Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present
Abstract
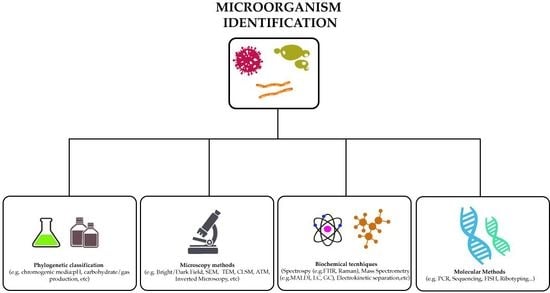
:1. Introduction
2. Methods to Identify Microorganisms
2.1. Historical Evolution of Microorganism Identification
2.2. Identification Methods Using Chromogenic Media
2.3. Microscopy Techniques
3. Biochemical Analytical Methods to Detect Microorganisms
3.1. Traditional Biochemical Methods
3.2. Mass Spectrometry-Based Methods
3.2.1. Liquid Chromatography: High Performance Liquid Chromatography (HPLC)-Based Methods
3.2.2. Gas Chromatography–Mass Spectrometry
3.2.3. Matrix-Assisted Laser Desorption/Ionization (MALDI)-Time-of-Flight (TOF)
3.3. Spectroscopic Methods
3.3.1. Infrared Spectroscopy (FTIR)
3.3.2. Raman Spectroscopy–Vibrational Spectroscopy
3.3.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
3.4. Electrokinetic Separation Methods
3.5. Microfluidic Chips
4. Molecular Methods Used to Detect Bacteria
4.1. 16S rRNA PCR-Sequencing
4.2. Real-Time PCR
4.3. Random Amplification of Polymorphic DNA–RAPD-PCR
4.4. Restriction Fragment Length Polymorphism–RFLP
4.5. Amplified Fragment Length Polymorphism–AFLP
4.6. Pulsed-Field Gel Electrophoresis–PFGE
4.7. Ribotyping
4.8. Whole-Genome Sequencing–WGS
4.9. MALDI-TOF-MS in Bacteria
5. Molecular Methods Used to Detect Yeasts
5.1. PCR
5.2. Quantitative Real Time-PCR
5.3. DNA Fingerprinting Methods
5.3.1. Pulsed Field Gel Electrophoresis (PFGE)
5.3.2. Restriction Fragment Length Polymorphisms (RFLP)
5.3.3. Fragment Length Polymorphisms (RFLP)
5.3.4. Random Amplified Polymorphic DNA (RAPD)
5.3.5. Amplified Fragment Length Polymorphism (AFLP)
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bisen, P.S.; Debnath, M.; Prasad, G.B. Microbes in Applied Research: Current Advances and Challenges; Bisen, P.S., Debnath, M., Prasad, G.B., Eds.; Wiley-Blackwell: Oxford, UK, 2012; p. 699. [Google Scholar]
- Prakash, O.; Verma, M.; Sharma, P.; Kumar, M.; Kumari, K.; Singh, A.; Kumari, H.; Jit, S.; Gupta, S.K.; Khanna, M.; et al. Polyphasic approach of bacterial classification—An overview of recent advances. Indian J. Microbiol. 2007, 47, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Manafi, M. Fluorogenic and chromogenic substrates in culture media and identification tests. Int. J. Food Microbiol. 1996, 31, 45–58. [Google Scholar] [CrossRef]
- Ramamurthy, T.; Ghosh, A.; Pazhani, G.P.; Shinoda, S. Current Perspectives on Viable but Non-Culturable (VBNC) Pathogenic Bacteria. Front. Public Heal. 2014, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Bochner, B.R. Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 2009, 33, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Castro-Escarpulli, G.; Alonso-Aguilar, N.M.; Rivera, G.; Bocanegra-Garcia, V.; Guo, X.; Jurez-Enrquez, S.R.; Luna-Herrera, J.; Martnez, C.M.; Guadalupe, A.-A.M. Identification and Typing Methods for theStudy of Bacterial Infections: A Brief Reviewand Mycobacterial as Case of Study. Arch. Clin. Microbiol. 2016, 7, 1–10. [Google Scholar]
- Lincoln, R.J.; Boxshall, G.A.; Clark, P.F. A Dictionary of Ecology, Evolution, and Systematics; Cambridge University Press: Cambridge, UK, 1998; p. 298. [Google Scholar]
- Wägele, J.W. Foundations of Phylogenetic Systematics; Verlag Dr. Friedrich Pfeil: Munich, Germany, 2005; p. 365. [Google Scholar]
- Guerra-García, J.M.; Espinosa, F.; García-Gómez, J.C. Trends in Taxonomy today: An overview about the main topics in Taxonomy. Zool. Baetica 2008, 19, 15–19. [Google Scholar]
- Godfray, H.C.J. Challenges for taxonomy. Nature 2002, 417, 17–19. [Google Scholar] [PubMed]
- Enghoff, H. What is taxonomy? An overview with myriapodological examples. Soil Org. 2009, 81, 441–451. [Google Scholar]
- Donelli, G.; Vuotto, C.; Mastromarino, P. Phenotyping and genotyping are both essential to identify and classify a probiotic microorganism. Microb. Ecol. Health Dis. 2013, 24, 1–8. [Google Scholar] [CrossRef]
- Yeung, P.S.M.; Sanders, M.E.; Kitts, C.L.; Cano, R.; Tong, P.S. Species-Specific Identification of Commercial Probiotic Strains. J. Dairy Sci. 2002, 85, 1039–1051. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Hugon, P.; Khelaifia, S.; Fournier, P.-E.; La Scola, B.; Raoult, D. The Rebirth of Culture in Microbiology through the Example of Culturomics to Study Human Gut Microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Skerman, V. A Guide to the Identification of the Genera of Bacteria, with Methods and Digests of Generic Characteristics, 2nd ed.; Williams & Wilkins Co.: Philadelphia, PA, USA, 1959. [Google Scholar]
- Steel, K.J. Microbial Identification. J. Gen. Microbiol. 1965, 40, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. (Eds.) Brock Biology of Microorganisms, 13th ed.; Pearson: London, UK, 2012; p. 1032. [Google Scholar]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Klenk, H.-P. Culturomics as Tool in Research and Service in Culture Collections. Available online: https://www.eccosite.org/wp-content/uploads/2015/07/HP-Klenk_ECCO-XXXIV.pdf (accessed on 12 February 2019).
