Enhanced Production of Fatty Acid Ethyl Ester with Engineered fabHDG Operon in Escherichia coli
Abstract
:1. Introduction
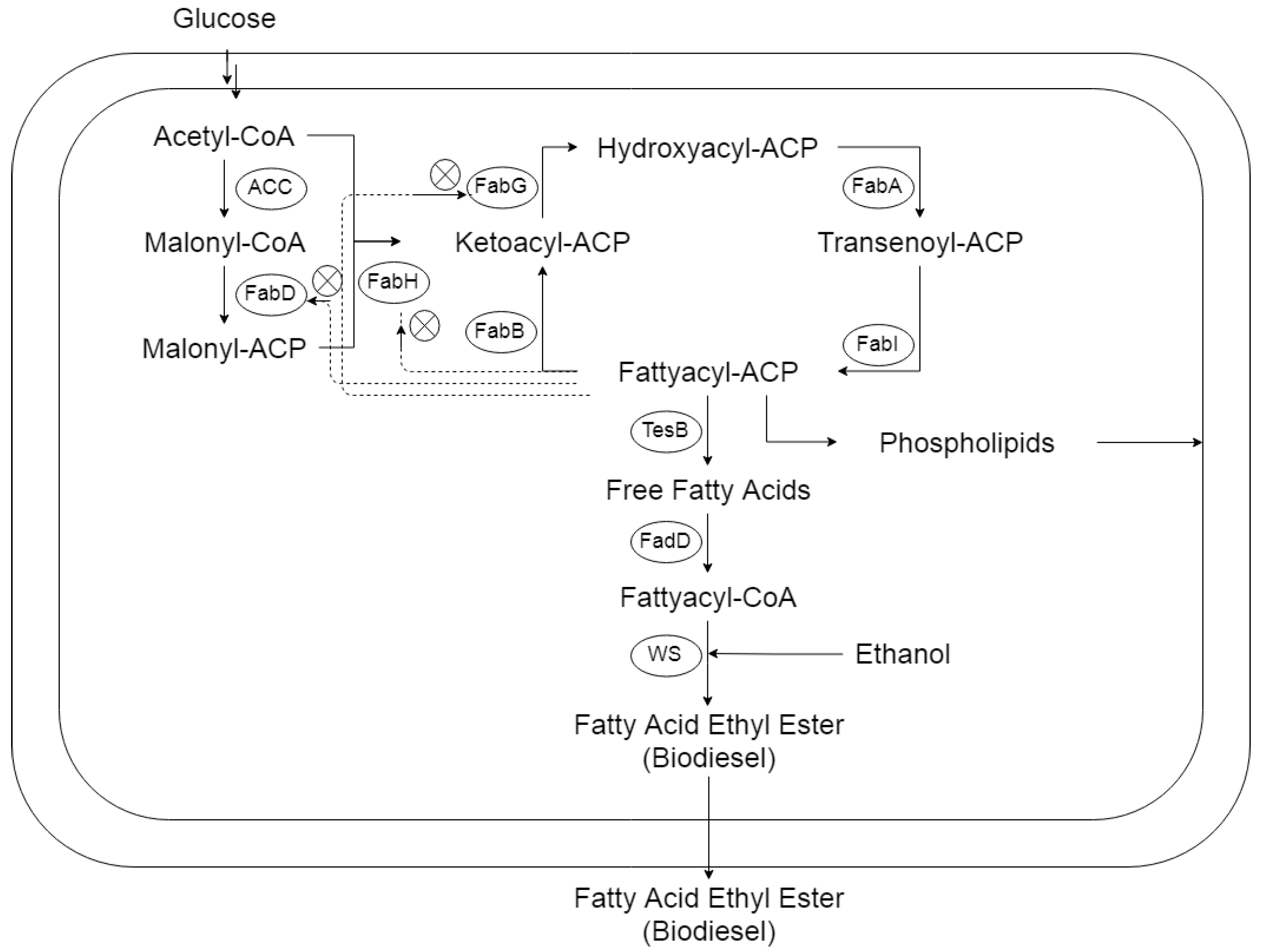
FAEE Production in Bacteria
2. Materials and Methods
2.1. Microbial Strains, Enzymes, and Chemicals
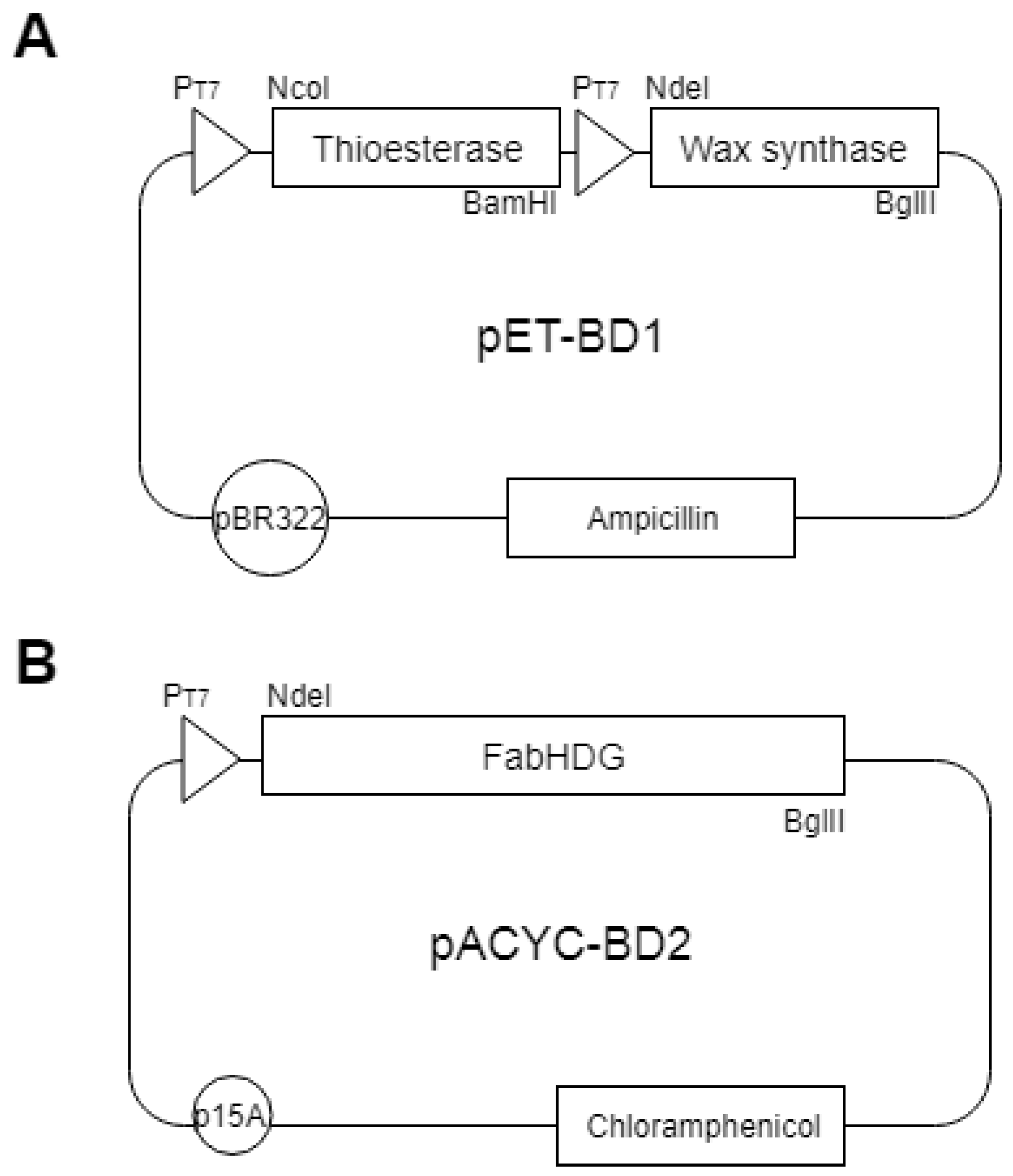
2.2. Construction of Plasmids Expressing Tes, and WS
2.3. Construction of Plasmids Expressing the FA Operon (fabHDG)
2.4. Media, Growth Conditions, and FAEE Analysis
3. Results and Discussion
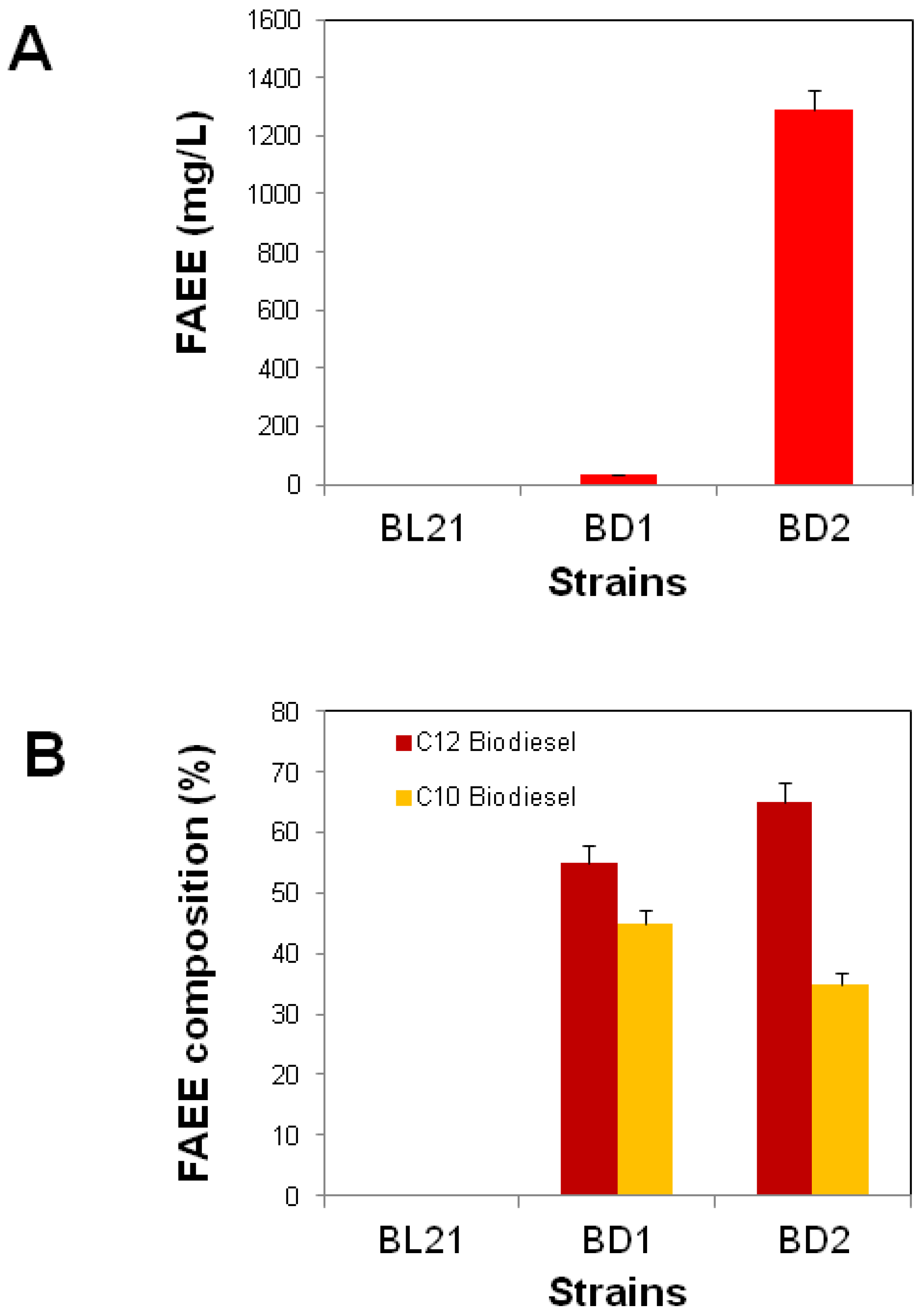
3.1. FAEE Production in E. coli with Plasmid Encoding TesB and WS
3.2. Improved FAEE Production with the Expression of the FA (fabHDG) Operon
Author Contributions
Funding
Conflicts of Interest
References
- Gronenberg, L.S.; Marcheschi, R.J.; Liao, J.C. Next generation biofuel engineering in prokaryotes. Curr. Opin. Chem. Biol. 2013, 17, 462–471. [Google Scholar] [CrossRef]
- Hong, J. Uncertainty propagation in life cycle assessment of biodiesel versus diesel: Global warming and non-renewable energy. Bioresour. Technol. 2012, 113, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Rahman, Z.; Nawab, J.; Sung, B.; Kim, S. A critical analysis of bio-hydrocarbon production in bacteria: Current challenges and future directions. Energies 2018, 11, 2663. [Google Scholar] [CrossRef]
- Rahman, Z.; Rashid, N.; Nawab, J.; Ilyas, M.; Sung, B.H.; Kim, S.C. Escherichia coli as a fatty acid and biodiesel factory: Current challenges and future directions. Environ. Sci. Pollut. Res. 2016, 23, 12007–12018. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Zhang, F.; Del Cardayre, S.B.; Keasling, J.D. Microbial engineering for the production of advanced biofuels. Nature 2012, 488, 320. [Google Scholar] [CrossRef]
