Changes in the Ultrastructure of Staphylococcus aureus Treated with Cationic Peptides and Chlorhexidine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptides
2.2. Microorganism and Growth Conditions
2.3. Effect of (KFF)3K and R9F2 Peptides on S. aureus Growth
2.4. Processing of the Samples for TEM Studies
3. Results
3.1. Viability of S. aureus Cells Incubated with R9F2 or (KFF)3K Peptides or Chlorhexidine
3.2. Ultrastructure of S. aureus Cells Incubated in Saline (Control)
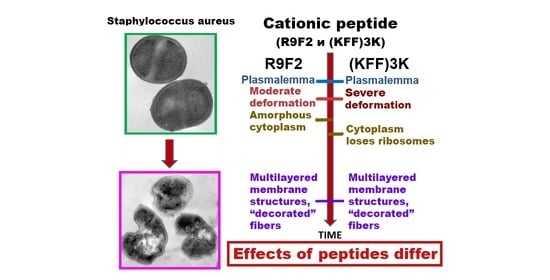
3.3. Ultrastructure of S. aureus Treated with Cationic Peptides
3.4. Ultrastructure of S. aureus Treated with Chlorhexidine
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stryjewski, T.P.; Zhang, F. Effect of Massachusetts health reform on chronic disease outcomes. Health Serv. Res. 2014, 49 (Suppl. 2), 2086–2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.Y.; Davis, J.S. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zouhir, A.; Jridi, T. Inhibition of methicillin-resistant Staphylococcus aureus (MRSA) by antimicrobial peptides (AMPs) and plant essential oils. Pharm. Biol. 2016, 54, 3136–3150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlova, Y.N.; Fomenko, N.V. Genetic and biochemical characterization of staphylococci occurring in Novosibirsk, Russia. Vavilovskii Zhurnal Genetiki i Selektsii 2017, 21, 952–958. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.D.; Won, H.S. Antimicrobial peptides for therapeutic applications: A review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [Green Version]
- Kamaruzzaman, N.F.; Tan, L.P. Antimicrobial Polymers: The Potential Replacement of Existing Antibiotics? Int. J. Mol. Sci. 2019, 20, 2747. [Google Scholar] [CrossRef] [Green Version]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Mwangi, J.; Hao, X. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G. Antimicrobial Peptides and Nanotechnology, Recent Advances and Challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Sun, E.; Belaner, C.R. Host defense (antimicrobial) peptides. Pept. Appl. Biomed. Biotechnol. Bioeng. 2018, 10, 253–285. [Google Scholar]
- Diehnelt, C.W. Peptide array based discovery of synthetic antimicrobial peptides. Front MicroBiol. 2013, 4, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scocchi, M.; Mardirossian, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Frankl, A.; Mari, M. Electron microscopy for ultrastructural analysis and protein localization in Saccharomyces cerevisiae. Microb. Cell 2015, 2, 412–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belley, A.; Harris, R. Ultrastructural effects of oritavancin on methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. Antimicrob. Agents Chemother. 2009, 53, 800–804. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Berditsch, M. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 2010, 54, 3132–3142. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.; Desai, S. Antibiofilm Activity and Synergistic Inhibition of Staphylococcus aureus Biofilms by Bactericidal Protein P128 in Combination with Antibiotics. Antimicrob. Agents Chemother. 2016, 60, 7280–7289. [Google Scholar]
- Kang, H.K.; Seo, C.H. Pse-T2, an Antimicrobial Peptide with High-Level, Broad-Spectrum Antimicrobial Potency and Skin Biocompatibility against Multidrug-Resistant Pseudomonas aeruginosa Infection. Antimicrob. Agents Chemother. 2018, 62, 12. [Google Scholar] [CrossRef] [Green Version]
- Amirkhanov, N.V.; Tikunova, N.V. Synthetic Antimicrobial Peptides. I. Antimicrobial Activity of the Amphiphilic and Non-Amphiphilic Cationic Peptides. Bioorgan. Chem. 2018, 44, 492–505. [Google Scholar] [CrossRef]
- Grigoreva, A.; Bardasheva, A. Changes in the Ultrastructure of Candida albicans Treated with Cationic Peptides. Microorganisms 2020, 8, 582. [Google Scholar] [CrossRef] [Green Version]
- Gow, N.A.R.; Latge, J.P. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 3. [Google Scholar]
- Anderson, R.C.; Haverkamp, R.G. Investigation of morphological changes to Staphylococcus aureus induced by ovine-derived antimicrobial peptides using TEM and AFM. FEMS Microbiol. Lett. 2004, 240, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Morita, D.; Sawada, H. Riccardin C derivatives cause cell leakage in Staphylococcus aureus. Biochim. Biophys. Acta 2015, 1848, 2057–2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Mao, R. Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 12124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, H. Antimicrobial Mechanism of pBD2 against Staphylococcus aureus. Molecules 2020, 25, 15. [Google Scholar] [CrossRef] [Green Version]
