Global Transcriptomic Responses of Roseithermus sacchariphilus Strain RA in Media Supplemented with Beechwood Xylan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Growth Profile
2.3. Enzyme Activities
2.4. RNA Extraction
2.5. Library Preparation and RNA Sequencing
2.6. Data Processing and Differentially Expressed Genes (DEGs) Analysis
3. Results and Discussion
3.1. Growth Profiles
3.2. Enzyme Activities
3.3. Technical Overview of RNA-Seq Data
3.4. Overview of Differential Expression (DEGs) Analysis
3.5. Transcription Factors
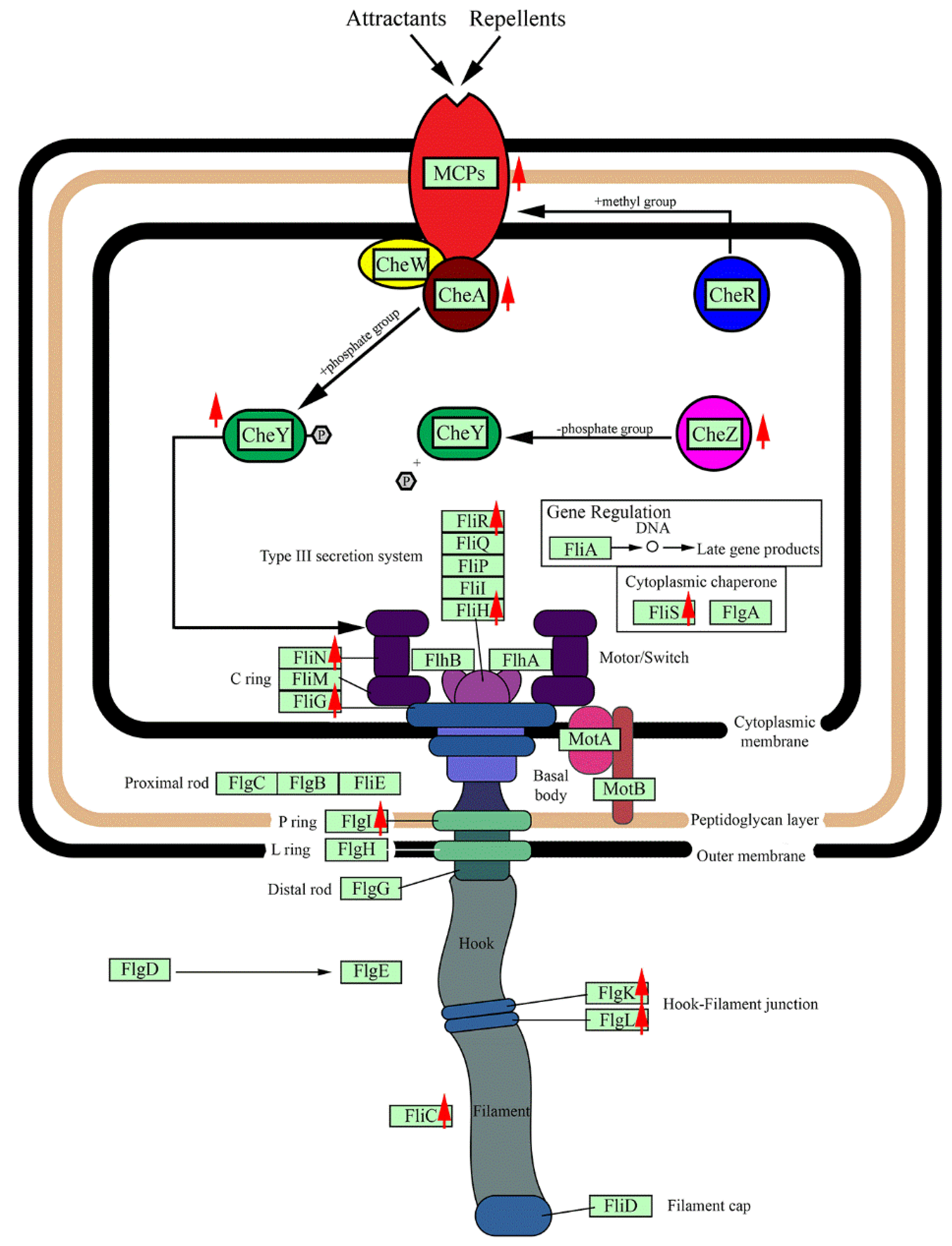
3.6. Chemotaxis and Cell Motility Response
3.7. CAZymes in R. sacchariphilus Strain RA
3.8. Sugar Transportation and Carbon Metabolism in R. sacchariphilus Strain RA
3.9. Possible Role of R. sacchariphilus Strain RA in the Environment
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McDonald, J.E.; Rooks, D.J.; McCarthy, A.J. Methods for the isolation of cellulose-degrading microorganisms. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 510, pp. 349–374. [Google Scholar]
- Carlos, C.; Fan, H.; Currie, C.R. Substrate shift reveals roles for members of bacterial consortia in degradation of plant cell wall polymers. Front. Microbiol. 2018, 9, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlik, A.; Wójcik, M.; Rułka, K.; Motyl-Gorzel, K.; Osińska-Jaroszuk, M.; Wielbo, J.; Marek-Kozaczuk, M.; Skorupska, A.; Rogalski, J.; Janusz, G. Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene. Int. J. Biol. Macromol. 2016, 92, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, R.; Bugg, T.D.H. Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: Oxidation of Mn (II) and polymeric lignin by Dyp1B. Arch. Biochem. Biophys. 2015, 574, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yuan, H.; Yang, Y.; Wang, R.; Wang, C.; Wei, X.; Chen, S.; Yu, J.; Ma, X. Enhanced Lignin Degradation in Tobacco Stalk Composting with Inoculation of White-Rot Fungi Trametes hirsuta and Pleurotus ostreatus. Waste Biomass Valorization 2019, 11, 1–11. [Google Scholar] [CrossRef]
- Sindhu, R.; Binod, P.; Pandey, A. Biological pretreatment of lignocellulosic biomass—An overview. Bioresour. Technol. 2016, 199, 76–82. [Google Scholar] [CrossRef]
- Alfredsson, G.A.; Kristjansson, J.K.; Hjorleifsdottir, S.; Stetter, K.O. Rhodothermus marinus, gen. nov., sp. nov., a thermophilic, halophilic bacterium from submarine hot springs in Iceland. Microbiology 1988, 134, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wang, M.; Gao, R.; Yu, X.; Chen, G. Synergistic effect of thermostable β-glucosidase TN0602 and cellulase on cellulose hydrolysis. 3 Biotech 2017, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Parisutham, V.; Chandran, S.-P.; Mukhopadhyay, A.; Lee, S.K.; Keasling, J.D. Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries. Bioresour. Technol. 2017, 239, 496–506. [Google Scholar] [CrossRef]
- Dodd, D.; Cann, I.K. Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy 2009, 1, 2–17. [Google Scholar] [CrossRef] [Green Version]
- Munoz, R.; Rosselló-Móra, R.; Amann, R. Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst. Appl. Microbiol. 2016, 39, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Park, M.-J.; Oh, J.H.; Yang, S.-H.; Kwon, K.K. Roseithermus sacchariphilus gen. nov., sp. nov. and proposal of Salisaetaceae fam. nov., representing new family in the order Rhodothermales. Int. J. Syst. Evol. Microbiol. 2019, 69, 1213–1219. [Google Scholar] [CrossRef]