- Colwell, R.R. Polyphasic taxonomy of the genus vibrio: Numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J. Bacteriol. 1970, 104, 410–433. [Google Scholar] [PubMed]
- Yang, F.; Chen, L.; Liu, Y.; Li, J.; Wang, L.; Chen, J. Identification of microorganisms producing lactic acid during solid-state fermentation of Maotai flavour liquor. J. Inst. Brew. 2019, 125, 171–177. [Google Scholar] [CrossRef]
- Dige, I.; Nilsson, H.; Kilian, M.; Nyvad, B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur. J. Oral Sci. 2007, 115, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, B.; Banada, P.P.; Hirleman, E.D.; Bhunia, A.K.; Robinson, J.P.; Rajwa, B. Feature extraction from light-scatter patterns of Listeria colonies for identification and classification. J. Biomed. Opt. 2006, 11, 034006. [Google Scholar] [CrossRef] [PubMed]
- Rajwa, B.; Dundar, M.M.; Akova, F.; Bettasso, A.; Patsekin, V.; Hirleman, E.D.; Bhunia, A.K.; Robinson, J.P. Discovering the unknown: Detection of emerging pathogens using a label-free light-scattering system. Cytometry. A 2010, 77, 1103–1112. [Google Scholar] [CrossRef]
- Obara, B.; Roberts, M.A.J.; Armitage, J.P.; Grau, V. Bacterial cell identification in differential interference contrast microscopy images. BMC Bioinformatics 2013, 14, 134. [Google Scholar] [CrossRef]
- Ahmed, W.M.; Bayraktar, B.; Bhunia, A.K.; Hirleman, E.D.; Robinson, J.P.; Rajwa, B. Classification of Bacterial Contamination Using Image Processing and Distributed Computing. IEEE J. Biomed. Heal. Informa. 2013, 17, 232–239. [Google Scholar] [CrossRef]
- Cardinale, M.; Kaiser, D.; Lueders, T.; Schnell, S.; Egert, M. Microbiome analysis and confocal microscopy of used kitchen sponges reveal massive colonization by Acinetobacter, Moraxella and Chryseobacterium species. Sci. Rep. 2017, 7, 5791. [Google Scholar] [CrossRef]
- Schuler, B.; Meyer, G.; Peña, D.; Mullins, O.C.; Gross, L. Unraveling the Molecular Structures of Asphaltenes by Atomic Force Microscopy. J. Am. Chem. Soc. 2015, 137, 9870–9876. [Google Scholar] [CrossRef]
- Beier, B.D.; Quivey, R.G.; Berger, A.J. Raman microspectroscopy for species identification and mapping within bacterial biofilms. AMB Express 2012, 2, 35. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Sabnis, R.W. Handbook of Fluorescent Dyes and Probes; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; p. 464. [Google Scholar]
- Nam, H.-M.; Srinivasan, V.; Gillespie, B.E.; Murinda, S.E.; Oliver, S.P. Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int. J. Food Microbiol. 2005, 102, 161–171. [Google Scholar] [CrossRef]
- Noble, R.; Fuhrman, J. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 1998, 14, 113–118. [Google Scholar] [CrossRef]
- Pierini, F.; Zembrzycki, K.; Nakielski, P.; Pawłowska, S.; Kowalewski, T.A. Atomic force microscopy combined with optical tweezers (AFM/OT). Meas. Sci. Technol. 2016, 27, 025904. [Google Scholar] [CrossRef]
- Xie, C.; Mace, J.; Dinno, M.A.; Li, Y.Q.; Tang, W.; Newton, R.J.; Gemperline, P.J. Identification of Single Bacterial Cells in Aqueous Solution Using Confocal Laser Tweezers Raman Spectroscopy. Anal. Chem. 2005, 77, 4390–4397. [Google Scholar] [CrossRef]
- Props, R.; Monsieurs, P.; Mysara, M.; Clement, L.; Boon, N. Measuring the biodiversity of microbial communities by flow cytometry. Methods Ecol. Evolut. 2016, 7, 1376–1385. [Google Scholar] [CrossRef]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of Microorganisms by Modern Analytical Techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef]
- Juste, A.; Thomma, B.; Lievens, B. Recent advances in molecular techniques to study microbial communities in food-associated matrices and processes. Food Microbiol. 2008, 25, 745–761. [Google Scholar] [CrossRef]
- O’Hara, C. Manual and Automated Instrumentation for Identification of Enterobacteriaceae and Other Aerobic Gram-Negative Bacilli. Clin. Microbiol. Rev. 2005, 18, 147–162. [Google Scholar] [CrossRef]
- Engvall, E. Quantitative enzyme immunoassay (ELISA) in microbiology. Med. Biol. 1977, 55, 193–200. [Google Scholar]
- Sutton, S. How do you decide which microbial identification system is best? Pharm. Microbiol. Forum News. 2007, 13, 4–13. [Google Scholar]
- Smith, P.B.; Tomfohrde, K.M.; Rhoden, D.L.; Balows, A. API System: A Multitube Micromethod for Identification of Enterobacteriaceae. Appl. Microbiol. 1972, 24, 449–452. [Google Scholar]
- Washington, J.A.; Yu, P.K.W.; Martin, W.J. Evaluation of Accuracy of Multitest Micromethod System for Identification of Enterobacteriaceae. Appl. Microbiol. 1971, 22, 267–269. [Google Scholar]
- Sandle, T. Biochemical and Modern Identification Techniques: Enterobacteriaceae, Coliforms, and Escherichia coli. In Encyclopedia of Food Microbiology; Batt, C., Tortorello, M., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 232–237. [Google Scholar]
- Varettas, K.; Mukerjee, C.; Schmidt, M. A comparative study of the BBL crystal enteric/nonfermenter identification system and the biomerieux API20E and API20NE identification systems after overnight incubation. Pathology 1995, 27, 358–361. [Google Scholar] [CrossRef]
- Funke, G.; Funke-Kissling, P. Evaluation of the New VITEK 2 Card for Identification of Clinically Relevant Gram-Negative Rods. J. Clin. Microbiol. 2004, 42, 4067–4071. [Google Scholar] [CrossRef]
- Puttaswamy, S.; Gupta, S.K.; Regunath, H.; Smith, L.P.; Sengupta, S. A Comprehensive Review of the Present and Future Antibiotic Susceptibility Testing (AST) Systems. Arch. Clin. Microbiol. 2018, 9, 83. [Google Scholar] [CrossRef]
- Ligozzi, M.; Bernini, C.; Bonora, M.G.; De Fatima, M.; Zuliani, J.; Fontana, R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Ferraro, M.J.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Klingler, J.M.; Stowe, R.P.; Obenhuber, D.C.; Groves, T.O.; Mishra, S.K.; Pierson, D.L. Evaluation of the Biolog automated microbial identification system. Appl. Environ. Microbiol. 1992, 58, 2089. [Google Scholar]
- Sandle, T.; Skinner, K.; Sandle, J.; Gebala, B.; Kothandaram, P. Evaluation of the GEN III OmniLog® ID System microbial identification system for the profiling of cleanroom bacteria. Eur. J. Parenter. Pharm. Sci. 2013, 18, 44–50. [Google Scholar]
- Fox, A. Mass Spectrometry: Identification and Biodetection, Lessons Learned and Future Developments. In Identification of Microorganisms by Mass Spectrometry; Pezzati, L., Targowski, P., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 23–37. [Google Scholar]
- Sauer, S.; Kliem, M. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 2010, 8, 74–82. [Google Scholar] [CrossRef]
- Matsuo, T. Introduction to Modern Biological Mass Spectrometry. J. Mass Spectrom. 2000, 130, 114–130. [Google Scholar] [CrossRef]
- Petrotchenko, E.V.; Borchers, C.H. Modern Mass Spectrometry-Based Structural Proteomics. In Proteomics in Biomedicine and Pharmacology; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 95, pp. 193–213. [Google Scholar]
- Sandrin, T.R.; Demirev, P.A. Characterization of microbial mixtures by mass spectrometry. Mass Spectrom. Rev. 2018, 37, 321–349. [Google Scholar] [CrossRef]
- Claydon, M.A.; Davey, S.N.; Edwards-Jones, V.; Gordon, D.B. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 1996, 14, 1584–1586. [Google Scholar] [CrossRef]
- Senes, C.; Saldan, N.; Costa, W.; Svidzinski, T.; Oliveira, C. Identification of Fusarium oxysporum Fungus in Wheat Based on Chemical Markers and Qualitative GC-MS Test. J. Braz. Chem. Soc. 2018, 1, 167–178. [Google Scholar] [CrossRef]
- Fernando, W.G.D.; Ramarathnam, R.; Krishnamoorthy, A.S.; Savchuk, S.C. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 2005, 37, 955–964. [Google Scholar] [CrossRef]
- Schauer, N.; Steinhauser, D.; Strelkov, S.; Schomburg, D.; Allison, G.; Moritz, T.; Lundgren, K.; Roessner-Tunali, U.; Forbes, M.G.; Willmitzer, L.; et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005, 579, 1332–1337. [Google Scholar] [CrossRef]
- Fox, A. Mass spectrometry for species or strain identification after culture or without culture: Past, present, and future. J. Clin. Microbiol. 2006, 44, 2677–2680. [Google Scholar] [CrossRef]
- Jang, K.S.; Kim, Y.H. Rapid and robust MALDI-TOF MS techniques for microbial identification: a brief overview of their diverse applications. J. Microbiol. 2018, 56, 209–216. [Google Scholar] [CrossRef]
- Haiko, J.; Savolainen, L.E.; Hilla, R.; Pätäri-sampo, A. Identification of urinary tract pathogens after 3-hours urine culture by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 129, 81–84. [Google Scholar] [CrossRef]
- Rahi, P.; Prakash, O.; Shouche, Y.S. Matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry (MALDI-TOF MS) based microbial identifications: Challenges and scopes for microbial ecologists. Front. Microbiol. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Sandle, T. Microbiological Identification Strategy for Pharmaceutical Microbiology. Available online: http://www.ivtnetwork.com/article/microbial-identification-strategy-pharmaceutical-microbiology (accessed on 10 May 2019).
