Molecular Characterization, Intra-Species Diversity and Abundance of Freshwater Plesiomonas shigelloides Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultural Isolation of P. shigelloides from River Water
2.2. DNA Extraction and Molecular Characterization of P. shigelloides
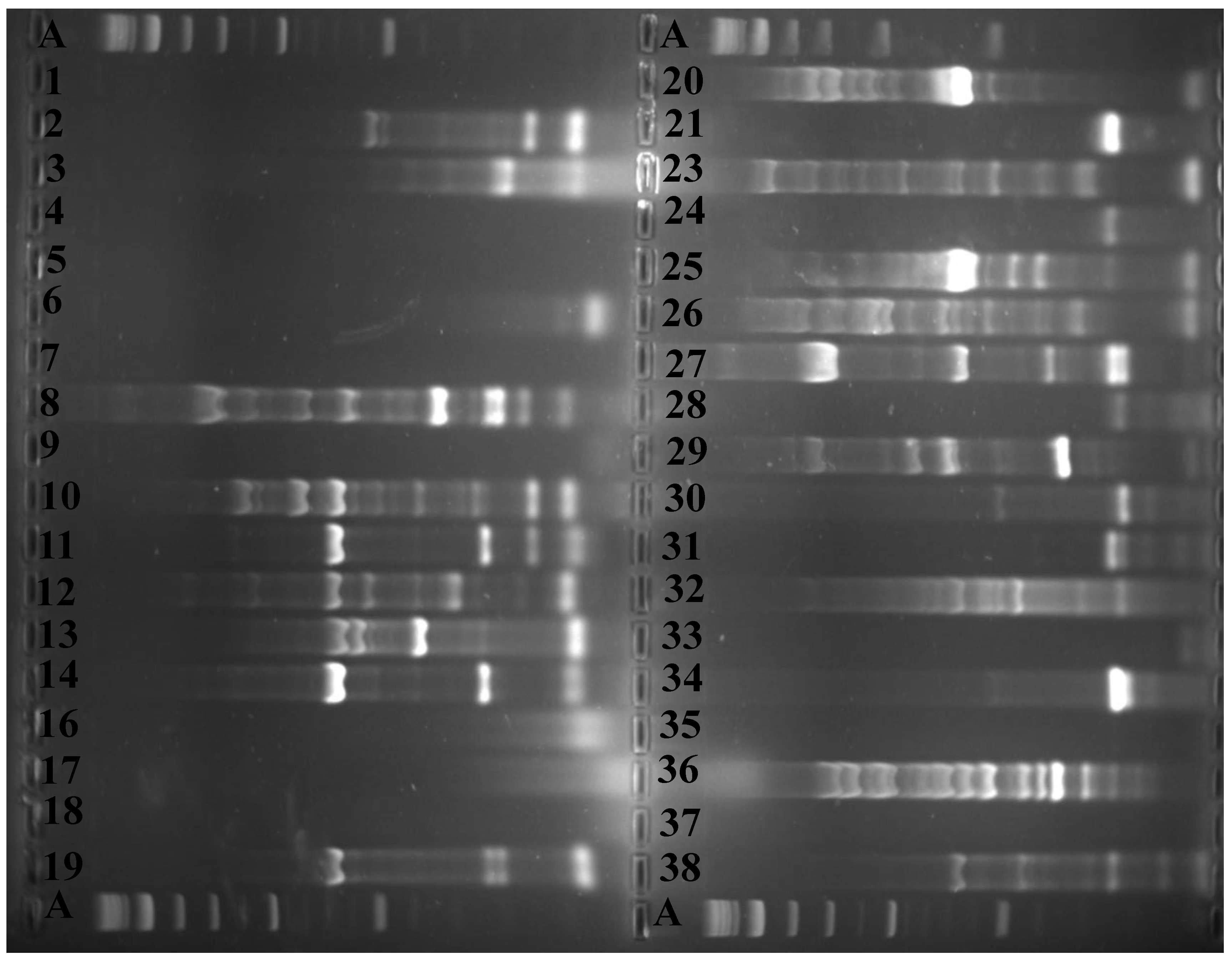
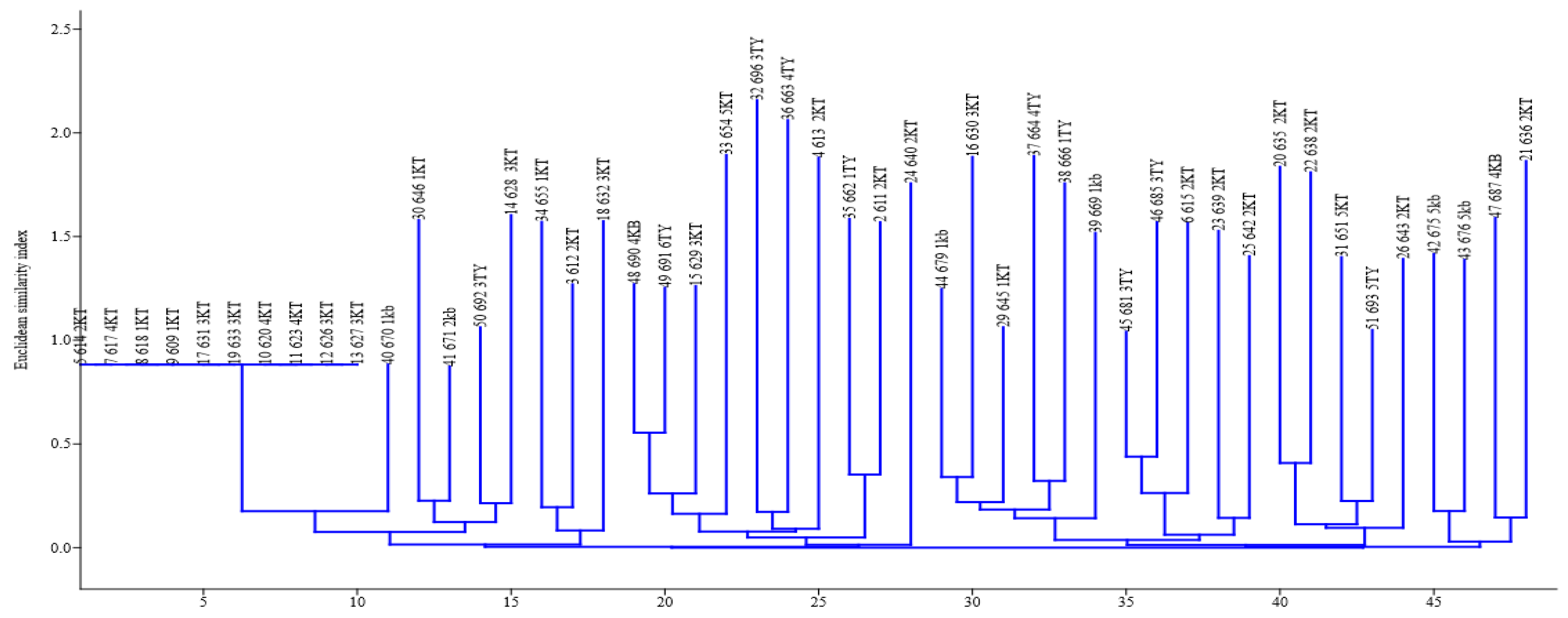
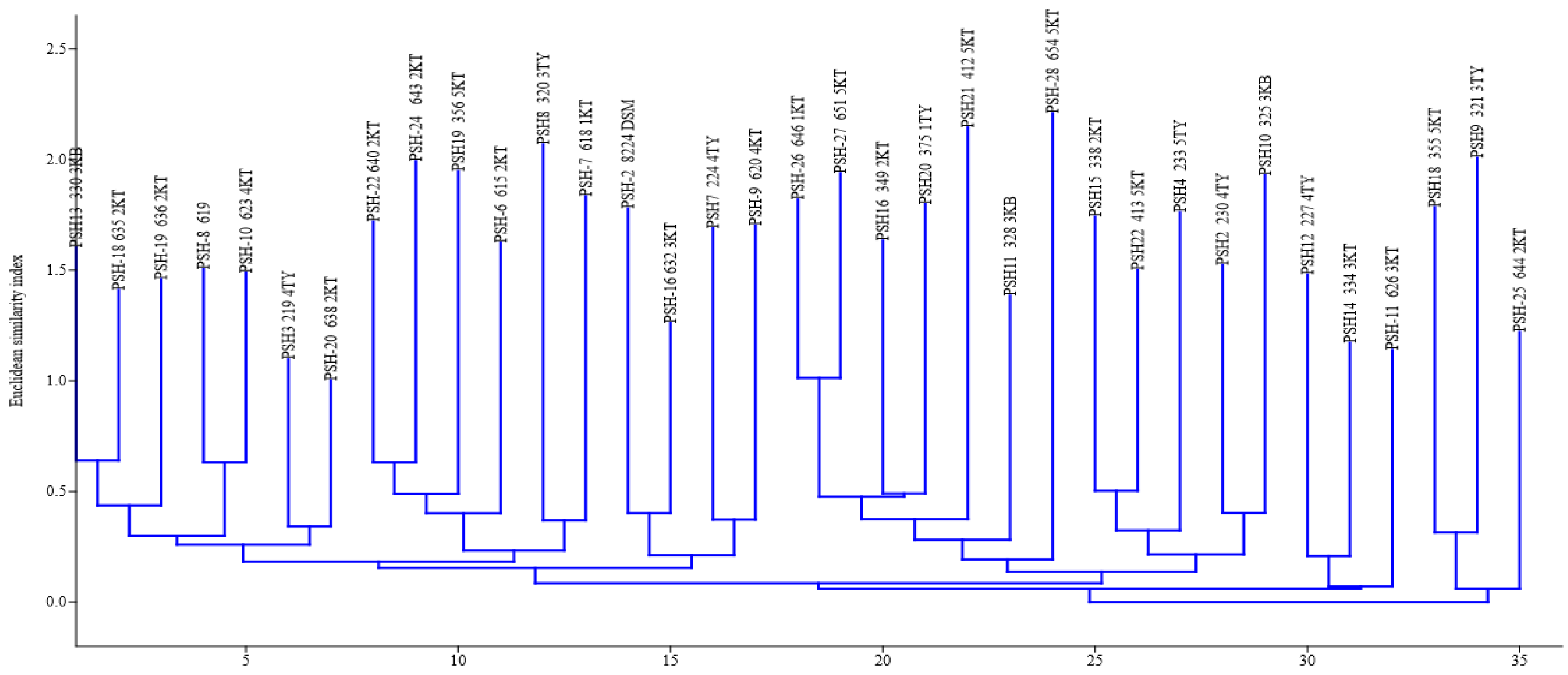
2.3. Fingerprinting, Fingerprint Treatment, and Computer-Assisted Analysis
2.4. Assessment of Intra-Species/Strain Diversity of P. shigelloides Isolates
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shimada, T.; Sakazaki, R. On the serology of Plesiomonas shigelloides. Jpn. J. Med. Sci. Biol. 1978, 31, 135–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldova, E.V.A. Serovars of Plesiomonas shigelloides. Zentralbl. Bakteriol. 1994, 281, 38–44. [Google Scholar] [CrossRef]
- Aldova, E.; Schubert, R.H.W. Serotyping of Plesiomonas shigelloides—A tool for understanding ecological relationships. Med. Microbiol. Lett. 1996, 5, 33–39. [Google Scholar]
- González-Rey, C.; Svenson, S.B.; Bravo, L.; Rosinsky, J.; Ciznar, I.; Krovacek, K. Specific detection of Plesiomonas shigelloides isolated from aquatic environments, animals and human diarrhoeal cases by PCR based on 23S rRNA gene. FEMS Immunol. Med. Microbiol. 2000, 29, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, J.; Gosselin, F.; Ismaïl, J.; Lorange, M.; Lior, H.; Woodward, D. Evaluation of commercial antisera for Shigella serogrouping. J. Clin. Microbiol. 1995, 33, 1997–2001. [Google Scholar] [CrossRef] [Green Version]
- Ekundayo, T.C.; Okoh, A.I. Plesiomonas shigelloides seventy years of systematics and taxonomy in perspective of the present-day diagnostic demands. Res. J. Med. Sci. 2017, 11, 103–113. [Google Scholar] [CrossRef]
- Xi, D.; Wang, X.; Ning, K.; Liu, Q.; Jing, F.; Guo, X.; Cao, B. O-antigen gene clusters of Plesiomonas shigelloides serogroups and its application in development of a molecular serotyping scheme. Front. Microbiol. 2019, 10, 741. [Google Scholar] [CrossRef]