- Irazazabal, L.N.; Porto, W.F. Fast and potent bactericidal membrane lytic activity of PaDBS1R1, a novel cationic antimicrobial peptide. Biochim. Biophys. Acta Biomembr. 2019, 1861, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, X. Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli. Commun. Biol. 2020, 3, 41. [Google Scholar] [CrossRef]
- Grigoreva, A.; Saranina, I. Fine mechanisms of the interaction of silver nanoparticles with the cells of Salmonella typhimurium and Staphylococcus aureus. Biometals 2013, 26, 479–488. [Google Scholar] [CrossRef]
- Singh, M.; Mukhopadhyay, K. C-terminal amino acids of alpha-melanocyte-stimulating hormone are requisite for its antibacterial activity against Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 1920–1929. [Google Scholar] [CrossRef] [Green Version]
- Matias, V.R.; Beveridge, T.J. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 2006, 188, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Auer, G.K.; Weibel, D.B. Bacterial Cell Mechanics. Biochemistry 2017, 56, 3710–3724. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuber, B.; Haenni, M. Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 2006, 188, 6652–6660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carson, C.F.; Mee, B.J. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.A.; Huttunen-Hennelly, H.E. Bioactivity and the first transmission electron microscopy immunogold studies of short de novo-designed antimicrobial peptides. Antimicrob. Agents Chemother. 2011, 55, 2137–2145. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, M.; Ohki, K. Morphology of defensin-treated Staphylococcus aureus. Infect. Immun. 1995, 63, 2886–2891. [Google Scholar] [CrossRef] [Green Version]
- Santhana Raj, L.; Hing, H.L. Mesosomes are a definite event in antibiotic-treated Staphylococcus aureus ATCC 25923. Trop. Biomed. 2007, 24, 105–109. [Google Scholar]
- Friedrich, C.L.; Moyles, D. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 2000, 44, 2086–2092. [Google Scholar] [CrossRef] [Green Version]
- Nagakubo, T.; Nomura, N. Cracking Open Bacterial Membrane Vesicles. Front Microbiol. 2019, 10, 3026. [Google Scholar] [CrossRef] [Green Version]
- Toyofuku, M.; Nomura, N. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Giesbrecht, P.; Kersten, T. Staphylococcal cell wall: Morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev 1998, 62, 1371–1414. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Gorr, S.U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinho, M.G.; Errington, J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 2003, 50, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Giesbrecht, P.; Labischinski, H. A special morphogenetic wall defect and the subsequent activity of “murosomes” as the very reason for penicillin-induced bacteriolysis in staphylococci. Arch Microbiol. 1985, 141, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rozek, A. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 2001, 276, 35714–35722. [Google Scholar] [CrossRef] [Green Version]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [Green Version]
- Hollmann, A.; Martinez, M. Antimicrobial Peptides: Interaction with Model and Biological Membranes and Synergism with Chemical Antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Chan, D.I.; Prenner, E.J. Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Biophys. Acta 2006, 1758, 1184–1202. [Google Scholar] [CrossRef] [Green Version]
- Mahlapuu, M.; Hakansson, J. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell Infect Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- Neelay, O.P.; Christian, A.P. Antimicrobial peptides interact with peptidoglycan. J. Mol. Struct. 2017, 1146, 329–336. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigor’eva, A.; Bardasheva, A.; Tupitsyna, A.; Amirkhanov, N.; Tikunova, N.; Pyshnyi, D.; Ryabchikova, E. Changes in the Ultrastructure of Staphylococcus aureus Treated with Cationic Peptides and Chlorhexidine. Microorganisms 2020, 8, 1991. https://doi.org/10.3390/microorganisms8121991
Grigor’eva A, Bardasheva A, Tupitsyna A, Amirkhanov N, Tikunova N, Pyshnyi D, Ryabchikova E. Changes in the Ultrastructure of Staphylococcus aureus Treated with Cationic Peptides and Chlorhexidine. Microorganisms. 2020; 8(12):1991. https://doi.org/10.3390/microorganisms8121991
Chicago/Turabian StyleGrigor’eva, Alina, Alevtina Bardasheva, Anastasiya Tupitsyna, Nariman Amirkhanov, Nina Tikunova, Dmitrii Pyshnyi, and Elena Ryabchikova. 2020. "Changes in the Ultrastructure of Staphylococcus aureus Treated with Cationic Peptides and Chlorhexidine" Microorganisms 8, no. 12: 1991. https://doi.org/10.3390/microorganisms8121991