- Dierig, A.; Frei, R.; Egli, A. The fast route to microbe identification: Matrix assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). Pediatr. Infect. Dis. J. 2015, 34, 97–99. [Google Scholar] [CrossRef]
- Dingle, T.C.; Butler-Wu, S.M. MALDI-TOF Mass Spectrometry for Microorganism Identification. Clin. Lab. Med. 2013, 33, 589–609. [Google Scholar] [CrossRef]
- Biswas, S.; Rolain, J.M. Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. J. Microbiol. Methods 2013, 92, 14–24. [Google Scholar] [CrossRef]
- Kłodzińska, E.; Buszewski, B. Electrokinetic detection and characterization of intact microorganisms. Anal. Chem. 2009, 81, 8–15. [Google Scholar] [CrossRef]
- Zhang, J.I.; Talaty, N.; Costa, A.B.; Xia, Y.; Tao, W.A.; Bell, R.; Callahan, J.H.; Cooks, R.G. Rapid direct lipid profiling of bacteria using desorption electrospray ionization mass spectrometry. Int. J. Mass Spectrom. 2011, 301, 37–44. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Goodacre, R. High-Throughput Microbial Characterizations Using Electrospray Ionization Mass Spectrometry and Its Role in Functional Genomics. In Identification of Microorganisms by Mass Spectrometry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; pp. 229–256. [Google Scholar]
- Lattanzio, V.; Solfrizzo, M.; Powers, S.; Visconti, A. Simultaneous determination of aflatoxins, ochratoxin A and Fusarium toxins in maize by liquid chromatography/ tandem mass spectrometry after multitoxin immunoaffinity cleanup. RAPID Commun. MASS Spectrom. 2007, 21, 3253–3261. [Google Scholar] [CrossRef]
- Meredith, S.A.; Smith, P.J.; Norman, J.; Wiesner, L. An LC–MS/MS method for the determination of ofloxacin in 20μL human plasma. J. Pharm. Biomed. Anal. 2012, 58, 177–181. [Google Scholar] [CrossRef]
- Warren, C.R. A liquid chromatography–mass spectrometry method for analysis of intact fatty-acid-based lipids extracted from soil. Eur. J. Soil Sci. 2018, 69, 791–803. [Google Scholar] [CrossRef]
- Van der Werf, M.J.; Overkamp, K.M.; Muilwijk, B.; Coulier, L.; Hankemeier, T. Microbial metabolomics: Toward a platform with full metabolome coverage. Anal. Biochem. 2007, 370, 17–25. [Google Scholar] [CrossRef]
- Bakhtiar, R.; Ramos, L.; Tse, F.L.S. HIGH-THROUGHPUT MASS SPECTROMETRIC ANALYSIS OF XENOBIOTICS IN BIOLOGICAL FLUIDS. J. Liq. Chromatogr. Relat. Technol. 2002, 25, 37–41. [Google Scholar] [CrossRef]
- Verhoeckx, K.C.M.; Bijlsma, S.; Jespersen, S.; Ramaker, R.; Verheij, E.R.; Witkamp, R.F.; van der Greef, J.; Rodenburg, R.J.T. Characterization of anti-inflammatory compounds using transcriptomics, proteomics, and metabolomics in combination with multivariate data analysis. Int. Immunopharmacol. 2004, 4, 1499–1514. [Google Scholar] [CrossRef]
- Smilde, A.K.; van der Werf, M.J.; Bijlsma, S.; van der Werff-van der Vat, B.J.C.; Jellema, R.H. Fusion of mass spectrometry-based metabolomics data. Anal. Chem. 2005, 77, 6729–6736. [Google Scholar] [CrossRef]
- Bajad, S.U.; Lu, W.; Kimball, E.H.; Yuan, J.; Peterson, C.; Rabinowitz, J.D. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 2006, 1125, 76–88. [Google Scholar] [CrossRef]
- Edwards, J.; Edwards, R.; Reid, K.; Kennedy, R. Effect of decreasing column inner diameter and use of off-line two-dimensional chromatography on metabolite detection in complex mixtures. J. Chromatogr. A 2007, 1172, 127–134. [Google Scholar] [CrossRef]
- Bruce, S.J.; Jonsson, P.; Antti, H.; Cloarec, O.; Trygg, J.; Marklund, S.L.; Moritz, T. Evaluation of a protocol for metabolic profiling studies on human blood plasma by combined ultra-performance liquid chromatography/mass spectrometry: From extraction to data analysis. Anal. Biochem. 2008, 372, 237–249. [Google Scholar] [CrossRef]
- Holčapek, M.; Jirásko, R.; Lísa, M. Recent developments in liquid chromatography-mass spectrometry and related techniques. J. Chromatogr. A 2012, 1259, 3–15. [Google Scholar] [CrossRef]
- Núñez, O.; Nakanishi, K.; Tanaka, N. Preparation of monolithic silica columns for high-performance liquid chromatography. J. Chromatogr. A 2008, 1191, 231–252. [Google Scholar] [CrossRef]
- Heideloff, C.; Bunch, D.; Wang, S. A novel HPLC method for quantification of 10 antiepileptic drugs or metabolites in serum/plasma using a monolithic column. Ther. Drug Monit. 2010, 32, 102–106. [Google Scholar] [CrossRef]
- Du, H.; Ren, J.; Wang, S. Rapid determination of three alkaloids from Lotus Plumule in human serum using an HPLC-DAD method with a short monolithic column. Food Chem. 2011, 129, 1320–1324. [Google Scholar] [CrossRef]
- Kadi, A.; Hefnawy, M.; Al-Majed, A.; Alonezi, S.; Asiri, Y.; Attia, S.; Abourashed, E.; El-Subbagh, H. Liquid chromatographic high-throughput analysis of the new ultra-short acting hypnotic “HIE-124” and its metabolite in mice serum using a monolithic silica column. Analyst 2011, 136, 591–597. [Google Scholar] [CrossRef]
- Heinisch, S.; Rocca, J.-L. Sense and nonsense of high-temperature liquid chromatography. J. Chromatogr. A 2009, 1216, 642–658. [Google Scholar] [CrossRef]
- Teutenberg, T. Potential of high temperature liquid chromatography for the improvement of separation efficiency—A review. Anal. Chim. Acta 2009, 643, 1–12. [Google Scholar] [CrossRef]
- Cunliffe, J.; Shen, J.; Wei, X.; Dreyer, D.; Hayes, R.; Clement, R. Implementation of high-temperature superficially porous technologies for rapid LC-MS/MS diastereomer bioanalysis. Bioanalysis 2011, 3, 735–743. [Google Scholar] [CrossRef]
- Plotas, P.; Anastasopoulos, C.; Makril, O.; Leotsinidis, M.; Georgakopoulos, C. A UPLC–MS Method for the Determination of Ofloxacin Concentrations in Aqueous Humor. Anal. Chem. Insights 2014, 9, 27–32. [Google Scholar]
- Mounier, J.; Le Blay, G.; Vasseur, V.; Le Floch, G.; Jany, J.L.; Barbier, G. Application of denaturing high-performance liquid chromatography (DHPLC) for yeasts identification in red smear cheese surfaces. Lett. Appl. Microbiol. 2010, 51, 18–23. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 2007, 8, 105. [Google Scholar] [CrossRef]
- Lommen, A.; van der Weg, G.; van Engelen, M.C.C.; Bor, G.; Hoogenboom, L.A.; Nielen, M.W. An untargeted metabolomics approach to contaminant analysis: Pinpointing potential unknown compounds. Anal. Chim. Acta 2007, 584, 43–49. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Umek, L.; Mendes, I.; Castro, C.C.; Fonseca, N.; Martins, R.; Silva-Ferreira, A.C.; Sampaio, P.; Pais, C.; Schuller, D. New integrative computational approaches unveil the Saccharomyces cerevisiae pheno-metabolomic fermentative profile and allow strain selection for winemaking. Food Chem. 2016, 211, 509–520. [Google Scholar] [CrossRef]
- Villas-bo, S.G.; Smedsgaard, J.; Nielsen, J.; Villas-Bôas, S.G.; Mas, S.; Akesson, M.; Villas-Boas, S.G. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Garcia, D.E.; Baidoo, E.E.; Benke, P.I.; Pingitore, F.; Tang, Y.J.; Villa, S.; Keasling, J.D. Separation and mass spectrometry in microbial metabolomics. Curr. Opin. Microbiol. 2008, 11, 233–239. [Google Scholar] [CrossRef]