- Antón, J.; Oren, A.; Benlloch, S.; Rodríguez-Valera, F.; Amann, R.; Rosselló-Mora, R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 2002, 52, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Makhdoumi-Kakhki, A.; Amoozegar, M.A.; Ventosa, A. Salinibacter iranicus sp. nov. and Salinibacter luteus sp. nov., isolated from a salt lake, and emended descriptions of the genus Salinibacter and of Salinibacter ruber. Int. J. Syst. Evol. Microbiol. 2012, 62, 1521–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Yoshizawa, S.; Kogure, K.; Yokota, A. Rubricoccus marinus gen. nov., sp. nov., of the family ‘Rhodothermaceae’, isolated from seawater. Int. J. Syst. Evol. Microbiol. 2011, 61, 2069–2072. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Song, J.; Yoshizawa, S.; Choi, A.; Cho, J.-C.; Kogure, K. Rubrivirga marina gen. nov., sp. nov., a member of the family Rhodothermaceae isolated from deep seawater. Int. J. Syst. Evol. Microbiol. 2013, 63, 2229–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisman, N.; Oren, A. Salisaeta longa gen. nov., sp. nov., a red, halophilic member of the Bacteroidetes. Int. J. Syst. Evol. Microbiol. 2009, 59, 2571–2574. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Zhou, Y.-X.; Zhao, L.-H.; Chen, G.-J.; Du, Z.-J. Longimonas halophila gen. nov., sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2015, 65, 2272–2276. [Google Scholar] [CrossRef]
- Xia, J.; Dunlap, C.A.; Flor-Weiler, L.; Rooney, A.P.; Chen, G.-J.; Du, Z.-J. Longibacter salinarum gen. nov., sp. nov., isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2016, 66, 3287–3292. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, D.Y.; Khijniak, T.V.; Galinski, E.A.; Kublanov, I.V. Natronotalea proteinilytica gen. nov., sp. nov. and Longimonas haloalkaliphila sp. nov., extremely haloalkaliphilic members of the phylum Rhodothermaeota from hypersaline alkaline lakes. Int. J. Syst. Evol. Microbiol. 2017, 67, 4161–4167. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.M.; Shahar, S.; Chan, K.-G.; Chong, C.S.; Amran, S.I.; Sani, M.H.; Zakaria, I.I.; Kahar, U.M. Current status and potential applications of underexplored prokaryotes. Microorganisms 2019, 7, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjornsdottir, S.H.; Blondal, T.; Hreggvidsson, G.O.; Eggertsson, G.; Petursdottir, S.; Hjorleifsdottir, S.; Thorbjarnardottir, S.H.; Kristjansson, J.K. Rhodothermus marinus: Physiology and molecular biology. Extremophiles 2006, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ara, K.Z.G.; Månberger, A.; Gabriško, M.; Linares-Pastén, J.A.; Jasilionis, A.; Friðjónsson, Ó.H.; Hreggviðsson, G.Ó.; Janeček, Š.; Karlsson, E.N. Characterization and diversity of the complete set of GH family 3 enzymes from Rhodothermus marinus DSM 4253. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sardari, R.R.R.; Kulcinskaja, E.; Ron, E.Y.C.; Björnsdóttir, S.; Friðjónsson, Ó.H.; Hreggviðsson, G.Ó.; Karlsson, E.N. Evaluation of the production of exopolysaccharides by two strains of the thermophilic bacterium Rhodothermus marinus. Carbohydr. Polym. 2017, 156, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ron, E.Y.C.; Plaza, M.; Kristjansdottir, T.; Sardari, R.R.R.; Bjornsdottir, S.H.; Gudmundsson, S.; Hreggvidsson, G.O.; Turner, C.; van Niel, E.W.J.; Nordberg-Karlsson, E. Characterization of carotenoids in Rhodothermus marinus. Microbiologyopen 2017, 7, e00536. [Google Scholar] [CrossRef] [Green Version]
- Ron, E.Y.C.; Sardari, R.R.R.; Anthony, R.; van Niel, E.W.J.; Hreggvidsson, G.O.; Nordberg-Karlsson, E. Cultivation technology development of Rhodothermus marinus DSM 16675. Extremophiles 2019, 23, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Dahlberg, L.; Holst, O.; Kristjansson, J.K. Thermostable xylanolytic enzymes from Rhodothermus marinus grown on xylan. Appl. Microbiol. Biotechnol. 1993, 40, 63–68. [Google Scholar] [CrossRef]
