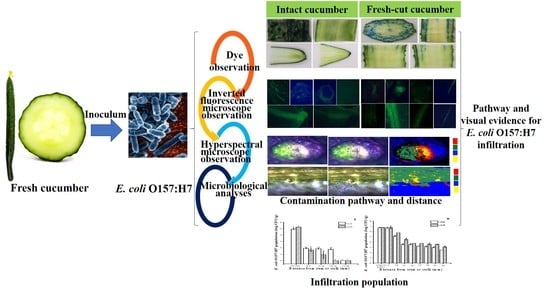
Elucidating Escherichia Coli O157:H7 Colonization and Internalization in Cucumbers Using an Inverted Fluorescence Microscope and Hyperspectral Microscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Colonization Mechanism in Intact Cucumbers
2.2. Colonization Mechanism in Crosscutting Cucumbers
2.3. Visualizing Colonization Mechanisms Using HMI
3. Materials and Methods
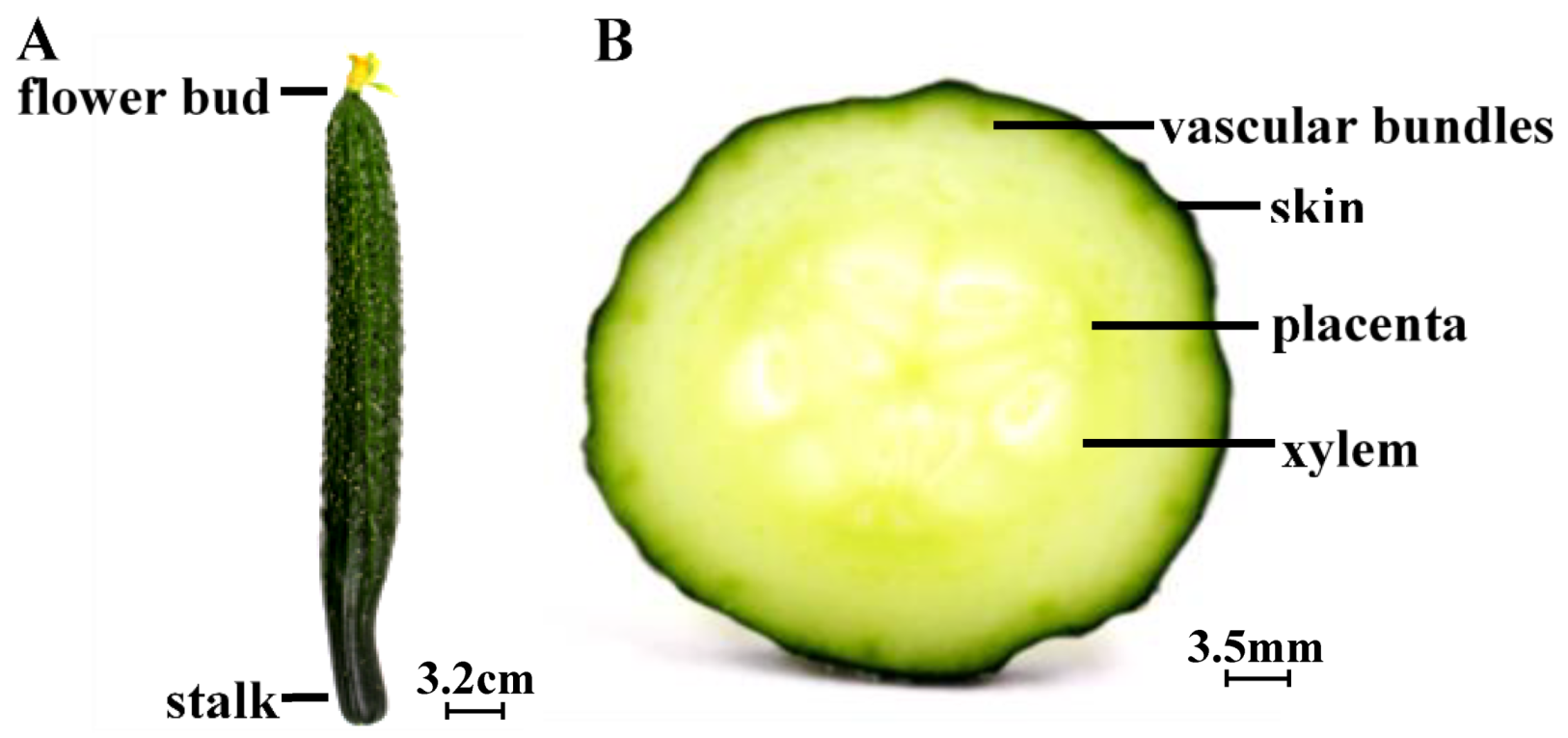
3.1. Cucumbers
3.2. Inoculum Preparation
3.3. Dye Infiltration into Cucumbers
3.4. Microbiological Analyses
3.4.1. Intact Cucumbers
3.4.2. Crosscutting Cucumbers
3.5. Visualizing E. coli O157:H7 Colonization Using an Inverted Fluorescence Microscope
3.6. Visualizing E. coli O157:H7 Colonization Using HMI
3.7. Statistical Analyses
4. Conclusions
5. Highlights
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bin, J.; Qiuli, Z.; Jinhua, Z.; Lipu, G.; Qing, W.; Wenqiang, G.; Junyan, S. Application of postharvest putrescine treatment to maintain the quality and increase the activity of antioxidative enzyme of cucumber. Sci. Hortic. 2018, 239, 210–215. [Google Scholar] [CrossRef]
- Tuffs, A. Outbreak of E. coli in Germany is linked to cucumbers from Spain. Br. Med. J. 2011, 342, d3394. [Google Scholar] [CrossRef] [PubMed]
- Franz, E.; Veenman, C.; Van Hoek, A.H.A.M.; de Roda Husman, A.; Blaak, H. Pathogenic Escherichia coli producing Extended-Spectrum β-Lactamases isolated from surface water and wastewater. Sci. R. 2015, 5, 14372. [Google Scholar] [CrossRef] [PubMed]
- de Quadros Rodrigues, R.; Loiko, M.R.; de Paula, C.M.D.; Hessel, C.T.; Jacxsens, L.; Uyttendaele, M.; Bender, R.J.; Tondo, E.C. Microbiological contamination linked to implementation of good agricultural practices in the production of organic lettuce in Southern Brazil. Food Control 2014, 42, 152–164. [Google Scholar] [CrossRef]
- Taştan, Ö.; Pataro, G.; Donsì, F.; Ferrari, G.; Baysal, T. Decontamination of fresh-cut cucumber slices by a combination of a modified chitosan coating containing carvacrol nanoemulsions and pulsed light. Int. J. Food Microbiol. 2017, 260, 75–80. [Google Scholar] [CrossRef]
- Cui, H.; Ma, C.; Lin, L. Co-loaded proteinase k/thyme oil liposomes for inactivation of Escherichia coli O157:H7 biofilms on cucumber. Food Funct. 2016, 7, 4030–4040. [Google Scholar] [CrossRef]
- Breidt, F.; Caldwell, J.M. Survival of Escherichia coli O157: H7 in cucumber fermentation brines. J. Food Sci. 2011, 76, M198–M203. [Google Scholar] [CrossRef]
- Sagdic, O.; Ozturk, I.; Tornuk, F. Inactivation of non-toxigenic and toxigenic Escherichia coli O157:H7 inoculated on minimally processed tomatoes and cucumbers: Utilization of hydrosols of lamiaceae spices as natural food sanitizers. Food Control 2013, 30, 7–14. [Google Scholar] [CrossRef]