- Christie, W.W. Gas chromatography-mass spectrometry methods for structural analysis of fatty acids. Lipids 1998, 33, 343–353. [Google Scholar] [CrossRef]
- Basile, F.; Voorhees, K.J.; Hadfield, T.L. Microorganism Gram-Type Differentiation Based on Pyrolysis-Mass Spectrometry of Bacterial Fatty Acid Methyl Ester Extracts. Appl. Environ. Microbiol. 1995, 61, 1534. [Google Scholar]
- Ishida, Y.; Kitagawa, K.; Nakayama, A.; Ohtani, H. Complementary analysis of lipids in whole bacteria cells by thermally assisted hydrolysis and methylation-GC and MALDI-MS combined with on-probe sample pretreatment. J. Anal. Appl. Pyrolysis 2006, 77, 116–120. [Google Scholar] [CrossRef]
- Connerth, M.; Grillitsch, K.; Köfeler, H.; Daum, G. Analysis of Lipid Particles from Yeast. Methods Mol. Biol. 2009, 579, 359–374. [Google Scholar]
- Carneiro, S.; Pereira, R.; Rocha, I. Yeast metabolomics: Sample preparation for a GC/MS-based analysis. Methods Mol. Biol. 2014, 1152, 197–207. [Google Scholar]
- Tambellini, N.P.; Zaremberg, V.; Turner, R.J.; Weljie, A.M. Evaluation of extraction protocols for simultaneous polar and non-polar yeast metabolite analysis using multivariate projection methods. Metabolites 2013, 3, 592–605. [Google Scholar] [CrossRef]
- Vasconcelos, B.; Teixeira, J.C.; Dragone, G.; Teixeira, J.A. Optimization of lipid extraction from the oleaginous yeasts Rhodotorula glutinis and Lipomyces kononenkoae. AMB Express 2018, 8, 126. [Google Scholar] [CrossRef]
- Halket, J.M.; Waterman, D.; Przyborowska, A.M.; Patel, R.K.P.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005, 56, 219–243. [Google Scholar] [CrossRef]
- Wittmann, C. Fluxome analysis using GC-MS. Microb. Cell Fact. 2007, 17, 1–17. [Google Scholar]
- Lu, H.; Liang, Y.; Dunn, W.B.; Shen, H.; Kell, D.B. Comparative evaluation of software for deconvolution of metabolomics data based on GC-TOF-MS. Trends Anal. Chem. 2008, 27, 215–227. [Google Scholar] [CrossRef]
- Koek, M.M.; Muilwijk, B.; van Stee, L.L.P.; Hankemeier, T. Higher mass loadability in comprehensive two-dimensional gas chromatography-mass spectrometry for improved analytical performance in metabolomics analysis. J. Chromatogr. A 2008, 1186, 420–429. [Google Scholar] [CrossRef]
- Mondello, L.; Tranchida, P.Q.; Dugo, P.; Dugo, G.; Farmaco-chimico, D. Comprehensive two-dimensional gas chromatography-mass spectrometry: A review. Mass Spectrom. Rev. 2008, 27, 101–124. [Google Scholar] [CrossRef]
- Jumtee, K.; Bamba, T.; Fukusaki, E. Fast GC-FID based metabolic fingerprinting of Japanese green tea leaf for its quality ranking prediction. J. Sep. Sci. 2009, 32, 2296–2304. [Google Scholar] [CrossRef]
- Sampath, R.; Hall, T.; Massire, C.; Li, F.; Blyn, L.; Eshoo, M.; Hofstadler, S.; Ecker, D. Rapid Identification of Emerging Infectious Agents Using PCR and Electrospray Ionization Mass Spectrometry. Ann. N. Y. Acad. Sci. 2007, 1102, 109–120. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Kell, D.B.; Goodacre, R. Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification. J. Am. Soc. Mass Spectrom. 2002, 13, 118–128. [Google Scholar]
- Smith, P.B.W.; Snyder, A.P.; Harden, C.S. Characterization of Bacterial Phospholipids by Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem. 1995, 67, 1824–1830. [Google Scholar] [CrossRef]
- Mariey, L.; Signolle, J.P.; Amiel, C.; Travert, J. Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Schmitt, M.; Popp, J. Raman spectroscopic identification of single yeast cells. J. Raman Spectrosc. 2005, 36, 377–379. [Google Scholar] [CrossRef]
- Benito, M.T.J.; Ojeda, C.; Rojas, F.; Jimaré Benito, M.T.; Bosch Ojeda, C.; Sanchez Rojas, F. Process Analytical Chemistry: Applications of Near Infrared Spectrometry in Environmental and Food Analysis: An Overview. Appl. Spectrosc. Rev. 2008, 43, 452–484. [Google Scholar] [CrossRef]
- Pavia, D.; Lampman, G.; Kriz, G. Introduction to Spectroscopy, 3rd ed.; Learning, T., Ed.; Thomson Learning: Stamford, CT, USA, 2001; p. 752. [Google Scholar]
- Amir, R.M.; Anjum, F.M.; Khan, M.I.; Khan, M.R.; Pasha, I.; Nadeem, M. Application of Fourier transform infrared (FTIR) spectroscopy for the identification of wheat varieties. J. Food Sci. Technol. 2013, 50, 1018–1023. [Google Scholar] [CrossRef]
- Wenning, M.; Scherer, S. Identification of microorganisms by FTIR spectroscopy: Perspectives and limitations of the method. Appl. Microbiol. Biotechnol. 2013, 97, 7111–7120. [Google Scholar] [CrossRef]
- Wenning, M.; Seiler, H.; Scherer, S. Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 2002, 68, 4717–4721. [Google Scholar] [CrossRef]
- Kosa, G.; Shapaval, V.; Kohler, A.; Zimmermann, B. FTIR spectroscopy as a unified method for simultaneous analysis of intra-and extracellular metabolites in high-throughput screening of microbial bioprocesses Microbial Cell Factories. Microb. Cell Fact. 2017, 16, 1–11. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Doglia, S.M. Fourier Transform Infrared Microespectroscopy of Complex Biological Systems: From Intact Cells to Whole Organisms. Methods Mol. Biol. 2012, 895, 85–100. [Google Scholar]
- Santos, C.; Fraga, M.E.; Kozakiewicz, Z.; Lima, N. Fourier transform infrared as a powerful technique for the identification and characterization of filamentous fungi and yeasts. Res. Microbiol. 2010, 161, 168–175. [Google Scholar] [CrossRef]
- Stöckel, S.; Kirchhoff, J.; Neugebauer, U.; Rösch, P.; Popp, J. The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman Spectrosc. 2016, 47, 89–109. [Google Scholar] [CrossRef]
- Walsh, T.J.; Wissel, M.C.; Grantham, K.J.; Petraitiene, R.; Petraitis, V.; Kasai, M.; Francesconi, A.; Cotton, M.P.; Hughes, J.E.; Greene, L.; et al. Molecular Detection and Species-Specific Identification of Medically Important Aspergillus Species by Real-Time PCR in Experimental Invasive Pulmonary Aspergillosis. J. Clin. Microbiol. 2011, 49, 4150–4157. [Google Scholar] [CrossRef]
- Lu, X.; Al-Qadiri, H.M.; Lin, M.; Rasco, B.A. Application of Mid-infrared and Raman Spectroscopy to the Study of Bacteria. Food Bioprocess Technol. 2011, 4, 919–935. [Google Scholar] [CrossRef]
- Kneipp, K.; Kneipp, H. Single Molecule Raman Scattering. Appl. Spectrosc. 2006, 60, 322A–334A. [Google Scholar] [CrossRef]
- Aston, D.E.; Lin, M.; Konkel, M.E.; Lu, X.; Rasco, B.A.; Jabal, J.M.F. Investigating Antibacterial Effects of Garlic (Allium sativum) Concentrate and Garlic-Derived Organosulfur Compounds on Campylobacter jejuni by Using Fourier Transform Infrared Spectroscopy, Raman Spectroscopy, and Electron Microscopy. Appl. Environ. Microbiol. 2011, 77, 5257–5269. [Google Scholar]
- Huang, W.E.; Li, M.; Jarvis, R.M.; Goodacre, R.; Banwart, S.A. Shining light on the microbial world the application of Raman microspectroscopy. Adv. Appl. Microbiol. 2010, 70, 153–186. [Google Scholar]