- González-Rey, C.; Siitonen, A.; Pavlova, A.; Ciznar, I.; Svenson, S.B.; Krovacek, K. Molecular evidence of Plesiomonas shigelloides as a possible zoonotic agent. Folia Microbiol. 2011, 56, 178. [Google Scholar] [CrossRef]
- Shigematsu, M.; Kaufmann, M.E.; Charlett, A.; Niho, Y.; Pitt, T.L. An epidemiological study of Plesiomonas shigelloides diarrhoea among Japanese travellers. Epidemiol. Infect. 2000, 125, 523–530. [Google Scholar] [CrossRef]
- Gu, W.; Levin, R.E. Factors affecting quantitative PCR assay of Plesiomonas shigelloides. Food Biotechnol. 2006, 20, 219–230. [Google Scholar] [CrossRef]
- Kaszowska, M.; Jachymek, W.; Lukasiewicz, J.; Niedziela, T.; Kenne, L.; Lugowski, C. The unique structure of complete lipopolysaccharide isolated from semi-rough Plesiomonas shigelloides O37 (strain CNCTC 39/89) containing (2S)-O-(4-oxopentanoic acid)-α-D-Glcp (α-D-Lenose). Carbohydr. Res. 2013, 378, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Kolínská, R.; Dřevínek, M.; Aldová, E.; Žemličková, H. Identification of Plesiomonas spp.: Serological and MALDI-TOF MS methods. Folia Microbiol. 2010, 55, 669–672. [Google Scholar] [CrossRef] [PubMed]
- González-Rey, C. ThesisStudies on Plesiomonas shigelloides Isolated from Different Environments. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2003; pp. 10–27. [Google Scholar]
- Giacometti, F.; Piva, S.; Vranckx, K.; De Bruyne, K.; Drigo, I.; Lucchi, A.; Manfreda, G.; Serraino, A. Application of MALDI-TOF MS for the subtyping of Arcobacter butzleri strains and comparison with their MLST and PFGE types. Int. J. Food Microbiol. 2018, 277, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Olive, D.M.; Bean, P. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 1999, 37, 1661–1669. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cangelosi, G.A.; Freeman, R.J.; Lewis, K.N.; Livingston-Rosanoff, D.; Shah, K.S.; Milan, S.J.; Goldberg, S.V. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J. Clin. Microbiol. 2004, 42, 2685–2693. [Google Scholar] [CrossRef] [Green Version]