- Zhou, B.; Luo, Y.; Bauchan, G.R.; Feng, H.; Stommel, J.R. Visualizing pathogen internalization pathways in fresh tomatoes using MicroCT and confocal laser scanning microscopy. Food Control 2018, 85, 276–282. [Google Scholar] [CrossRef]
- Macarisin, D.; Patel, J.; Bauchan, G.; Giron, J.A.; Ravishankar, S. Effect of spinach cultivar and bacterial adherence factors on survival of Escherichia coli O157:H7 on spinach leaves. J. Food Prot. 2013, 76, 1829–1837. [Google Scholar] [CrossRef]
- Mitra, R.; Cuesta-Alonso, E.; Wayadande, A.; Talley, J.; Gilliland, S.; Fletcher, J. Effect of route of introduction and host cultivar on the colonization, internalization, and movement of the human pathogen Escherichia coli O157:H7 in spinach. J. Food Prot. 2009, 72, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.K.; Berger, C.N.; Feys, B.; Knutton, S.; Pallen, M.J.; Frankel, G. Enterohemorrhagic Escherichia coli exploits espa filaments for attachment to salad leaves. Appl. Environ. Microbiol. 2008, 74, 2908–2914. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Newman, M.C.; Archbold, D.D.; Hamilton-Kemp, T.R. Survival of Escherichia coli O157:H7 on strawberry fruit and reduction of the pathogen population by chemical agents. J. Food Pro. 2001, 64, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Yuk, H.G.; Bartz, J.A.; Schneider, K.R. The effectiveness of sanitizer treatments in inactivation of Salmonella spp. from bell pepper, cucumber, and strawberry. J. Food Sci. 2010, 71, M95–M99. [Google Scholar] [CrossRef]
- Klerks, M.M.; Franz, E.; Van Gent-Pelzer, M.; Zijlstra, C.; Van Bruggen, A.H.C. Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 2007, 1, 620–631. [Google Scholar] [CrossRef]
- Park, B.; Seo, Y.; Yoon, S.C.; Hinton, A., Jr.; Windham, W.R.; Lawrence, K.C. Hyperspectral microscope imaging methods to classify gram-positive and gram-negative foodborne pathogenic bacteria. Trans. Am. Soc. Agric. Biol. Eng. 2015, 58, 5–16. [Google Scholar] [CrossRef]
- Yoon, S.C.; Windham, W.; Ladely, S.; Heitschmidt, J.; Lawrence, K.; Park, B.; Narang, N.; Cray, W. Hyperspectral imaging for differentiating colonies of non-O157 Shiga-toxin producing Escherichia coli (STEC) serogroups on spread plates of pure cultures. J. Near Infrared Spectrosc. 2013, 21, 81–95. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, W.; He, H.; Nie, J.; Zhao, J.; Ren, G.; Li, Y.; Zhang, D.; Pan, J.; Cai, R. Tuberculate fruit gene Tu encodes a C2H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 2014, 78, 1034–1046. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Ding, L.; Yan, S.; Liu, M.; Jiang, L.; Wang, Q.; Yan, L.; Liu, R.; Zhang, X.; et al. Phloem transcriptome signatures underpin the physiological differentiation of the pedicel, stalk and fruit of cucumber (Cucumis sativus L.). Plant Cell Physiol. 2015, 57, 19–34. [Google Scholar] [CrossRef]