- Jarvis, R.M.; Goodacre, R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 2004, 76, 565–579. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- Loo, J.A.; Udseth, H.R.; Smith, R.D. Peptide and protein analysis by electrospray ionization-mass spectrometry and capillary electrophoresis-mass spectrometry. Anal. Biochem. 1989, 179, 404–412. [Google Scholar] [CrossRef]
- Soga, T.; Ueno, Y.; Naraoka, H.; Ohashi, Y.; Tomita, M.; Nishioka, T. Simultaneous Determination of Anionic Intermediates for Bacillus subtilis Metabolic Pathways by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry. Anal. Chem. 2002, 74, 2233–2239. [Google Scholar] [CrossRef]
- Terabe, S.; Markuszewski, M.; Inoue, N.; Otsuka, K.; Nishioka, T. Capillary electrophoretic techniques toward the metabolome analysis. Pure Appl. Chem. 2001, 73, 1563–1572. [Google Scholar] [CrossRef]
- Kobayashi, J.; Shirao, M.; Nakazawa, H. Simultaneous Determination of Anions and Cations in Mineral Water by Capillary Electrophoresis with a Chelating Agent. J. Liq. Chromatogr. Relat. Technol. 1998, 21, 1445–1456. [Google Scholar] [CrossRef]
- Shirao, M.; Furuta, R.; Suzuki, S.; Nakasawa, H.; Fujita, S.; Maruyama, T. Determination of organic-acids in urine by capillary zone electrophoresis. J. Chromatogr. A 1994, 680, 247–251. [Google Scholar] [CrossRef]
- Soga, T.; Heiger, D.N. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2000, 72, 1236–1241. [Google Scholar] [CrossRef]
- Cohen, A.S.; Terabe, S.; Smith, J.A.; Karger, B.L. High-performance capillary electrophoretic separation of bases, nucleosides, and oligonucleotides: retention manipulation via micellar solutions and metal additives. Anal. Chem. 1987, 59, 1021–1027. [Google Scholar] [CrossRef]
- Schreiner, M.; Razzazi, E.; Luf, W. Determination of watersoluble vitamins in soft drinks and vitamin supplements using capillary electrophoresis. Food/Nahrung 2003, 47, 243–247. [Google Scholar] [CrossRef]
- Carru, C.; Zinellu, A.; Galistu, F.; Sotgia, S.; Usai, M.; Pes, G.; Deiana, L. Ultra rapid capillary electrophoresis method for total plasma thiols measurement. Clin. Chem. 2003, 49, 36. [Google Scholar]
- Soga, T.; Heiger, D.N. Simultaneous determination of monosaccharides in glycoproteins by capillary electrophoresis. Anal. Biochem. 1998, 261, 73–78. [Google Scholar] [CrossRef]
- Perret, D.; Birch, A.; Ross, G. Capillary electrophoresis for peptides, including neuropeptides. Biochem. Soc. Trans. 1994, 22, 127–131. [Google Scholar] [CrossRef]
- Armstrong, D.W.; Schulte, G.; Schneiderheinze, J.M.; Westenberg, D.J. Separating microbes in the manner of molecules. 1. Capillary electrokinetic approaches. Anal. Chem. 1999, 71, 5465–5469. [Google Scholar] [CrossRef]
- Girod, M.; Armstrong, D.W. Monitoring the migration behavior of living microorganisms in capillary electrophoresis using laser-induced fluorescence detection with a charge-coupled device imaging system. Electrophoresis 2002, 23, 2048–2056. [Google Scholar] [CrossRef]
- Armstrong, D.W.; Girod, M.; He, L.; Rodriguez, M.A.; Wei, W.; Zheng, J.; Yeung, E.S. Mechanistic aspects in the generation of apparent ultrahigh efficiencies for colloidal (microbial) electrokinetic separations. Anal. Chem. 2002, 74, 5523–5530. [Google Scholar] [CrossRef]
- Lantz, A.W.; Bao, Y.; Armstrong, D.W. Single-cell detection: Test of microbial contamination using capillary electrophoresis. Anal. Chem. 2007, 79, 1720–1724. [Google Scholar] [CrossRef]
- Saenton, S.; Lee, H.; Gao, Y.S.; Ranville, J.F.; Williams, S.K.R. Evaluation of different field-flow fractionation techniques for separating bacteria. Sep. Sci. Technol. 2000, 35, 1761–1775. [Google Scholar] [CrossRef]
- Desai, M.J.; Armstrong, D.W. Separation, identification, and characterization of microorganisms by capillary electrophoresis. Microbiol. Mol. Biol. Rev. 2003, 67, 38–51. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Tehranirokh, M.; Kouzani, A.Z.; Francis, P.S.; Kanwar, J.R. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 2013, 7, 051502. [Google Scholar] [CrossRef]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of Pathogenic Microorganisms by Microfluidics Based Analytical Methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, D.; Cai, G.; Xiong, Y.; Li, Y.; Wang, M.; Huo, H.; Lin, J. Fast and sensitive detection of foodborne pathogen using electrochemical impedance analysis, urease catalysis and microfluidics. Biosens. Bioelectron. 2016, 86, 770–776. [Google Scholar] [CrossRef]
- Ölcer, Z.; Esen, E.; Ersoy, A.; Budak, S.; Sever Kaya, D.; Yağmur Gök, M.; Barut, S.; Üstek, D.; Uludag, Y. Microfluidics and nanoparticles based amperometric biosensor for the detection of cyanobacteria (Planktothrix agardhii NIVA-CYA 116) DNA. Biosens. Bioelectron. 2015, 70, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Buchsbaum, S.F.; Wu, T.-T.; Hsieh, K.; Xiao, Y.; Sun, R.; Soh, H.T. Genetic Analysis of H1N1 Influenza Virus from Throat Swab Samples in a Microfluidic System for Point-of-Care Diagnostics. J. Am. Chem. Soc. 2011, 133, 9129–9135. [Google Scholar] [CrossRef]
- Hsieh, K.; Ferguson, B.S.; Eisenstein, M.; Plaxco, K.W.; Soh, H.T. Integrated Electrochemical Microsystems for Genetic Detection of Pathogens at the Point of Care. Acc. Chem. Res. 2015, 48, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Oblath, E.A.; Henley, W.H.; Alarie, J.P.; Ramsey, J.M. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva. Lab Chip 2013, 13, 1325. [Google Scholar] [CrossRef] [PubMed]
- Trindade, M.A.G.; Martins, C.A.; Angnes, L.; Herl, T.; Raith, T.; Matysik, F.-M. New Electrochemical Flow-Cell Configuration Integrated into a Three-Dimensional Microfluidic Platform: Improving Analytical Application in the Presence of Air Bubbles. Anal. Chem. 2018, 90, 10917–10926. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, C.; Mauk, M.G.; Yin, K.; Kadimisetty, K. Interfacing Pathogen Detection with Smartphones for Point-of-Care Applications. Anal. Chem. 2018, 91, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Walper, S.A.; Lasarte Aragonés, G.; Sapsford, K.E.; Brown, C.W.; Rowland, C.E.; Breger, J.C.; Medintz, I.L. Detecting Biothreat Agents: From Current Diagnostics to Developing Sensor Technologies. ACS Sensors 2018, 3, 1894–2024. [Google Scholar] [CrossRef]
- Cao, Q.; Mahalanabis, M.; Chang, J.; Carey, B.; Hsieh, C.; Stanley, A.; Odell, C.A.; Mitchell, P.; Feldman, J.; Pollock, N.R.; et al. Microfluidic chip for molecular amplification of influenza a RNA in human respiratory specimens. PLoS ONE 2012, 7, 1–11. [Google Scholar] [CrossRef]
- Sun, Y.; Dhumpa, R.; Bang, D.D.; Høgberg, J.; Handberg, K.; Wolff, A. A lab-on-a-chip device for rapid identification of avian influenza viral RNA by solid-phase PCR. Lab Chip 2011, 11, 1457–1463. [Google Scholar] [CrossRef]
- Feng, X.; Liu, B.-F.; Li, J.; Liu, X. Advances in coupling microfluidic chips to mass spectrometry. Mass Spectrom. Rev. 2015, 34, 535–557. [Google Scholar] [CrossRef]
- Lee, J.; Musyimi, H.K.; Soper, S.A.; Murray, K.K. Development of an Automated Digestion and Droplet Deposition Microfluidic Chip for MALDI-TOF MS. J. Am. Soc. Mass Spectrom. 2008, 19, 964–972. [Google Scholar] [CrossRef]