- Halldórsdóttir, S.; Thorolfsdottir, E.; Spilliaert, R.; Johansson, M.; Thorbjarnardottir, S.; Palsdottir, A.; Hreggvidsson, G.; Kristjansson, J.; Holst, O.; Eggertsson, G. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulase of glycosyl hydrolase family 12. Appl. Microbiol. Biotechnol. 1998, 49, 277–284. [Google Scholar] [CrossRef]
- Goh, K.M.; Chan, K.-G.; Lim, S.W.; Liew, K.J.; Chan, C.S.; Shamsir, M.S.; Ee, R.; Adrian, T.-G.-S. Genome analysis of a new Rhodothermaceae strain isolated from a hot spring. Front. Microbiol. 2016, 7, 1109. [Google Scholar] [CrossRef]
- Liew, K.J.; Ngooi, C.Y.; Shamsir, M.S.; Sani, R.K.; Chong, C.S.; Goh, K.M. Heterologous expression, purification and biochemical characterization of a new endo-1,4-β-xylanase from Rhodothermaceae bacterium RA. Protein Expr. Purif. 2019, 164, 105464. [Google Scholar] [CrossRef]
- Teo, S.C.; Liew, K.J.; Shamsir, M.S.; Chong, C.S.; Bruce, N.C.; Chan, K.-G.; Goh, K.M. Characterizing a halo-tolerant GH10 xylanase from Roseithermus sacchariphilus strain RA and its CBM-truncated variant. Int. J. Mol. Sci. 2019, 20, 2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manelius, Å.; Dahlberg, L.; Holst, O. Some properties of a thermostable β-xylosidase from Rhodothermus marinus. Appl. Biochem. Biotechnol. 1994, 44, 39–48. [Google Scholar] [CrossRef]
- Gomes, J.; Gomes, I.; Terler, K.; Gubala, N.; Ditzelmüller, G.; Steiner, W. Optimisation of culture medium and conditions for α-L-arabinofuranosidase production by the extreme thermophilic eubacterium Rhodothermus marinus. Enzyme Microb. Technol. 2000, 27, 414–422. [Google Scholar] [CrossRef]
- Politz, O.; Krah, M.; Thomsen, K.K.; Borriss, R. A highly thermostable endo-(1, 4)-β-mannanase from the marine bacterium Rhodothermus marinus. Appl. Microbiol. Biotechnol. 2000, 53, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-m.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Chan, K.-G.; Ee, R.; Hong, K.-W.; Urbieta, M.S.; Donati, E.R.; Shamsir, M.S.; Goh, K.M. Effects of physiochemical factors on prokaryotic biodiversity in Malaysian circumneutral hot springs. Front. Microbiol. 2017, 8, 1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, K.J.; Teo, S.C.; Shamsir, M.S.; Sani, R.K.; Chong, C.S.; Chan, K.-G.; Goh, K.M. Complete genome sequence of Rhodothermaceae bacterium RA with cellulolytic and xylanolytic activities. 3 Biotech 2018, 8, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, K.A.; Summers, R.S.; Cook, S.M. Development and experimental validation of the composition and treatability of a new synthetic bathroom greywater (SynGrey). Environ. Sci. Water Res. Technol. 2017, 3, 1120–1131. [Google Scholar] [CrossRef]
- Kahar, U.M.; Sani, M.H.; Chan, K.-G.; Goh, K.M. Immobilization of α-Amylase from Anoxybacillus sp. SK3-4 on ReliZyme and Immobead Supports. Molecules 2016, 21, 1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, A.; Hu, Y.; Li, J.; Wang, W.; Zhang, M.; Guan, S. Characterization of a recombinant β-xylosidase of GH43 family from Bacteroides ovatus strain ATCC 8483. Biocatal. Biotransformation 2019, 38, 1–7. [Google Scholar] [CrossRef]
- Sinha, S.K.; Datta, S. β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Appl. Microbiol. Biotechnol. 2016, 100, 8399–8409. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Osman, A.; Hitzler, W.E.; Ameur, A.; Provost, P. Differential expression analysis by RNA-Seq reveals perturbations in the platelet mRNA transcriptome triggered by pathogen reduction systems. PLoS ONE 2015, 10, e0133070. [Google Scholar] [CrossRef]