- Xia, X.; Luo, Y.; Yang, Y.; Vinyard, B.; Schneider, K.; Meng, J. Effects of tomato variety, temperature differential, and post-stem removal time on internalization of Salmonella enterica serovar Thompson in tomatoes. J. Food Pro. 2012, 75, 297–303. [Google Scholar] [CrossRef]
- Berger, C.N.; Shaw, R.K.; Ruiz-Perez, F.; Nataro, J.P.; Henderson, I.R.; Pallen, M.J.; Frankel, G. Interaction of enteroaggregative Escherichia coli with salad leaves. Environ. Microbiol. R. 2010, 1, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Warning, A.; Datta, A.K.; Bartz, J.A. Mechanistic understanding of temperature-driven water and bacterial infiltration during hydrocooling of fresh produce. Postharvest Biol. Technol. 2016, 118, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Currie, C.G.; Mackie, S.; Tree, J.; McAteer, S.; McKendrick, I.; McNeilly, T.N.; Roe, A.; La Ragione, R.M.; Woodward, M.J.; et al. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell Microbiol. 2010, 11, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Payvandi, S.; Daly, K.R.; Jones, D.L.; Talboys, P.; Zygalakis, K.C.; Roose, T. A mathematical model of water and nutrient transport in xylem vessels of a wheat plant. Bull. Math. Biol. 2014, 76, 566–596. [Google Scholar] [CrossRef]
- Richards, J.A. Remote Sensing Digital Image Analysis: An Introduction; Springer: Berlin, Germany, 2013. [Google Scholar] [CrossRef]
- Manjunatha, M.; Anurag, R.K. Effect of modified atmosphere packaging and storage conditions on quality characteristics of cucumber. J. Food Sci. Technol. 2014, 51, 3470–3475. [Google Scholar] [CrossRef]
- Dan, H.; Okuhara, K.; Kohyama, K. Characterization of mechanical stress distributions in a cross section of cucumber fruits: Bisector reference line represents tissue anatomy. J. Sci. Food Agric. 2010, 85, 785–790. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Y.; Tao, D.; Wang, S.; Li, C.; Zheng, F.; Wu, Z. Reduction of Escherichia coli O157: H7, Listeria monocytogenes, and Naturally Present Microbe Counts on Lettuce using an Acid Mixture of Acetic and Lactic Acid. Microorganisms 2019, 7, 373. [Google Scholar] [CrossRef]
- Gutierrez, M.E.; Janes, M.E.; Torrico, D.D.; Carabante, K.M.; Prinyawiwatkul, W. Assessment of the ability of five culture media for the detection of\r, Escherichia coli\r, O157. Int. J. Food Sci. Technol. 2016, 51, 1910–1915. [Google Scholar] [CrossRef]
- Kazue, T.; Matute, C.M.; Hassan, A.N.; Frank, J.F. Comparison of the attachment of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella Typhimurium, and Pseudomonas fluorescens to lettuce leaves. J. Food Prot. 2000, 63, 1433–1437. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Zhao, X.; Meng, Z.; Zou, W. Using hyperspectral imaging technology to identify diseased tomato leaves. Spie/cos Photonics Asia. Int. Soc. Opt. Photon. 2016, 10030, 1003011. [Google Scholar] [CrossRef]