- Nawabi, P.; Bauer, S.; Kyrpides, N.; Lykidis, A. Engineering Escherichia coli for biodiesel production utilizing a bacterial fatty acid methyltransferase. Appl. Environ. Microbiol. 2011, 77, 8052–8061. [Google Scholar] [CrossRef]
- Bautista, L.F.; Vicente, G.; Rodriguez, R.; Pacheco, M. Optimisation of FAME production from waste cooking oil for biodiesel use. Biomass Bioenergy 2009, 33, 862–872. [Google Scholar] [CrossRef]
- Antolin, G.; Tinaut, F.V.; Briceno, Y.; Castano, V.; Perez, C.; Ramirez, A.I. Optimisation of biodiesel production by sunflower oil transesterification. Bioresour. Technol. 2002, 83, 111–114. [Google Scholar] [CrossRef]
- Sathesh-Prabu, C.; Shin, K.S.; Kwak, G.H.; Jung, S.-K.; Lee, S.K. Microbial Production of Fatty Acid via Metabolic Engineering and Synthetic Biology. Biotechnol. Bioprocess. Eng. 2019, 24, 23–40. [Google Scholar] [CrossRef]
- Wierzbicki, M.; Niraula, N.; Yarrabothula, A.; Layton, D.S.; Trinh, C.T. Engineering an Escherichia coli platform to synthesize designer biodiesels. J. Biotechnol. 2016, 224, 27–34. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Stölting, T.; Steinbüchel, A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 2006, 152, 2529–2536. [Google Scholar] [CrossRef] [PubMed]
- Elbahloul, Y.; Steinbüchel, A. Pilot-scale production of fatty acid ethyl esters by an engineered Escherichia coli strain harboring the p (Microdiesel) plasmid. Appl. Environ. Microbiol. 2010, 76, 4560–4565. [Google Scholar] [CrossRef] [PubMed]