- Yang, P.; Wang, B.; Bian, X.; Zhang, Y.S.; Lan, Y.; Qiao, L.; Liu, B.; Zhang, W. Microfluidic Air Sampler for Highly Efficient Bacterial Aerosol Collection and Identification. Anal. Chem. 2016, 88, 11504–11512. [Google Scholar]
- Zhang, R.Q.; Liu, S.L.; Zhao, W.; Zhang, W.P.; Yu, X.; Li, Y.; Li, A.J.; Pang, D.W.; Zhang, Z.L. A simple point-of-care microfluidic immunomagnetic fluorescence assay for pathogens. Anal. Chem. 2013, 85, 2645–2651. [Google Scholar] [CrossRef]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef]
- Sabat, A.J.; van Zanten, E.; Akkerboom, V.; Wisselink, G.; van Slochteren, K.; de Boer, R.F.; Hendrix, R.; Friedrich, A.W.; Rossen, J.W.A.; Kooistra-Smid, A.M.D. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification—Increased discrimination of closely related species. Sci. Rep. 2017, 7, 3434. [Google Scholar] [CrossRef]
- Daims, H.; Wagner, M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 2007, 75, 237–248. [Google Scholar] [CrossRef]
- Cerqueira, L.; Azevedo, N.; Almeida, C.; Jardim, T.; Keevil, C.; Vieira, M. DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence in situ Hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944–1960. [Google Scholar] [CrossRef]
- Spratt, D.A. Significance of bacterial identification by molecular biology methods. Endod. Top. 2004, 9, 5–14. [Google Scholar] [CrossRef]
- Höfling, J.F.; Rosa, E.A.; Baptista, M.J.; Spolidório, D.M. New strategies on molecular biology applied to microbial systematics. Rev. Inst. Med. Trop. Sao Paulo 1997, 39, 345–352. [Google Scholar] [CrossRef]
- Galluzzi, L.; Magnani, M.; Saunders, N.; Harms, C.; Bruce, I.J. Current molecular techniques for the detection of microbial pathogens. Sci. Prog. 2007, 90, 29–50. [Google Scholar] [CrossRef]
- Emerson, D.; Agulto, L.; Liu, H.; Liu, L. Identifying and Characterizing Bacteria in an Era of Genomics and Proteomics. Bioscience 2008, 58, 925–936. [Google Scholar] [CrossRef]
- Fouad, A.F.; Barry, J.; Caimano, M.; Clawson, M.; Zhu, Q.; Carver, R.; Hazlett, K.; Radolf, J.D. PCR-Based Identification of Bacteria Associated with Endodontic Infections. J. Clin. Microbiol. 2002, 40, 3223–3231. [Google Scholar] [CrossRef]
- La Scola, B.; Gundi, V.A.K.B.; Khamis, A.; Raoult, D. Sequencing of the rpoB Gene and Flanking Spacers for Molecular Identification of Acinetobacter Species. J. Clin. Microbiol. 2006, 44, 827–832. [Google Scholar] [CrossRef]
- Adekambi, T.; Drancourt, M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 2004, 54, 2095–2105. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.P.; Petti, C.A. Detection and Identification of Microorganisms by Gene Amplification and Sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Rådström, P.; Knutsson, R.; Wolffs, P.; Lövenklev, M.; Löfström, C. Pre-PCR Processing: Strategies to Generate PCR-Compatible Samples. Mol. Biotechnol. 2004, 26, 133–146. [Google Scholar] [CrossRef]
- Yamamoto, Y. MINIREVIEWS PCR in Diagnosis of Infection: Detection of Bacteria in Cerebrospinal Fluids. Clin. Diagn. Lab. Immunol. 2002, 9, 508–514. [Google Scholar]
- Kai, S.; Matsuo, Y.; Nakagawa, S.; Kryukov, K.; Matsukawa, S.; Tanaka, H.; Iwai, T.; Imanishi, T.; Hirota, K.; Matsuo, C.Y.; et al. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION TM nanopore sequencer. FEBS Open. 2019, 9, 548–557. [Google Scholar] [CrossRef]
- Fredricks, D.N.; Fiedler, T.L.; Marrazzo, J.M. Molecular Identification of Bacteria Associated with Bacterial Vaginosis. N. Engl. J. Med. 2005, 353, 1899–1911. [Google Scholar] [CrossRef]
- Järvinen, A.-K.; Laakso, S.; Piiparinen, P.; Aittakorpi, A.; Lindfors, M.; Huopaniemi, L.; Piiparinen, H.; Mäki, M. Rapid identification of bacterial pathogens using a PCR- and microarray-based assay. BMC Microbiol. 2009, 9, 161. [Google Scholar] [CrossRef]
- Taponen, S.; Salmikivi, L.; Simojoki, H.; Koskinen, M.T.; Pyörälä, S. Real-time polymerase chain reaction-based identification of bacteria in milk samples from bovine clinical mastitis with no growth in conventional culturing. J. Dairy Sci. 2009, 92, 2610–2617. [Google Scholar] [CrossRef]
- Höppener-Ogawa, S.; Leveau, J.H.J.; Smant, W.; van Veen, J.A.; de Boer, W. Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems. Appl. Environ. Microbiol. 2007, 73, 4191–4197. [Google Scholar] [CrossRef]
- Clifford, R.J.; Milillo, M.; Prestwood, J.; Quintero, R.; Zurawski, D.V.; Kwak, Y.I.; Waterman, P.E.; Lesho, E.P.; Mc Gann, P. Detection of Bacterial 16S rRNA and Identification of Four Clinically Important Bacteria by Real-Time PCR. PLoS ONE 2012, 7, e48558. [Google Scholar] [CrossRef]
- Melendez, J.H.; Frankel, Y.M.; An, A.T.; Williams, L.; Price, L.B.; Wang, N.-Y.; Lazarus, G.S.; Zenilman, J.M. Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin. Microbiol. Infect. 2010, 16, 1762–1769. [Google Scholar] [CrossRef]
- Huijsdens, X.W.; Linskens, R.K.; Mak, M.; Meuwissen, S.G.M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 2002, 40, 4423–4427. [Google Scholar] [CrossRef]
- Jung, J.Y.; Yoon, H.K.; An, S.; Lee, J.W.; Ahn, E.-R.; Kim, Y.-J.; Park, H.-C.; Lee, K.; Hwang, J.H.; Lim, S.-K. Rapid oral bacteria detection based on real-time PCR for the forensic identification of saliva. Sci. Rep. 2018, 8, 10852. [Google Scholar] [CrossRef]
- Tamburro, M.; Ripabelli, G. High Resolution Melting as a rapid, reliable, accurate and cost-effective emerging tool for genotyping pathogenic bacteria and enhancing molecular epidemiological surveillance: a comprehensive review of the literature. Ann. Ig 2017, 29, 293–316. [Google Scholar] [CrossRef]
- Iacumin, L.; Ginaldi, F.; Manzano, M.; Anastasi, V.; Reale, A.; Zotta, T.; Rossi, F.; Coppola, R.; Comi, G. High resolution melting analysis (HRM) as a new tool for the identification of species belonging to the Lactobacillus casei group and comparison with species-specific PCRs and multiplex PCR. Food Microbiol. 2015, 46, 357–367. [Google Scholar] [CrossRef]
- Jones, C.; Kortenkamp, A. RAPD library fingerprinting of bacterial and human DNA: applications in mutation detection. Teratog. Carcinog. Mutagen. 2000, 20, 49–63. [Google Scholar] [CrossRef]
- Baker, J.C.; Crumley, R.E.; Eckdahl, T.T. Laboratory Exercises Random Amplified Polymorphic DNA PCR in the Microbiology Teaching Laboratory. Biochem. Mol. Biol. Educ. 2016, 8, 391–396. [Google Scholar]
- Saxena, S.; Verma, J.; Shikha, R.; Modi, D. RAPD-PCR and 16S rDNA phylogenetic analysis of alkaline protease producing bacteria isolated from soil of India: Identification and detection of genetic variability. J. Genet. Eng. Biotechnol. 2014, 12, 27–35. [Google Scholar] [CrossRef]
- Reale, A.; Tremonte, P.; Succi, M.; Sorrentino, E.; Coppola, R. Exploration of lactic acid bacteria ecosystem of sourdoughs from the Molise region. Ann. Microbiol. 2005, 55, 17–22. [Google Scholar]
- Abdollahniya, D.; Hosseini, S.M.; Baghbaderani, K.B.; Mordadi, A.; Reza, M. Identification of Lactobacillus Species Isolated FromTraditional Dairy Products Using RAPD-PCR. Avicenna J. Clin. Microbiol. Infect. 2018, 5, 7–13. [Google Scholar]