- Gunalan, K.; Sá, J.M.; Barros, R.R.M.; Anzick, S.L.; Caleon, R.L.; Mershon, J.P.; Kanakabandi, K.; Paneru, M.; Virtaneva, K.; Martens, C. Transcriptome profiling of Plasmodium vivax in Saimiri monkeys identifies potential ligands for invasion. Proc. Natl. Acad. Sci. USA 2019, 116, 7053–7061. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Ai, C.; Kong, L. CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018, 45, 489–504. [Google Scholar] [CrossRef]
- Darbani, B.; Stewart, C.N., Jr. Reproducibility and reliability assays of the gene expression-measurements. J. Biol. Res. 2014, 21, 3. [Google Scholar] [CrossRef] [Green Version]
- Jia, K.; Wang, G.; Liang, L.; Wang, M.; Wang, H.; Xu, X. Preliminary transcriptome analysis of mature biofilm and planktonic cells of Salmonella Enteritidis exposure to acid stress. Front. Microbiol. 2017, 8, 1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoroshkin, M.S.; Leyn, S.A.; Van Sinderen, D.; Rodionov, D.A. Transcriptional regulation of carbohydrate utilization pathways in the Bifidobacterium Genus. Front. Microbiol. 2016, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Evangelopoulos, D.; Gupta, A.; Lack, N.A.; Maitra, A.; Ten Bokum, A.M.C.; Kendall, S.; Sim, E.; Bhakta, S. Characterisation of a putative AraC transcriptional regulator from Mycobacterium smegmatis. Tuberculosis 2014, 94, 664–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüller, A.; Slater, A.W.; Norambuena, T.; Cifuentes, J.J.; Almonacid, L.I.; Melo, F. Computer-based annotation of putative AraC/XylS-family transcription factors of known structure but unknown function. BioMed Res. Int. 2012, 2012, 103132. [Google Scholar] [CrossRef] [PubMed]
- Schleif, R. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010, 34, 779–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Yip, C.B.; Geddes, B.A.; Oresnik, I.J.; Hynes, M.F. Glycerol utilization by Rhizobium leguminosarum requires an ABC transporter and affects competition for nodulation. Microbiology 2012, 158, 1369–1378. [Google Scholar] [CrossRef]
- Stetz, M.A.; Carter, M.V.; Wand, A.J. Optimized expression and purification of biophysical quantities of Lac repressor and Lac repressor regulatory domain. Protein Expr. Purif. 2016, 123, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Suvorova, I.A.; Korostelev, Y.D.; Gelfand, M.S. GntR family of bacterial transcription factors and their DNA binding motifs: Structure, positioning and co-evolution. PLoS ONE 2015, 10, e0132618. [Google Scholar] [CrossRef]
- Derr, P.; Boder, E.; Goulian, M. Changing the specificity of a bacterial chemoreceptor. J. Mol. Biol. 2006, 355, 923–932. [Google Scholar] [CrossRef]
- Webre, D.J.; Wolanin, P.M.; Stock, J.B. Bacterial chemotaxis. Curr. Biol. 2003, 13, R47–R49. [Google Scholar] [CrossRef] [Green Version]
- Bi, S.; Sourjik, V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 2018, 45, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Broeker, J.; Mechelke, M.; Baudrexl, M.; Mennerich, D.; Hornburg, D.; Mann, M.; Schwarz, W.H.; Liebl, W.; Zverlov, V.V. The hemicellulose-degrading enzyme system of the thermophilic bacterium Clostridium stercorarium: Comparative characterisation and addition of new hemicellulolytic glycoside hydrolases. Biotechnol. Biofuels 2018, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez, C.; Reyes-Sosa, F.M.; Díez, B. Enzymatic hydrolysis of biomass from wood. Microb. Biotechnol. 2016, 9, 149–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suto, M.; Tomita, F. Induction and catabolite repression mechanisms of cellulase in fungi. J. Biosci. Bioeng. 2001, 92, 305–311. [Google Scholar] [CrossRef]