- Wieser, M.; Busse, H.J. Rapid identification of Staphylococcus epidermidis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1087–1093. [Google Scholar] [CrossRef] [Green Version]
- Dunne, W.M.; Wang, W. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J. Clin. Microbiol. 1997, 35, 388–392. [Google Scholar] [CrossRef] [Green Version]
- Lanoot, B.; Vancanneyt, M.; Dawyndt, P.; Cnockaert, M.; Zhang, J.; Huang, Y.; Liu, Z.; Swings, J. BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst. Appl. Microbiol. 2004, 27, 84–92. [Google Scholar]
- Willems, R.J.; Top, J.; Braak van den, N.; van Belkum, A.; Endtz, H.; Mevius, D.; Stobberingh, E.; Van den Bogaard, A.; van Embden, J.D. Host specificity of vancomycin-resistant Enterococcus faecium. J. Infect. Dis. 2000, 182, 816–823. [Google Scholar] [CrossRef] [Green Version]
- Bedendo, J.; Pignatari, A.C.C. Typing of Enterococcus faecium by polymerase chain reaction and pulsed field gel electrophoresis. Braz. J. Med. Biol. Res. 2000, 33, 1269–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrity, G.M.; Winters, M.; Searles, D.B. Taxonomic outline of the prokaryotic genera. In Bergey’s Manual of Systematic Bacteriology; Garrity, G.M., Ed.; Springer-Verlag: New York, NY, USA, 2001; pp. 1–39. [Google Scholar]
- Xia, F.Q.; Liu, P.N.; Zhou, Y.H. Meningoencephalitis caused by Plesiomonas shigelloides in a Chinese neonate: Case report and literature review. Ital. J. Pediatr. 2015, 41, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.K.; Zhang, X.C.; Hack, J.B. Plesiomonas shigelloides meningitis in an adult in the ED. Am. J. Emerg. Med. 2016, 34, 1329.e1. [Google Scholar] [CrossRef] [PubMed]
- Novoa-Farías, O.; Frati-Munari, A.C.; Peredo, M.A.; Flores-Juárez, S.; Novoa-García, O.; Galicia-Tapia, J.; Romero-Carpio, C.E. Susceptibilidad de las bacterias aisladas de infecciones gastrointestinales agudas a la rifaximina y otros agentes antimicrobianos en México. Rev. Gastroenterol. México. 2016, 81, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hustedt, J.W.; Ahmed, S. Plesiomonas shigelloides Periprosthetic Knee Infection After Consumption of Raw Oysters. Am. J. Orthop. (Belle Mead, NJ) 2017, 46, E32–E34. [Google Scholar] [PubMed]