- Reale, A.; Di Renzo, T.; Succi, M.; Tremonte, P.; Coppola, R.; Sorrentino, E. Microbiological and Fermentative Properties of Baker’s Yeast Starter Used in Breadmaking. J. Food Sci. 2013, 78, M1224–M1231. [Google Scholar] [CrossRef]
- Mitani, N.; Koizumi, A.; Sano, R.; Masutani, T.; Murakawa, K.; Mikasa, K.; Okamoto, Y. Molecular typing of methicillin-resistant Staphylococcus aureus by PCR-RFLP and its usefulness in an epidemiological study of an outbreak. Jpn. J. Infect. Dis. 2005, 58, 250–252. [Google Scholar]
- Bukholm, G.; Tannaes, T.; Kjelsberg, A.B.B.; Smith-Erichsen, N. An outbreak of multidrug-resistant Pseudomonas aeruginosa associated with increased risk of patient death in an intensive care unit. Infect. Control Hosp. Epidemiol. 2002, 23, 441–446. [Google Scholar] [CrossRef]
- Parizad, E.G.; Parizad, E.G.; Valizadeh, A. The Application of Pulsed Field Gel Electrophoresis in Clinical Studies. J. Clin. DIAGNOSTIC Res. 2016, 10, 1–4. [Google Scholar] [CrossRef]
- Taneja, N.; Sangar, G.; Chowdhury, G.; Ramamurthy, T.; Mishra, A.; Singh, M.; Sharma, M. Molecular epidemiology of Vibrio cholerae causing outbreaks & sporadic cholera in northern India. Indian J. Med. Res. 2012, 136, 656–663. [Google Scholar]
- Bouchet, V.; Huot, H.; Goldstein, R. Molecular Genetic Basis of Ribotyping. Clin. Microbiol. Rev. 2008, 21, 262–273. [Google Scholar] [CrossRef]
- Bidet, P.; Lalande, V.; Salauze, B.; Burghoffer, B.; Avesani, V.; Delmée, M.; Rossier, A.; Barbut, F.; Petit, J.C. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 2000, 38, 2484–2487. [Google Scholar]
- Janezic, S.; Strumbelj, I.; Rupnik, M. Use of modified PCR ribotyping for direct detection of Clostridium difficile ribotypes in stool samples. J. Clin. Microbiol. 2011, 49, 3024–3025. [Google Scholar] [CrossRef]
- Quainoo, S.; Coolen, J.P.M.; van Hijum, S.A.F.T.; Huynen, M.A.; Melchers, W.J.G.; van Schaik, W.; Wertheim, H.F.L. Whole-Genome Sequencing of Bacterial Pathogens: The Future of Nosocomial Outbreak Analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef]
- McGann, P.; Bunin, J.L.; Snesrud, E.; Singh, S.; Maybank, R.; Ong, A.C.; Kwak, Y.I.; Seronello, S.; Clifford, R.J.; Hinkle, M.; et al. Real time application of whole genome sequencing for outbreak investigation—What is an achievable turnaround time? Diagn. Microbiol. Infect. Dis. 2016, 85, 277–282. [Google Scholar] [CrossRef]
- Edwards-Jones, V.; Claydon, M.A.; Walker, J.; Fox, A.J.; Evason, D.J.; Gordon, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef]
- Badiee, P.; Badali, H.; Abastabar, M.; Safa, A.; Hadipour, M.; Yazdani, H.; Heshmat, F.; Mohammadi, R. Use of restriction fragment length polymorphism to identify Candida species, related to onychomycosis. Adv. Biomed. Res. 2015, 4, 95. [Google Scholar] [CrossRef]
- Posteraro, B.; Efremov, L.; Leoncini, E.; Amore, R.; Posteraro, P.; Ricciardi, W.; Sanguinetti, M. Are the Conventional Commercial Yeast Identification Methods Still Helpful in the Era of New Clinical Microbiology Diagnostics? A Meta-Analysis of Their Accuracy. J. Clin. Microbiol. 2015, 53, 2439–2450. [Google Scholar] [CrossRef]
- Zhang, J.; Hung, G.-C.; Nagamine, K.; Li, B.; Tsai, S.; Lo, S.-C. Development of Candida-Specific Real-Time PCR Assays for the Detection and Identification of Eight Medically Important Candida Species. Microbiol. Insights 2016, 9, 21–28. [Google Scholar] [CrossRef]
- Criseo, G.; Scordino, F.; Romeo, O. Current methods for identifying clinically important cryptic Candida species. J. Microbiol. Methods 2015, 111. [Google Scholar] [CrossRef]
- Schönian, G.; Tietz, H.J.; Thanos, M.; Gräser, Y. [Application of molecular biological methods for diagnosis and epidemiology of human fungal infections]. Mycoses 1996, 39, 73–80. [Google Scholar] [CrossRef]
- Farber, J.M. An Introduction to the Hows and Whys of Molecular Typing. J. Food Prot. 1996, 59, 1091–1101. [Google Scholar] [CrossRef]
- Lackner, M.; Lass-Flörl, C. Commercial Molecular Tests for Fungal Diagnosis from a Practical Point of View. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: New York, NY, USA, 2017; Volume 1508, pp. 85–105. [Google Scholar]
- Madhavan, P.; Jamal, F.; Chong, P.P. Laboratory Isolation and Identification of Candida Species. J. Appl. Sci. 2011, 11, 2870–2877. [Google Scholar] [CrossRef]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and Nonmolecular Diagnostic Methods for Invasive Fungal Infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef]
- Ragheb, S.M.; Jimenez, L. Polymerase Chain Reaction/Rapid Methods Are Gaining a Foothold in Developing Countries. PDA J. Pharm. Sci. Technol. 2014, 68, 239–255. [Google Scholar] [CrossRef]
- Arastehfar, A.; Fang, W.; Pan, W.; Liao, W.; Yan, L.; Boekhout, T. Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect. Dis. 2018, 18, 480. [Google Scholar] [CrossRef]
- Costa, M.V.; Landgraf, T.N.; Corrêa, P.C.; Souza, I.E.L.; Fernandes, F.F.; Panunto-Castelo, A.; Costa, M.V.; Landgraf, T.N.; Corrêa, P.C.; Souza, I.E.L.; et al. Quantitation of pulmonary fungal burden in Paracoccidioides brasiliensis-infected mice by real-time PCR. Rev. Inst. Med. Trop. Sao Paulo 2018, 61, e02. [Google Scholar] [CrossRef]
- Soll, D.R. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 2000, 13, 332–370. [Google Scholar] [CrossRef]
- Murayama, S.Y.; Yamaguchi, H.; Makimura, K. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 1994, 40, 358–364. [Google Scholar]
- Chen, Y.C.; Eisner, J.D.; Kattar, M.M.; Rassoulian-Barrett, S.L.; LaFe, K.; Yarfitz, S.L.; Limaye, A.P.; Cookson, B.T. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 2000, 38, 2302–2310. [Google Scholar]
- De Barros Lopes, M.; Soden, A.; Martens, A.L.; Henschke, P.A.; Langridge, P. Differentiation and species identification of yeasts using PCR. Int. J. Syst. Bacteriol. 1998, 48, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Dalla-Costa, L.M.; Morello, L.G.; Conte, D.; Pereira, L.A.; Palmeiro, J.K.; Ambrosio, A.; Cardozo, D.; Krieger, M.A.; Raboni, S.M. Comparison of DNA extraction methods used to detect bacterial and yeast DNA from spiked whole blood by real-time PCR. J. Microbiol. Methods 2017, 140, 61–66. [Google Scholar] [CrossRef]