- Gao, J.; Qian, Y.; Wang, Y.; Qu, Y.; Zhong, Y. Production of the versatile cellulase for cellulose bioconversion and cellulase inducer synthesis by genetic improvement of Trichoderma reesei. Biotechnol. Biofuels 2017, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Schuerg, T.; Prahl, J.-P.; Gabriel, R.; Harth, S.; Tachea, F.; Chen, C.-S.; Miller, M.; Masson, F.; He, Q.; Brown, S.; et al. Xylose induces cellulase production in Thermoascus aurantiacus. Biotechnol. Biofuels 2017, 10, 271. [Google Scholar] [CrossRef]
- Fatokun, E.; Nwodo, U.; Okoh, A. Classical optimization of cellulase and xylanase production by a marine Streptomyces species. Appl. Sci. 2016, 6, 286. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.H.; Liu, X.; Zhan, T.; He, J. Production of cellulase by Trichoderma reesei from pretreated straw and furfural residues. RSC Adv. 2018, 8, 36233–36238. [Google Scholar] [CrossRef] [Green Version]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, M.G.; Cetinić, I.; Chaves, J.E.; Mannino, A. The adsorption of dissolved organic carbon onto glass fiber filters and its effect on the measurement of particulate organic carbon: A laboratory and modeling exercise. Limnol. Oceanogr. Methods 2018, 16, 356–366. [Google Scholar] [CrossRef] [PubMed]
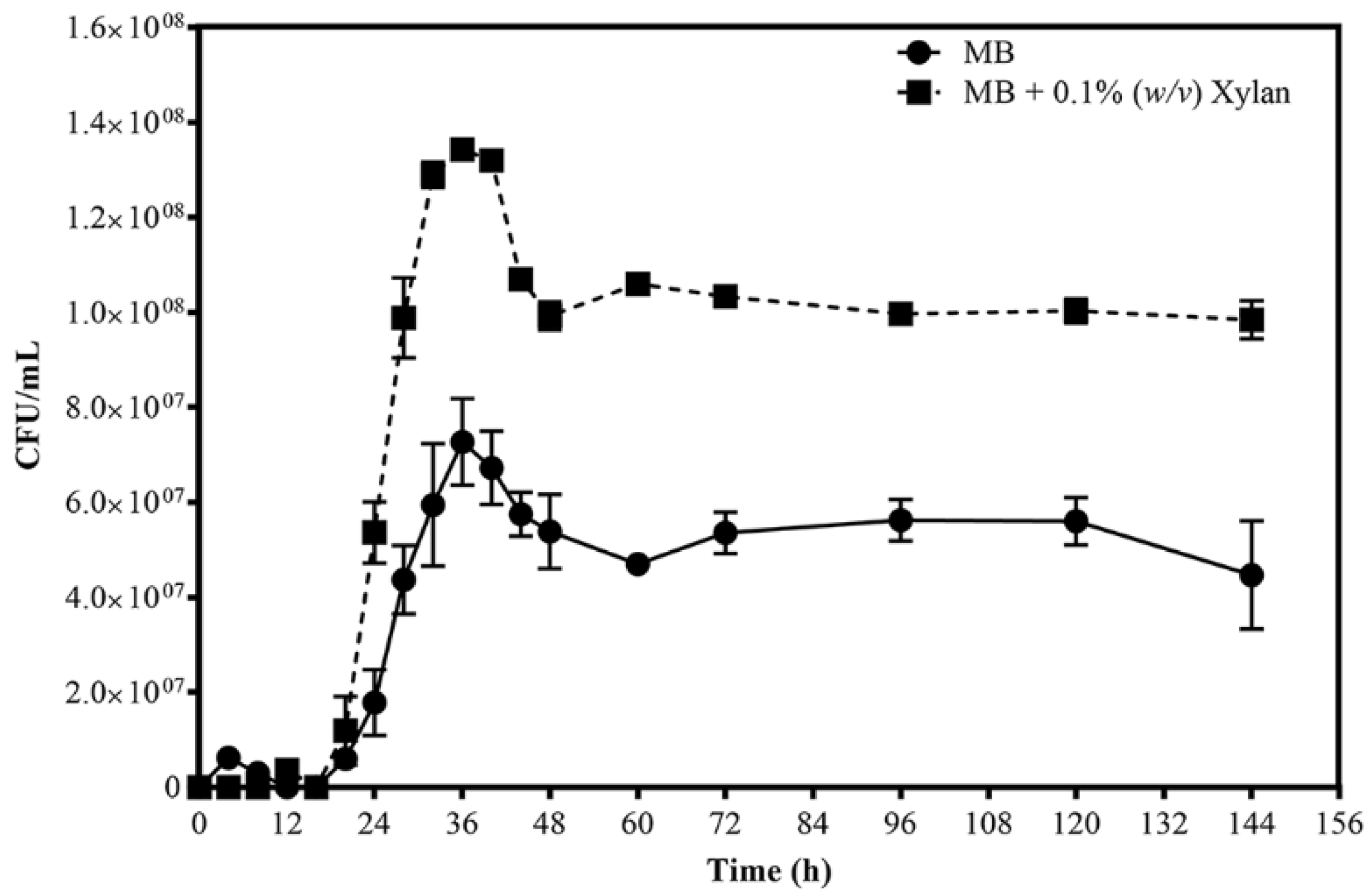

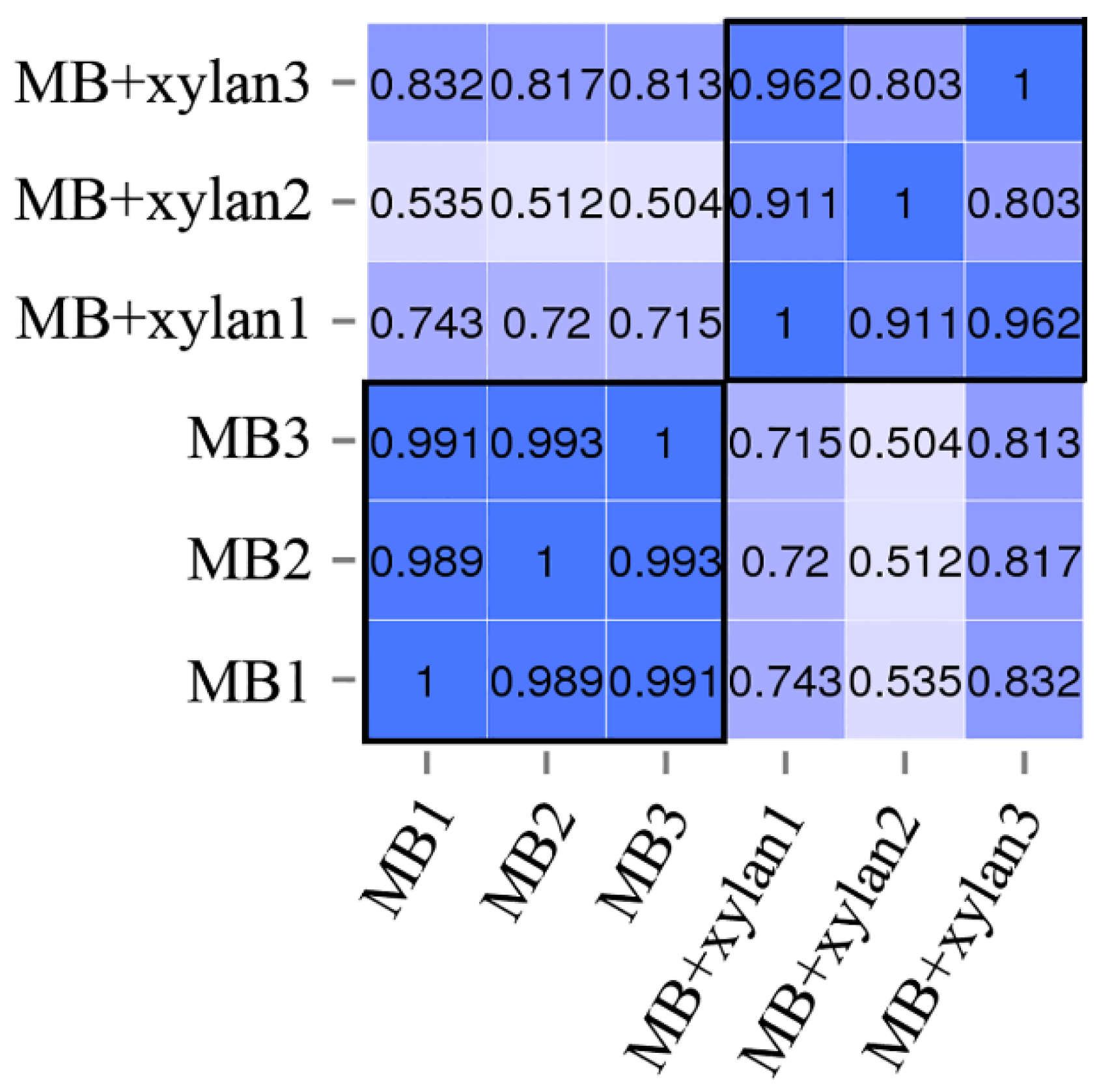


| Strain 1 | Origin | Opt. Temp; Opt. pH; Opt. NaCl | Hydrolytic Activity 2 | Genome Size (Mb)/Status | 16S rRNA 3 (%) | ANI 4 (%) | Taxonomy Ref. 5 | Cellulase/Hemicellulase Ref. 6 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| A | C | H | ||||||||
| RA | Saline hot spring | 50 °C; pH 7; 2% (w/v) | + | + | + | 4.65 (Complete) | 100 | 100 | [30] | [31,32] |
| Rssac | Sea port | 55 °C; pH 7; 2–4% (w/v) | ND | + | ND | 4.81 (Complete) | 99.3 | 96.2 | [13] | NA |
| Rmar | Hydrothermal vent | 65 °C; pH 6.5–7; 2% (w/v) | + | + | + | 3.39 (Complete) | 87.5 | 73.3 | [7] | [24,28,29,33,34,35] |
| Srub | Saltern crystallizer pond | 37–47 °C; pH 6.5–8; 20–30% (w/v) | + | ND | ND | 3.59 (Complete) | 85.6 | 71.9 | [14] | NA |
| Sira | Salt Lake | 37 °C; pH 7.5; 17% (w/v) | - | ND | ND | 3.41 (Draft) | 83.1 | 70.0 | [15] | NA |
| Slon | Dead/Red sea water | 37–46 °C; pH 6.5–8.5; 10–12% (w/v) | + | ND | ND | 3.19 (Draft) | 85.7 | 71.5 | [18] | NA |
| Lhalo | Marine solar saltern | 37–42 °C; pH 7.5–8; 6–8% (w/v) | + | - | ND | 3.73 (Draft) | 81.9 | 69.1 | [19] | NA |
| Lsali | Marine solar saltern | 40 °C; pH 7.5–8; 8–12% (w/v) | + | - | ND | 4.41 (Draft) | 82.4 | 69.3 | [20] | NA |
| Npro | Hypersaline alkaline lake | 37 °C; pH 9.5–9.8; 14.6–17.5% (w/v) | + | - | - | ND | 83.8 | ND | [21] | NA |
| Rcmar | Sea water | 20–30 °C; pH 5–9; 2% (w/v) | ND | ND | ND | 4.43 (Draft) | 81.5 | 69.5 | [16] | NA |
| Rvmar | Deep sea water | 25–30 °C; pH 6–8; 1–5% (w/v) | ND | ND | ND | 4.98 (Draft) | 81.3 | 69.9 | [17] | NA |
| Sample 1 | Raw Reads | Q20 (%) 2 | Clean Reads 3 | Total Mapped Reads 4 | Uniquely Mapped (%) | Multiple Mapped (%) | |
|---|---|---|---|---|---|---|---|
| MB | 1 | 21,980,340 | 97.81 | 21,497,336 | 21,402,185 (99.56%) | 98.40 | 1.16 |
| 2 | 22,412,010 | 97.91 | 21,857,496 | 21,728,678 (99.41%) | 98.18 | 1.23 | |
| 3 | 21,996,694 | 97.86 | 21,478,716 | 21,381,885 (99.55%) | 98.36 | 1.19 | |
| MB+xylan | 1 | 23,769,554 | 97.61 | 23,307,532 | 23,022,398 (98.78%) | 97.69 | 1.09 |
| 2 | 25,168,712 | 97.58 | 24,610,454 | 24,377,051 (99.05%) | 97.96 | 1.09 | |
| 3 | 22,425,294 | 97.69 | 21,935,340 | 21,597,320 (98.46%) | 97.30 | 1.16 | |
| Gene ID and Annotated Names | FC and Gene Regulation Status |
|---|---|
| AraC family (regulates carbon catabolism, stress responses, and virulence) | |
| AWN76_004845 AraC family transcriptional regulator | ~ |
| AWN76_005395 AraC family transcriptional regulator | 1.52➚ |