- Wouafo, M.; Pouillot, R.; Kwetche, P.F.; Tejiokem, M.C.; Kamgno, J.; Fonkoua, M.C. An acute foodborne outbreak due to Plesiomonas shigelloides in Yaounde, Cameroon. Foodborne Pathog. Dis. 2006, 3, 209–211. [Google Scholar] [CrossRef]
- Wang, S.; Duan, H.; Zhang, W.; Li, J.W. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 2007, 51, 8–13. [Google Scholar] [CrossRef] [Green Version]
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Public Health England. Identification of Vibrio and Aeromonas Species. UK Standards for Microbiology Investigations. ID 19 Issue 3. 2015. Available online: https://www.gov.uk/ukstandards-for-microbiology-investigations-smi-quality-andconsistency-in-clinical-laboratories (accessed on 6 January 2020).
- Salerno, A.; Delétoile, A.; Lefevre, M.; Ciznar, I.; Krovacek, K.; Grimont, P.; Brisse, S. Recombining population structure of Plesiomonas shigelloides (Enterobacteriaceae) revealed by multilocus sequence typing. J. Bacteriol. 2007, 189, 7808–7818. [Google Scholar] [CrossRef] [Green Version]
- Versalovic, J.; Schneider, M.; De Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST-palaeontological statistics, ver. 1.89. Palaeontol. Electron 2001, 4, 1–9. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Petrušić, M.; Vidaković, D.O.; Lazić, S.; Radnović, D.; Knežević, P. Prevalence and genetic variability of Plesiomonas shigelloides in temperate climate surface waters of the Pannonian Plain. Arch. Biol. Sci. 2018, 70, 099–108. [Google Scholar] [CrossRef]
- Mondino, S.S.B.D.; Nunes, M.P.; Ricciardi, I.D. Occurrence of Plesiomonas shigelloides in water environments of Rio de Janeiro city. Memórias Inst. Oswaldo Cruz 1995, 90, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Kwaga, J.K.P.; Adesiyun, A.A.; Abdullahi, S.U.; Bello, C.S.S. Prevalence of salmonellae, shigellae and Plesiomonas shigelloides in dogs in Zaria, Nigeria. Br. Vet. J. 1989, 145, 174–177. [Google Scholar] [CrossRef]