- Steen, E.J.; Kang, Y.; Bokinsky, G.; Hu, Z.; Schirmer, A.; McClure, A.; Del Cardayre, S.B.; Keasling, J.D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010, 463, 559. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, K.; Jackowski, S.; Rock, C.O.; Cronan, J.E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Mol. Biol. Rev. 1993, 57, 522–542. [Google Scholar]
- Payoe, R.; Fahlman, R.P. Dependence of RelA-mediated (p) ppGpp formation on tRNA identity. Biochemistry 2018, 50, 3075–3083. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, H.; Jiang, X.; Ai, G.; Yu, B.; Zhu, K. Biosynthesis of butenoic acid through fatty acid biosynthesis pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 1795–1804. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Agrawal, A.; San, K.-Y. Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab. Eng. 2011, 13, 713–722. [Google Scholar] [CrossRef]
- Voelker, T.A.; Davies, H.M. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 1994, 176, 7320–7327. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, L.; Liu, Q.; Qin, W.; Yang, J.; Cao, Y.; Jiang, X.; Zhao, G.; Xian, M. Boosting the free fatty acid synthesis of Escherichia coli by expression of a cytosolic Acinetobacter baylyi thioesterase. Biotechnol. Biofuels 2012, 5, 76. [Google Scholar] [CrossRef]
- Liao, J.C.; Mi, L.; Pontrelli, S.; Luo, S. Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 2016, 14, 288. [Google Scholar] [CrossRef]
- Davis, M.S.; Solbiati, J.; Cronan, J.E. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 2000, 275, 28593–28598. [Google Scholar] [CrossRef] [PubMed]
- My, L.; Rekoske, B.; Lemke, J.J.; Viala, J.P.; Gourse, R.L.; Bouveret, E. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J. Bacteriol. 2013, 195, 3784–3795. [Google Scholar] [CrossRef] [PubMed]
- Geiger, O. Biogenesis of Fatty Acids, Lipids and Membranes; Springer: Berlin, Germany, 2018. [Google Scholar]
