobia show induction of major chaperone genes upon acid shock. Microb. Ecol. 2013, 65, 145–153. [Google Scholar] [CrossRef]
- Li, D.F.; Voigt, T.B.; Kent, A.D. Plant and soil effects on bacterial communities associated with Miscanthus × giganteus rhizosphere and rhizomes. Glob. Chang. Biol. Bioenergy 2016, 8, 183–193. [Google Scholar] [CrossRef]
- Abd Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Shurigin, V.V.; Hashem, A.; Abd Allah, E.F. Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under salt stress. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Berg, G.; Lindstrom, K.; Rasanen, L.A. Co-inoculation of Pseudomonas spp. with Rhizobium improves growth and symbiotic performance of fodder galega (Galega orientalis Lam.). Eur. J. Soil Biol. 2010, 46, 269–272. [Google Scholar] [CrossRef]
- Zhao, L.F.; Xu, Y.J.; Ma, Z.Q.; Deng, Z.S.; Shan, C.J.; Wei, G.H. Colonization and plant growth promoting characterization of endophytic Pseudomonas chlororaphis strain Zong1 isolated from Sophora alopecuroides root nodules. Braz. J. Microbiol. 2013, 44, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Abd Allah, E.F.; Alqarawi, A.A.; Ai-Huqail, A.A.; Wirth, S.; Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Li, J.H.; Wang, E.T.; Chen, W.F.; Chen, W.X. Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang province of China. Soil Biol. Biochem. 2008, 40, 238–246. [Google Scholar] [CrossRef]
- Bai, Y.M.; Zhou, X.M.; Smith, D.L. Enhanced soybean plant growth resulting from coinoculation of Bacillus strains with Bradyrhizobium japonicum. Crop. Sci. 2003, 43, 1774–1781. [Google Scholar] [CrossRef]
- Hungria, M.; Nogueira, M.A.; Araujo, R.S. Co-inoculation of soybeans and common beans with rhizobia and azospirilla: Strategies to improve sustainability. Biol. Fertil. Soils 2013, 49, 791–801. [Google Scholar] [CrossRef]
- Rajendran, G.; Sing, F.; Desai, A.J.; Archana, G. Enhanced growth and nodulation of pigeon pea by co-inoculation of Bacillus strains with Rhizobium spp. Bioresour. Technol. 2008, 99, 4544–4550. [Google Scholar] [CrossRef]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How Auxin and Cytokinin Phytohormones Modulate Root Microbe Interactions. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, H.; Choi, D.; Hwang, I. Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells 2012, 34, 117–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.B.; Charles, T.C.; Glick, B.R. PV expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 2004, 70, 5891–5897. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.B.; Guinel, F.C.; Glick, B.R. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 2003, 69, 4396–4402. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Oliveira, S.; Alho, L. Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 2012, 55, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Rossi, M.J. The Role of Rhizobial ACC Deaminase in the Nodulation Process of Leguminous Plants. Int. J. Agron. 2016. [Google Scholar] [CrossRef]
- Jorge, G.L.; Kisiala, A.; Morrison, E.; Aoki, M.; Nogueira, A.P.O.; Emery, R.J.N. Endosymbiotic Methylobacterium oryzae mitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environ. Exp. Bot. 2019, 162, 525–540. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, M.A.; Khan, A.L.; Waqas, M.; Shahzad, R.; Kim, A.Y.; Kang, S.M.; Lee, I.J. Bacterial endophytes from arid land plants regulate endogenous hormone content and promote growth in crop plants: An example of Sphingomonas sp. and Serratia marcescens. J. Plant. Interact. 2017, 12, 31–38. [Google Scholar] [CrossRef]
- Mrabet, M.; Mnasri, B.; Romdhane, S.B.; Laguerre, G.; Aouani, M.E.; Mhamdi, R. Agrobacterium strains isolated from root nodules of common bean specifically reduce nodulation by Rhizobium gallicum. FEMS Microbiol. Ecol. 2006, 56, 304–309. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; López-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
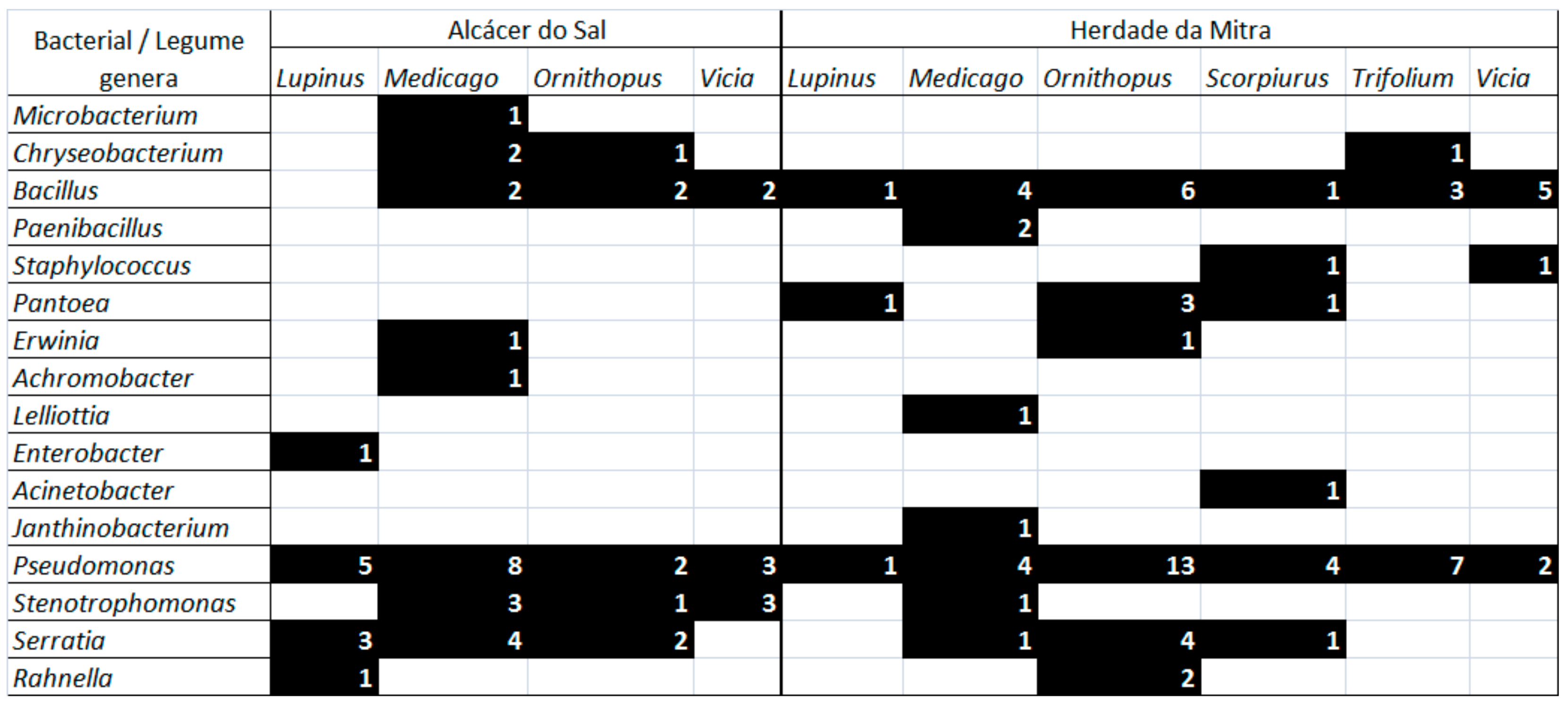


| Origin | Plant Species | Number of Isolates |
|---|---|---|
| Herdade da Mitra | Trifolium sp. | 4 |
| Lupinus luteus L. | 3 | |
| Ornithopus compressus L. | 26 | |
| Scorpiurus muricatus L. | 4 | |
| Trifolium subterraneum L. | 5 | |
| Medicago polymorpha L. | 14 | |
| Scorpiurus sulcatus L. | 5 | |
| Trifolium tomentosum L. | 2 | |
| Ornithopus pinnatus (Mill.) Druce | 3 | |
| Vicia sativa L. | 8 | |
| Alcácer do Sal | Ornithopus compressus L. | 2 |
| Lupinus luteus L. | 4 | |
| Ornithopus sativus Brot. | 6 | |
| Lupinus angustifolius L. | 6 | |
| Medicago polymorpha L. | 22 | |
| Vicia sativa L. | 8 | |
| Total | 122 |
| Bacterial Genera | Number of Isolates | IAA Production | Cellulase Production | Siderophore Production | Phosphate Solubilization |
|---|---|---|---|---|---|
| Microbacterium | 1 | 0 | 1 | 0 | 0 |
| Chryseobacterium | 4 | 4 | 1 | 2 | 0 |
| Bacillus | 26 | 15 | 15 | 1 | 2 |