- Arai, T.; Ikejima, N.; Itoh, T.; Sakai, S.; Shimada, T.; Sakazaki, R. A survey of Plesiomonas shigelloides from aquatic environments, domestic animals, pets and humans. Epidemiol. Infect. 1980, 84, 203–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.S.; Alam, M.J.; Khan, S.I. Distribution of Plesiomonas shigelloides in various components of pond ecosystems in Dhaka, Bangladesh. Microbiol. Immunol. 1991, 35, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Bülger, B. Occurrence of Plesiomonas shigelloides and relationship with faecal pollution in Nilufer Stream, Bursa, Turkey. Turk. Electron J. Biotechnol. 2004, 2, 22–29. [Google Scholar]
- Holmberg, S.D.; Farmer, J.J., III. Aeromonas hydrophila and Plesiomonas shigelloides as causes of intestinal infections. Rev. Infect. Dis. 1984, 6, 633–639. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Kinoshita, Y.; Shimada, T.; Sakazaki, R. Two epidemics of diarrhoeal disease possibly caused by Plesiomonas shigelloides. Epidemiol. Infect. 1978, 80, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Penn, R.G.; Giger, D.K.; Knoop, F.C.; Preheim, L.C. Plesiomonas shigelloides overgrowth in the small intestine. J. Clin. Microbiol. 1982, 15, 869–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huq, M.I.; Islam, M.R. Microbiological and clinical studies in diarrhoea due to Plesiomonas shigelloides. Indian J. Med Res. 1983, 77, 793–797. [Google Scholar]
- Krovacek, K.; Eriksson, L.M.; González-Rey, C.; Rosinsky, J.; Ciznar, I. Isolation, biochemical and Serological characterisation of Plesiomonas shigelloides from freshwater in Northern Europe. Comp. Immunol. Microbiol. Infect. Dis. 2000, 23, 45–51. [Google Scholar] [CrossRef]
- Aldova, E.; Melter, O.; Chýle, P.; Slosarek, M.; Kodym, P. Plesiomonas shigelloides in water and fish. Cent. Eur. J. Public Health 1999, 7, 172–175. [Google Scholar] [PubMed]