| AWN76_009110 AraC family transcriptional regulator | 0.26➘ |
| AWN76_012545 AraC family transcriptional regulator | ~ |
| AWN76_014955 AraC family transcriptional regulator | ~ |
| DeoR family (regulates sugar catabolism, regulates aga operon for n-acetyl galactosamine transport and metabolism) | |
| AWN76_014420 transcriptional regulator AgaR | 1.94➚ |
| AWN76_014295 DeoR family transcriptional regulator | 1.93➚ |
| LacI family (regulates sugar/lactose catabolism) | |
| AWN76_006400 LacI family transcriptional regulator | 1.54➚ |
| AWN76_010965 LacI family transcriptional regulator | ~ |
| AWN76_004120 LacI family transcriptional regulator | 2.02➚ |
| AWN76_016215 LacI family transcriptional regulator | ~ |
| AWN76_013990 LacI family transcriptional regulator | 2.49➚ |
| AWN76_003445 LacI family transcriptional regulator | 0.56➘ |
| AWN76_012325 LacI family transcriptional regulator | ~ |
| GntR family (regulates carbohydrate transport and metabolism; transcriptional repressor for pyruvate dehydrogenase complex) | |
| AWN76_002320 GntR family transcriptional regulator | 0.37➘ |
| AWN76_018420 transcriptional regulator PdhR | 0.39➘ |
| AWN76_008220 transcriptional regulator PdhR | 0.41➘ |
| AWN76_003490 GntR family transcriptional factor | 2.37➚ |
| Gene ID and Annotated Names | FC and Gene Regulation Status |
|---|---|
| Chemotaxis pathway | |
| AWN76_009955 methyl-accepting chemotaxis protein MCP | 1.90➚ |
| AWN76_011645 methyl-accepting chemotaxis protein MCP | ~ |
| AWN76_016375 methyl-accepting chemotaxis protein MCP | 2.98➚ |
| AWN76_017480 methyl-accepting chemotaxis protein MCP | ~ |
| AWN76_017530 methyl-accepting chemotaxis protein MCP | 3.29➚ |
| AWN76_017495 sensor kinase CheA | 2.32 ➚ |
| AWN76_017520 chemotaxis protein methyltransferase CheR | 0.45➘ |
| AWN76_017540 purine-binding chemotaxis protein CheW | ~ |
| AWN76_008370 chemotaxis protein CheY | 2.29 ➚ |
| AWN76_016800 chemotaxis protein CheY | 3.70➚ |
| AWN76_017505 chemotaxis protein CheY | ~ |
| AWN76_017515 chemotaxis protein CheY | 0.40➘ |
| AWN76_017500 chemotaxis protein CheZ | 2.20➚ |
| Flagella assembly | |
| AWN76_013275 flagellar motor protein MotB | ~ |
| AWN76_017405 RNA polymerase sigma factor FliA | 0.51➘ |
| AWN76_017420 flagellar biosynthesis protein FlhF | ~ |
| AWN76_017415 flagellar protein FliS | 2.61➚ |
| AWN76_017425 flagellar biosynthesis protein FlhA | ~ |
| AWN76_017430 flagellar biosynthetic protein FlhB | ~ |
| AWN76_017435 flagellar biosynthetic protein FliR | 2.31➚ |
| AWN76_017440 flagellar biosynthetic protein FliQ | ~ |
| AWN76_017445 flagellar biosynthetic protein FliP | 0.65➘ |
| AWN76_017455 flagellar motor switch protein FliN | 3.20➚ |
| AWN76_017460 flagellar hook-basal body complex protein FliM | ~ |
| AWN76_017465 flagellar FliL protein | ~ |
| AWN76_017470 flagellar motor protein MotB | ~ |
| AWN76_017475 flagellar motor protein MotA | ~ |
| AWN76_017545 flagellar hook protein FlgE | ~ |
| AWN76_017555 flagellar hook assembly protein FlgD | ~ |
| AWN76_017575 flagellum-specific ATP synthase | 3.70➚ |
| AWN76_017580 flagellar assembly protein FliH | 3.59➚ |
| AWN76_017585 flagellar motor switch protein FliG | 3.53➚ |
| AWN76_017595 flagellar hook-basal body complex protein FliE | ~ |
| AWN76_017605 flagellar basal body rod protein FlgC | ~ |
| AWN76_017610 flagellar biosynthesis protein FlgB | ~ |
| AWN76_017635 flagellar hook protein FliD | 0.59➘ |
| AWN76_017640 flagellar protein FliS | ~ |
| AWN76_017650 flagellin FliC | 5.20➚ |
| AWN76_017655 flagellin FliC | 2.15➚ |
| AWN76_017660 flagellin FliC | ~ |
| AWN76_017675flagellar hook-associated protein 3 FlgL | 1.91➚ |