- Rahman, Z.; Sung, B.H.; Yi, J.-Y.; Bui, L.M.; Lee, J.H.; Kim, S.C. Enhanced production of n-alkanes in Escherichia coli by spatial organization of biosynthetic pathway enzymes. J. Biotechnol. 2014, 192, 187–191. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Lennen, R.M.; Pfleger, B.F. Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 2012, 30, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Durfee, T.; Hansen, A.-M.; Zhi, H.; Blattner, F.R.; Jin, D.J. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 2008, 190, 1084–1096. [Google Scholar] [CrossRef]
- Saraf, S.; Thomas, B. Influence of feedstock and process chemistry on biodiesel quality. Process. Saf. Environ. Prot. 2007, 85, 360–364. [Google Scholar] [CrossRef]
- Ruppe, A.; Fox, J.M. Analysis of Interdependent Kinetic Controls of Fatty Acid Synthases. Acs Catal. 2018, 8, 11722–11734. [Google Scholar] [CrossRef]
- He, L.; Xiao, Y.; Gebreselassie, N.; Zhang, F.; Antoniewicz, M.R.; Tang, Y.J.; Peng, L. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol. Bioeng. 2014, 111, 575–585. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef]
- Zhang, F.; Ouellet, M.; Batth, T.S.; Adams, P.D.; Petzold, C.J.; Mukhopadhyay, A.; Keasling, J.D. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab. Eng. 2012, 14, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Royce, L.A.; Liu, P.; Stebbins, M.J.; Hanson, B.C.; Jarboe, L.R. The damaging effects of short chain fatty acids on Escherichia coli membranes. Appl. Microbiol. Biotechnol. 2013, 97, 8317–8327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Items | Description | Reference |
|---|---|---|
| Strains | ||