| Paenibacillus | 2 | 2 | 2 | 1 | 0 |
| Staphylococcus | 2 | 2 | 0 | 2 | 1 |
| Pantoea | 5 | 5 | 1 | 0 | 1 |
| Erwinia | 2 | 2 | 0 | 0 | 1 |
| Achromobacter | 1 | 0 | 0 | 0 | 0 |
| Lelliottia | 1 | 1 | 0 | 0 | 1 |
| Enterobacter | 1 | 0 | 0 | 0 | 0 |
| Acinetobacter | 1 | 0 | 0 | 0 | 0 |
| Janthinobacterium | 1 | 0 | 0 | 1 | 0 |
| Pseudomonas | 49 | 16 | 30 | 28 | 19 |
| Stenotrophomonas | 8 | 5 | 1 | 3 | 0 |
| Serratia | 15 | 11 | 0 | 1 | 1 |
| Rahnella | 3 | 3 | 1 | 0 | 1 |
| Total | 122 | 66 | 52 | 39 | 27 |
| Origin Site | Origin Legume Species | Endophytic Bacterial Isolate | Maximal Concentration Tolerated | ||
|---|---|---|---|---|---|
| NaCl (mM) | Mn (mM) | Al (mM) | |||
| Herdade da Mitra | Trifolium sp. | Bacillus sp. A1 | 1275 | 0.1 | 0.1 |
| Lupinus luteus | Bacillus sp. B2 | 850 | 2.5 | 2.5 | |
| Lupinus luteus | Pantoea sp. B3 | 1275 | 1 | 2.5 | |
| Scorpiurus muricatus | Pseudomonas sp. D4 | 850 | 2.5 | 1 | |
| Trifolium subterraneum | Bacillus sp. F5 | 850 | 2.5 | 2.5 | |
| Medicago polymorpha | Serratia sp. G1 | 85 | 0.5 | 2.5 | |
| Medicago polymorpha | Paenibacillus sp. G2 | 850 | 1 | 2.5 | |
| Trifolium tomentoseum | Pseudomonas sp. I2 | 1275 | 2.5 | 0.1 | |
| Ornithopus compressus | Pseudomonas sp. L13 | 850 | 2.5 | 2.5 | |
| Vicia sativa | Bacillus sp. M4 | 850 | 2.5 | 2.5 | |
| Alcácer do Sal | Lupinus angustifolius | Pseudomonas sp. Q1 | 850 | 2.5 | 0.1 |
| Lupinus angustifolius | Pseudomonas sp. Q2 | 85 | 2.5 | 1 | |
| Lupinus angustifolius | Serratia sp. Q5 | 850 | 2.5 | 1 | |
| Lupinus angustifolius | Rahnella sp. Q6 | 1700 | 2.5 | 2.5 | |
| Medicago polymorpha | Erwinia sp. T5 | 1700 | 2.5 | 0.05 | |
| Cicer arietinum * | Kosakonia sp. MH5 * | 850 * | 1 * | 2.5 | |
| Condition | Treatment | Biomass | NDW | NN |
|---|---|---|---|---|
| Control | Positive Control | 2.561 ± 0.102 | 0 | 0 |
| LMS-1 | 1.370 ± 0.040 | 0.143 ± 0.004 | 79.8 ± 7.4 | |
| LMS-1 + D4 | 1.378 ± 0.036 | 0.134 ± 0.003 | 58.2 ± 3.0 ** | |
| LMS-1+ Q1 | 1.549 ± 0.036 | 0.164 ± 0.022 | 69.3 ± 5.5 | |
| LMS-1 + MH5 | 1.552 ± 0.097 | 0.173 ± 0.019 | 70.5 ± 4.4 | |
| LMS-1 + L13 | 1.575 ± 0.075 ** | 0.167 ± 0.018 | 58.3 ± 1.0 ** | |
| Ca36 | 1.295 ± 0.048 | 0.195 ± 0.015 | 84.2 ± 4.7 | |
| Ca36 + D4 | 1.298 ± 0.029 | 0.185 ± 0.020 | 60.8 ± 6.1 ** | |
| Ca36 + Q1 | 1.204 ± 0.081 | 0.198 ± 0.016 | 69.5 ± 8.8 | |
| Ca36 + MH5 | 1.269 ± 0.057 | 0.198 ± 0.010 | 72.3 ± 14.1 | |
| Ca36 + L13 | 1.363 ± 0.037 | 0.196 ± 0.011 | 78 ± 8.3 | |