| AWN76_017680 flagellar hook-associated protein FlgK | 2.00➚ |
| AWN76_017695 flagellar basal body P-ring protein FlgI | 2.82➚ |
| AWN76_017700 flagellar basal body L-ring protein FlgH | ~ |
| AWN76_017705 flagella basal body P-ring formation protein FlgA | ~ |
| AWN76_017710 flagellar basal-body rod protein FlgG | ~ |
| AWN76_017715 flagellar basal-body rod protein FlgF | 0.62➘ |
| Gene ID and Annotated Names | FC and Gene Regulation Status | |
|---|---|---|
| Family | Cellulolytic GHs | |
| GH3 | AWN76_006445 β-glucosidase (1) | 5.30➚ |
| GH44 | AWN76_008195 hypothetical protein (2) | 1.45➚ |
| GH3 | AWN76_008215 β-glucosidase (3) | 3.43➚ |
| GH9 | AWN76_008290 cellulase (4) | 1.99➚ |
| GH5 | AWN76_009395 glycoside hydrolase (5) | 1.78➚ |
| GH9 | AWN76_010685 glycoside hydrolase family 9 (6) | ~ |
| Family | Hemicellulolytic GHs | |
| GH78 | AWN76_002810 α-l-rhamnosidase | 2.53➚ |
| GH92 | AWN76_002955 α-1,2-mannosidase | ~ |
| GH10 | AWN76_003690 endo-1,4-β-xylanase (7) | ~ |
| GH31 | AWN76_004235 glycoside hydrolase family 31 | ~ |
| GH31 | AWN76_008190 α-xylosidase | ~ |
| GH10 | AWN76_008205 endo-1,4-β-xylanase (8) | 1.66➚ |
| GH67 | AWN76_008230 α-glucuronidase (9) | 1.70➚ |
| GH106 | AWN76_008320 α-l-rhamnosidase (10) | 3.28➚ |
| GH78 | AWN76_009025 α-l-rhamnosidase | 1.60➚ |
| GH16 | AWN76_009940 glycoside hydrolase family 16 | ~ |
| GH29 | AWN76_010630 α-l-fucosidase | ~ |
| GH78 | AWN76_012010 α-l-rhamnosidase | 2.61➚ |
| GH43 | AWN76_012335 β-xylosidase (11) | ~ |
| GH51 | AWN76_012350 α-d-arabinofuranosidase (12) | 2.74➚ |
| GH113 | AWN76_013895 β-mannase (13) | 3.31➚ |
| GH130 | AWN76_014035 glycosidase | 4.00➚ |
| GH130 | AWN76_014055 glycosidase | 3.90➚ |
| GH2 | AWN76_014570 glycoside hydrolase family 2 | ~ |
| GH154 | AWN76_017060 hypothetical protein | ~ |
| GH53 | AWN76_017855 endo-1,4-β-galactanase (14) | 2.43➚ |
| Gene ID and Annotated Names | FC and Gene Regulation Status | |
|---|---|---|
| Family | AAs | |
| AA3 | AWN76_001955 GMC family oxidoreductase | 0.44➘ |
| AA3 | AWN76_003120 GMC family oxidoreductase | 0.43➘ |
| AA12 | AWN76_005825 sorbosone dehydrogenase | ~ |
| AA3 | AWN76_007025 GMC family oxidoreductase | 1.88➚ |
| AA3 | AWN76_007050 patatin-like phospholipase family protein | 2.70➚ |
| AA12 | AWN76_011490 sorbosone dehydrogenase | ~ |
| AA3 | AWN76_011750 GMC family oxidoreductase | 0.60➘ |
| AA2 | AWN76_014060 catalase/peroxidase HPI | 0.36➘ |
| Gene ID and Annotated Names | FC and Gene Regulation Status |
|---|---|
| Glycolysis pathway | |
| AWN76_000290 triose-phosphate isomerase | 1.49➚ |
| AWN76_000410 pyruvate kinase | 0.49➘ |
| AWN76_001275 fructose-bisphosphate aldolase | 0.15➘ |
| AWN76_003230 phosphoglycerate kinase | ~ |
| AWN76_003235 glyceraldehyde-3-phosphate dehydrogenase | 0.26➘ |
| AWN76_004260 enolase | 0.49➘ |
| AWN76_004270 glucokinase | ~ |
| AWN76_005185 glucose/mannose-6-phosphate isomerase | ~ |
| AWN76_005345 6-phosphofructokinase | ~ |
| AWN76_007495 phosphoglycerate mutase | ~ |
| AWN76_007770 fructose-1,6-bisphosphatase I | 0.56➘ |
| AWN76_009045 6-phosphofructokinase | ~ |
| AWN76_012860 glucose/mannose-6-phosphate isomerase | ~ |
| AWN76_013745 6-phosphofructokinase | ~ |
| AWN76_014560 galactose mutarotase | 0.65➘ |
| AWN76_017730 polyphosphate glucokinase | ~ |
| AWN76_018330 phosphoglucomutase | ~ |
| Tricarboxylic acid (TCA) cycle | |
| AWN76_001060 pyruvate-ferredoxin/flavodoxin oxidoreductase | 3.04➚ |
| AWN76_003860 class II fumarate hydratase | 0.39➘ |
| AWN76_004160 malate dehydrogenase | 0.22➘ |
| AWN76_004295 succinyl-CoA synthetase β subunit | 0.47➘ |
| AWN76_004595 dihydrolipoyl dehydrogenase | 0.39➘ |
| AWN76_004770 NADP-dependent isocitrate dehydrogenas | ~ |