- Siripatrawan, U.; Makino, Y.; Kawagoe, Y.; Oshita, S. Rapid detection of Escherichia coli contamination in packaged fresh spinach using hyperspectral imaging. Talanta 2011, 85, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.J.; Monteiro, S.T.; Schneider, S. Evaluating classification techniques for mapping vertical geology using field-based hyperspectral sensors. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3066–3080. [Google Scholar] [CrossRef]
- Sankaran, S.; Ehsani, R.; Etxeberria, E. Mid-infrared spectroscopy for detection of Huanglongbing (greening) in citrus leaves. Talanta 2010, 83, 574–581. [Google Scholar] [CrossRef] [PubMed]







| Symbol of Cucumber Tissue | Description | Stomata Training Samples (Points) | Vascular Bundle Training Samples (Points) |
|---|---|---|---|
| I1 | Infiltrated area | 218 | 200 |
| U1 | Uninfiltrated area | 937 | 3123 |
| U2 | Dark and uninfiltrated area | 1008 | 1884 |
| U3 | Bright and uninfiltrated area | 169 | 140 |
| Total | 2332 | 5347 |
| Stomata | Vascular Bundles | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| I1 | U1 | U2 | U3 | I1 | U1 | U2 | U3 | ||
| Mean angle | 0.1659 | 0.2670 | 0.1460 | 0.1931 | 0.0928 | 0.0757 | 0.0645 | 0.1205 | |
| Standard deviation | 0.0474 | 0.0726 | 0.0631 | 0.0482 | 0.0516 | 0.0554 | 0.0308 | 0.0637 | |
| PA | 85.78 | 72.25 | 76.88 | 88.17 | 45.5 | 94.04 | 100 | 90.71 | |
| UA | 75.71 | 100 | 99.74 | 83.71 | 91.92 | 97.25 | 98.64 | 39.94 | |
| OA | 76.67 | 94.24 | |||||||
| Kappa | 67.78 | 89.37 | |||||||
| Inter-class | I1 | - | 0.1221 | 0.5102 | 0.0208 | - | - | - | - |
| U1 | - | - | 0.5568 | 0.0693 | 0.0614 | - | - | - | |
| U2 | - | - | - | 0.2633 | 0.4779 | 0.5393 | - | - | |
| U3 | - | - | - | -- | 0.0407 | 0.1021 | 0.2796 | - | |
| J–M distance | I1 | - | 1.986 | 2.000 | 1.884 | - | - | - | - |
| U1 | - | - | 1.939 | 1.999 | 1.980 | - | - | - | |
| U2 | - | - | - | 2.000 | 1.964 | 1.999 | - | - | |
| U3 | - | - | - | - | 1.868 | 1.984 | 1.995 | - | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Wang, D.; Ma, Y.; Guan, H.; Liang, H.; Zhao, X. Elucidating Escherichia Coli O157:H7 Colonization and Internalization in Cucumbers Using an Inverted Fluorescence Microscope and Hyperspectral Microscopy. Microorganisms 2019, 7, 499. https://doi.org/10.3390/microorganisms7110499
Sun Y, Wang D, Ma Y, Guan H, Liang H, Zhao X. Elucidating Escherichia Coli O157:H7 Colonization and Internalization in Cucumbers Using an Inverted Fluorescence Microscope and Hyperspectral Microscopy. Microorganisms. 2019; 7(11):499. https://doi.org/10.3390/microorganisms7110499
Chicago/Turabian StyleSun, Yeting, Dan Wang, Yue Ma, Hongyang Guan, Hao Liang, and Xiaoyan Zhao. 2019. "Elucidating Escherichia Coli O157:H7 Colonization and Internalization in Cucumbers Using an Inverted Fluorescence Microscope and Hyperspectral Microscopy" Microorganisms 7, no. 11: 499. https://doi.org/10.3390/microorganisms7110499