| Salt | Positive Control | 1.605 ± 0.179 # | 0 | 0 |
| LMS-1 | 0.923 ± 0.019 | 0.083 ± 0.006 | 54.6 ± 9.1 | |
| LMS-1 + D4 | 0.925 ± 0.075 | 0.076 ± 0.012 | 78.3 ± 3.1 | |
| LMS-1 + Q1 | 1.036 ± 0.037 ** | 0.088 ± 0.007 | 70.8 ± 8.4 | |
| LMS-1 + MH5 | 1.259 ± 0.102 ** | 0.104 ± 0.009 | 98.5 ± 10.2 ** | |
| LMS-1 + L13 | 1.151 ± 0.080 ** | 0.093 ± 0.012 | 89.7 ± 4.8 ** | |
| Ca36 | 0.787 ± 0.043 | 0.083 ± 0.004 | 68.0 ± 12.4 | |
| Ca36 + D4 | 1.087 ± 0.128 ** | 0.128 ± 0.014 ** | 104.5 ± 20.7 | |
| Ca36 + Q1 | 0.997 ± 0.045 ** | 0.199 ± 0.006 ** | 100.2 ±17.3 | |
| Ca36 + MH5 | 0.768 ± 0.048 | 0.093 ± 0.004 | 76.0 ± 18.3 | |
| Ca36 + L13 | 0.989 ± 0.047 ** | 0.110 ± 0.004 ** | 72.6 ± 10.4 | |
| Mn | Positive Control | 2.031 ± 0.143 # | 0 | 0 |
| LMS-1 | 0.575 ± 0.044 | 0.043 ± 0.005 | 52.0 ± 9.8 | |
| LMS-1 + D4 | 0.685 ± 0.080 | 0.048 ± 0.008 | 53.7 ± 6.3 | |
| LMS-1 + Q1 | 0.695 ± 0.067 | 0.042 ± 0.010 | 52.5 ± 3.4 | |
| LMS-1 + MH5 | 0.500± 0.044 | 0.036 ± 0.005 | 59.0 ± 9.4 | |
| LMS-1 + L13 | 0.572 ± 0.064 | 0.041 ± 0.008 | 54.0 ± 15.3 | |
| Ca36 | 0.688 ± 0.043 | 0.089 ± 0.003 | 50 ± 2.6 | |
| Ca36 + D4 | 0.528 ± 0.041 ** | 0.060 ± 0.009 ** | 31.8 ± 3.2 ** | |
| Ca36 + Q1 | 0.972 ± 0.035 ** | 0.120 ± 0.005 ** | 47.0 ± 4.9 | |
| Ca36 + MH5 | 0.636 ± 0.013 | 0.100 ± 0.004 | 61.0 ± 11.9 | |
| Ca36 + L13 | 0.807 ± 0.082 | 0.112 ± 0.009 ** | 60.2 ± 5.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brígido, C.; Menéndez, E.; Paço, A.; Glick, B.R.; Belo, A.; Félix, M.R.; Oliveira, S.; Carvalho, M. Mediterranean Native Leguminous Plants: A Reservoir of Endophytic Bacteria with Potential to Enhance Chickpea Growth under Stress Conditions. Microorganisms 2019, 7, 392. https://doi.org/10.3390/microorganisms7100392
Brígido C, Menéndez E, Paço A, Glick BR, Belo A, Félix MR, Oliveira S, Carvalho M. Mediterranean Native Leguminous Plants: A Reservoir of Endophytic Bacteria with Potential to Enhance Chickpea Growth under Stress Conditions. Microorganisms. 2019; 7(10):392. https://doi.org/10.3390/microorganisms7100392
Chicago/Turabian StyleBrígido, Clarisse, Esther Menéndez, Ana Paço, Bernard R. Glick, Anabela Belo, Maria R. Félix, Solange Oliveira, and Mário Carvalho. 2019. "Mediterranean Native Leguminous Plants: A Reservoir of Endophytic Bacteria with Potential to Enhance Chickpea Growth under Stress Conditions" Microorganisms 7, no. 10: 392. https://doi.org/10.3390/microorganisms7100392