- Medema, G.; Schets, C. Occurrence of Plesiomonas shigelloides in surface water: Relationship with faecal pollution and trophic state. Zent. Hyg. Umweltmed. Int. J. Hyg. Environ. Med. 1993, 194, 3. [Google Scholar]
- Gonzalez-Rey, C.; Svenson, S.B.; Eriksson, L.M.; Ciznar, I.; Krovacek, K. Unexpected finding of the” tropical” bacterial pathogen Plesiomonas shigelloides from lake water north of the Polar Circle. Polar Biol. 2003, 26, 495–499. [Google Scholar] [CrossRef]
- Prabhu, V.; Isloor, S.; Balu, M.; Suryanarayana, V.V.S.; Rathnamma, D. Genotyping by ERIC-PCR of Escherichia coli isolated from bovine mastitis cases. Indian J. Biotechnol. 2010, 9, 298–301. [Google Scholar]
- Ramazanzadeh, R.; Zamani, S.; Zamani, S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)–PCR technique in Sanandaj hospitals. Iran. J. Microbiol. 2013, 5, 126. [Google Scholar]
- Ekundayo, T.C.; Okoh, A.I. A global bibliometric analysis of Plesiomonas-related research (1990–2017). PloS ONE 2018, 13, e0207655. [Google Scholar] [CrossRef] [Green Version]
- Ekundayo, T.C.; Okoh, A.I. Modelling the effects of physicochemical variables and anthropogenic activities as ecological drivers of Plesiomonas shigelloides distribution and freshwaters quality. Sci. Total Environ. 2019, 682, 765–778. [Google Scholar] [CrossRef]



| Sites | Coordinates | Name |
|---|---|---|
| 1KB | S32°35.429′ E027°21.277′ | Stutt. Game Reserve * |
| 1KT | S32°32.891′ E026°45.835′ | Seymour |
| 1TY | S32°36.683′; E022°57.612′ | Hogsberg |
| 2KB | S32°35.249′ E027°23.916′ | StuttTW1 * |
| 2KT | S32°33.870′ E026°43.252′ | Katberg |
| 2TY | S32°36.652′ E026°54.564′ | Hillfoot * |
| 3KB | S32°35.278′ E027°24.35′ | StuttVBridge1 * |
| 3KT | S32°32.450′ E026°40.568′ | Balfour |
| 3TY | S32°38.374′ E026°56.163′ | Kayalethu |
| 4KB | S32°35.190′ E027°25.415′ | StuttEbBridge * |
| 4KT | S32°42.748′ E026°35.682′ | Blinkwater |
| 4TY | S32°40.980′ E026°54.080′ | Binfield |
| 5KB | S32°35.853′ E027°27.114′ | StuttFbridge * |