- Gonzlez-Mendoza, D.; Argumedo-Delira, R.; Morales-Trejo, A.; Pulido-Herrera, A.; Cervantes-Daz, L.; Grimaldo-Juarez, O.; Alarcn, A. A rapid method for isolation of total DNA from pathogenic filamentous plant fungi. Genet. Mol. Res. 2010, 9, 162–166. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS. In PCR Protocols; Elsevier, Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Carvalho, A.; Costa-De-Oliveira, S.; Martins, M.L.; Pina-Vaz, C.; Rodrigues, A.G.; Ludovico, P.; Rodrigues, F. Multiplex PCR identification of eight clinically relevant Candida species. Med. Mycol. 2007, 45, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.; Fernandez, V.; Holmström, K.O.; Claesson, B.E.B.; Enroth, H. Rapid identification of pathogenic yeast isolates by real-time PCR and two-dimensional melting-point analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Ağca, H.; Dalyan Cilo, B.; Özmerdiven, G.E.; Sağlam, S.; Ener, B. [Development of a real-time polymerase chain reaction method for the identification of Candida species]. Mikrobiyol. Bul. 2015, 49, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Alnuaimi, A.D.; Wiesenfeld, D.; O’Brien-Simpson, N.M.; Reynolds, E.C.; Peng, B.; McCullough, M.J. The development and validation of a rapid genetic method for species identification and genotyping of medically important fungal pathogens using high-resolution melting curve analysis. Mol. Oral Microbiol. 2014, 29, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Rapid and Accurate Identification of Candida albicans and Candida dubliniensis by Real-Time PCR and Melting Curve Analysis. Med. Princ. Pract. 2018, 27, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and accurate molecular identification of the emerging multidrug resistant pathogen Candida auris. J. Clin. Microbiol. 2017, 24, 24. [Google Scholar] [CrossRef]
- Leach, L.; Zhu, Y.; Chaturvedi, S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2018, 56, e01223-17. [Google Scholar] [CrossRef]
- Merz, W.G. Candida albicans strain delineation. Clin. Microbiol. Rev. 1990, 3, 321–334. [Google Scholar] [CrossRef]
- Doi, M.; Homma, M.; Chindamporn, A.; Tanaka, K. Estimation of chromosome number and size by pulsed-field gel electrophoresis (PFGE) in medically important Candida species. J. Gen. Microbiol. 1992, 138, 2243–2251. [Google Scholar] [CrossRef]
- Shokohi, T.; Hashemi Soteh, M.; Pouri, Z.S.; Hedayati, M.; Mayahi, S. Identification of Candida species using PCR-RFLP in cancer patients in Iran. Indian J. Med. Microbiol. 2010, 28, 147. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, S.; Duran, N.; Duran, G.; Eryilmaz, N.; Aslan, H.; Önlen, C.; Özer, B. Identification of medically important Candida species by polymerase chain reaction-restriction fragment length polymorphism analysis of the rDNA ITS1 and ITS2 regions. Indian J. Med. Microbiol. 2017, 35, 535. [Google Scholar] [PubMed]
- Yiş, R.; Doluca, M. Identification of Candida species by restriction enzyme analysis. Turkish J. Med. Sci. 2018, 48, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Sadrossadati, S.Z.; Ghahri, M.; Imani Fooladi, A.A.; Sayyahfar, S.; Beyraghi, S.; Baseri, Z. Phenotypic and genotypic characterization of Candida species isolated from candideamia in Iran. Curr. Med. Mycol. 2018, 4, 14–20. [Google Scholar] [CrossRef]
- Bartelli, T.F.; Ferreira, R.C.; Colombo, A.L.; Briones, M.R.S. Intraspecific comparative genomics of Candida albicans mitochondria reveals non-coding regions under neutral evolution. Infect. Genet. Evol. 2013, 14, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-S.; Meyer, S.A. Characterization of Mitochondrial DNA in Various Candida Species: Isolation, Restriction Endonuclease Analysis, Size, and Base Composition. Int. J. Syst. Bacteriol. 1991, 41, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Reiss, E.; Tanaka, K.; Bruker, G.; Chazalet, V.; Coleman, D.; Debeaupuis, J.P.; Hanazawa, R.; Latgé, J.P.; Lortholary, J.; Makimura, K.; et al. Molecular diagnosis and epidemiology of fungal infections. Med. Mycol. 1998, 36, 249–257. [Google Scholar] [PubMed]
- Bautista-Muñoz, C.; Boldo, X.M.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Identification of Candida spp. by randomly amplified polymorphic DNA analysis and differentiation between Candida albicans and Candida dubliniensis by direct PCR methods. J. Clin. Microbiol. 2003, 41, 414–420. [Google Scholar] [CrossRef]
- Bonfim-Mendonca, P.D.S.; Fiorini, A.; Shinobu-Mesquita, C.S.; Baeza, L.C.; Fernandez, M.A.; Svidzinski, T.I.E. MOLECULAR TYPING OF Candida albicans ISOLATES FROM HOSPITALIZED PATIENTS. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 385–391. [Google Scholar] [CrossRef]
- Borst, A.; Theelen, B.; Reinders, E.; Boekhout, T.; Fluit, A.C.; Savelkoul, P.H.M. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 2003, 41, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Hensgens, L.A.M.; Tavanti, A.; Mogavero, S.; Ghelardi, E.; Senesi, S. AFLP genotyping of Candida metapsilosis clinical isolates: Evidence for recombination. Fungal Genet. Biol. 2009, 46, 750–758. [Google Scholar] [CrossRef]
- Prakash, A.; Sharma, C.; Singh, A.; Kumar Singh, P.; Kumar, A.; Hagen, F.; Govender, N.P.; Colombo, A.L.; Meis, J.F.; Chowdhary, A. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin. Microbiol. Infect. 2016, 22, 277.e1–277.e9. [Google Scholar] [CrossRef]
- Vigentini, I.; Antoniani, D.; Roscini, L.; Comasio, A.; Galafassi, S.; Picozzi, C.; Corte, L.; Compagno, C.; Dal Bello, F.; Cardinali, G.; et al. Candida milleri species reveals intraspecific genetic and metabolic polymorphisms. Food Microbiol. 2014, 42, 72–81. [Google Scholar] [CrossRef] [PubMed]
- López-Ribot, J.L.; McAtee, R.K.; Kirkpatrick, W.R.; Perea, S.; Patterson, T.F. Comparison of DNA-based typing methods to assess genetic diversity and relatedness among Candida albicans clinical isolates. Rev. Iberoam. Micol. 2000, 17, 49–54. [Google Scholar] [PubMed]
- Ruiz Gaitán, A.C.; Moret, A.; López Hontangas, J.L.; Molina, J.M.; Aleixandre López, A.I.; Cabezas, A.H.; Mollar Maseres, J.; Arcas, R.C.; Gómez Ruiz, M.D.; Chiveli, M.Á.; et al. Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Rev. Iberoam. Micol. 2017, 34, 23–27. [Google Scholar] [CrossRef] [PubMed]

| Type | Basis | References |
|---|---|---|
| Indirect | - Conventional methods. Isolation and culture of microorganisms and the determination of their various phenotypic characteristics | [3] |
| Direct | - Culture-independent. May be used to identify specific microbes in a mixed population as well as identify non-culturable microbes. For example, microscopic techniques are powerful tools used in the identification of microorganisms by visualization of the characteristic structures and for organisms in the VBNC (viable but not culturable) state. | [4] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco-Duarte, R.; Černáková, L.; Kadam, S.; S. Kaushik, K.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 2019, 7, 130. https://doi.org/10.3390/microorganisms7050130
Franco-Duarte R, Černáková L, Kadam S, S. Kaushik K, Salehi B, Bevilacqua A, Corbo MR, Antolak H, Dybka-Stępień K, Leszczewicz M, et al. Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms. 2019; 7(5):130. https://doi.org/10.3390/microorganisms7050130
Chicago/Turabian StyleFranco-Duarte, Ricardo, Lucia Černáková, Snehal Kadam, Karishma S. Kaushik, Bahare Salehi, Antonio Bevilacqua, Maria Rosaria Corbo, Hubert Antolak, Katarzyna Dybka-Stępień, Martyna Leszczewicz, and et al. 2019. "Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present" Microorganisms 7, no. 5: 130. https://doi.org/10.3390/microorganisms7050130