| BL21 | E. coli str. B F–ompT gal dcm lon hsdSB(rB–mB–) λ (DE3 [lacI lacUV5−T7p07 ind1 sam7 nin5]) [malB+]K-12 (λS) | [25] |
| BD1 | BL21 (DE3) + pET-BD1 | This study |
| BD2 | BD1 + pACYC-BD2 | This study |
| Plasmids | ||
| pET Duet-1 | pBR322 origin.; Ampr; | Novagen, Inc. |
| pACYC Duet-1 | p15A origin.; Cmr | Novagen, Inc. |
| pET-BD1 | pET Duet-1.; pBR322 origin.; Ampr; PT7−. (tesB) PT7 (ws) | This study |
| pACYC-BD2 | pACYC Duet-1.; p15A origin.; Cmr.; PT7− (fabHDG) | This study |
| Primers | ||
| TES-F (NcoI) | TTCCATGGATGGCCACCACCTCTTTAGCT | This study |
| TES-R (BamHI) | ATGAGGATCCTTACACCCTCGGTTCTGCGG | This study |
| WS-F (NdeI) | CCAACATATGATGAAGATGAAGAGTTTGATTTA | This study |
| WS-R (BglII) | GGTAAGATCTTTAATTGGCTGTTTTAAT | This study |
| FAB-F (NdeI) | CCAACATATGATGTATACGAAGATTATTGG | This study |
| FAB-R (BglI1) | GGTAAGATCTTTTCAGACCATGTACATCCC | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, Z.; Sung, B.H.; Nawab, J.; Siddiqui, M.F.; Ali, A.; Geraldi, A.; Kim, S.C. Enhanced Production of Fatty Acid Ethyl Ester with Engineered fabHDG Operon in Escherichia coli. Microorganisms 2019, 7, 552. https://doi.org/10.3390/microorganisms7110552
Rahman Z, Sung BH, Nawab J, Siddiqui MF, Ali A, Geraldi A, Kim SC. Enhanced Production of Fatty Acid Ethyl Ester with Engineered fabHDG Operon in Escherichia coli. Microorganisms. 2019; 7(11):552. https://doi.org/10.3390/microorganisms7110552
Chicago/Turabian StyleRahman, Ziaur, Bong Hyun Sung, Javed Nawab, Muhammad Faisal Siddiqui, Abid Ali, Almando Geraldi, and Sun Chang Kim. 2019. "Enhanced Production of Fatty Acid Ethyl Ester with Engineered fabHDG Operon in Escherichia coli" Microorganisms 7, no. 11: 552. https://doi.org/10.3390/microorganisms7110552