| 5KT | S32°46.753′ E026°37.250′ | Fort Beaufort |
| 5TY | S32°43.223′ E026°51.646′ | Melani |
| Sites | Coordinates | Name | Isolates | PCR+(%) | PCR+/211 × 100 |
|---|---|---|---|---|---|
| 1KB | S32°35.429′ E027°21.277′ | Stutt.Game Reserve * | 23 | 07(30.44) | 3.32 |
| 1KT | S32°32.891′ E026°45.835′ | Seymour | 89 | 12(13.5) | 5.69 |
| 1TY | S32°36.683′; E022°57.612′ | Hogsberg | 09 | 08(88.9) | 3.79 |
| 2KB | S32°35.249′ E027°23.916′ | StuttTW1 * | 30 | 06(20.0) | 2.84 |
| 2KT | S32°33.870′ E026°43.252′ | Katberg | 65 | 26(40.0) | 12.32 |
| 2TY | S32°36.652′ E026°54.564′ | Hillfoot * | 10 | 03 (30.0) | 1.42 |
| 3KB | S32°35.278′ E027°24.35′ | StuttVBridge1 * | 97 | 15(15.5) | 7.11 |
| 3KT | S32°32.450′ E026°40.568′ | Balfour | 30 | 20(66.7) | 9.48 |
| 3TY | S32°38.374′ E026°56.163′ | Kayalethu | 19 | 09(47.4) | 4.27 |
| 4KB | S32°35.190′ E027°25.415′ | StuttEbBridge * | 42 | 14(33.33) | 6.64 |
| 4KT | S32°42.748′ E026°35.682′ | Blinkwater | 54 | 13(24.1) | 6.16 |
| 4TY | S32°40.980′ E026°54.080′ | Binfield | 59 | 18(30.5) | 8.53 |
| 5KB | S32°35.853′ E027°27.114′ | StuttFbridge * | 23 | 07(30.4) | 3.32 |
| 5KT | S32°46.753′ E026°37.250′ | Fort Beaufort | 119 | 38(31.9) | 18.01 |
| 5TY | S32°43.223′ E026°51.646′ | Melani | 79 | 19(24.1) | 9.01 |
| Total | 748 | 211(28.21) | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekundayo, T.; Okoh, A. Molecular Characterization, Intra-Species Diversity and Abundance of Freshwater Plesiomonas shigelloides Isolates. Microorganisms 2020, 8, 1081. https://doi.org/10.3390/microorganisms8071081
Ekundayo T, Okoh A. Molecular Characterization, Intra-Species Diversity and Abundance of Freshwater Plesiomonas shigelloides Isolates. Microorganisms. 2020; 8(7):1081. https://doi.org/10.3390/microorganisms8071081
Chicago/Turabian StyleEkundayo, Temitope, and Anthony Okoh. 2020. "Molecular Characterization, Intra-Species Diversity and Abundance of Freshwater Plesiomonas shigelloides Isolates" Microorganisms 8, no. 7: 1081. https://doi.org/10.3390/microorganisms8071081