| AWN76_006135 isocitrate dehydrogenase (NAD+) | 0.59➘ |
| AWN76_006890 pyruvate dehydrogenase E2 component | 0.56➘ |
| AWN76_006895 pyruvate dehydrogenase E1 subunit β | 0.27➘ |
| AWN76_006900 pyruvate dehydrogenase E1 subunit α | 0.16➘ |
| AWN76_006920 NADP-dependent isocitrate dehydrogenase | 0.19➘ |
| AWN76_008605 2-oxoglutarate dehydrogenase E2 component | 0.46➘ |
| AWN76_009515 succinate-CoA ligase subunit α | 0.43➘ |
| AWN76_009950 2-oxoglutarate dehydrogenase E1 component | 0.40➘ |
| AWN76_011245 citrate synthase | 0.57➘ |
| AWN76_011250 succinate dehydrogenase cytochrome b subunit | 0.37➘ |
| AWN76_011255 succinate dehydrogenase | 0.52➘ |
| AWN76_011260 succinate dehydrogenase flavoprotein subunit | 0.51➘ |
| AWN76_011265 succinate dehydrogenase iron–sulfur subunit | 0.46➘ |
| AWN76_014835 citrate synthase | 0.28➘ |
| AWN76_014900 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit β | 0.64➘ |
| AWN76_017390 pyruvate dehydrogenase E2 component | ~ |
| AWN76_018190 aconitate hydratase | 0.35➘ |
| Pentose phosphate pathway (PPP) | |
| AWN76_002240 6-phosphogluconolactonase | 2.36➚ |
| AWN76_002245 glucose-6-phosphate dehydrogenase | ~ |
| AWN76_002250 6-phosphogluconate dehydrogenase | 2.72➚ |
| AWN76_005185 glucose/mannose-6-phosphate isomerase | ~ |
| AWN76_005350 6-phosphogluconate dehydrogenase | 0.47➘ |
| AWN76_006030 transketolase | ~ |
| AWN76_008645 ribose-phosphate pyrophosphokinase | 0.37➘ |
| AWN76_008995 fructose-6-phosphate aldolase | 0.40➘ |
| AWN76_010760 d-arabino 3-hexulose 6-phosphate aldehyde lyase | 0.62➘ |
| AWN76_012150 ribulose-phosphate 3-epimerase | ~ |
| AWN76_012860 glucose-6-phosphate isomerase | ~ |
| AWN76_016655 ribose-5-phosphate isomerase A | 1.86➚ |
| Other genes involved in carbon metabolism | |
| AWN76_000185 aminomethyltransferase | 0.64➘ |
| AWN76_002260 bifunctional methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase | ~ |
| AWN76_002310 phosphoenolpyruvate carboxykinase (ATP) | 0.34➘ |
| AWN76_002820 methylmalonyl-CoA mutase | ~ |
| AWN76_003760 glycerate 2-kinase | 2.74➚ |
| AWN76_003920 d-3-phosphoglycerate dehydrogenase/2-oxoglutarate reductase | 2.04➚ |
| AWN76_004480 phosphoenolpyruvate carboxylase | 0.62➘ |
| AWN76_004595 dihydrolipoyl dehydrogenase | 0.39➘ |
| AWN76_005090 acetyl-CoA carboxylase carboxyltransferase subunit β | 0.26➘ |
| AWN76_005450 zinc-binding alcohol dehydrogenase family protein | 0.63➘ |
| AWN76_005870 3-hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase family protein | 0.51➘ |
| AWN76_006190 acetate-CoA ligase | 0.37➘ |
| AWN76_006195 acetyl-CoA C-acyltransferase | |
| AWN76_006780 acetyl-coenzyme A synthetase | 1.98➚ |
| AWN76_007665 glycine cleavage system protein GcvH | 0.39➘ |
| AWN76_007670 acetyl-CoA carboxylase, biotin carboxylase | ~ |
| AWN76_007675 acetyl-CoA carboxylase, biotin carboxyl carrier protein | 0.53➘ |
| AWN76_008645 ribose-phosphate pyrophosphokinase | 0.37➘ |
| AWN76_009505 phosphoglycerate dehydrogenase | ~ |
| AWN76_009860 aldehyde dehydrogenase | ~ |
| AWN76_009995 enoyl-CoA hydratase | ~ |
| AWN76_010345 methylenetetrahydrofolate reductase | 0.48➘ |
| AWN76_010475 acetyl-CoA carboxylase carboxyltransferase subunit α | ~ |
| AWN76_010955 bifunctional 4-hydroxy-2-oxoglutarate aldolase/2-dehydro-3-deoxy-phosphogluconate aldolase | 2.34➚ |
| AWN76_011330 acyl-CoA carboxylase subunit β | ~ |
| AWN76_012245 l-lactate dehydrogenase | ~ |
| AWN76_013500 Threonine dehydratase | 2.04➚ |
| AWN76_014285 glycine dehydrogenase (aminomethyl-transferring) | 0.50➘ |
| AWN76_014810 serine hydroxymethyltransferase | 0.57➘ |
| AWN76_016000 aldehyde dehydrogenase family protein | 2.22➚ |
| AWN76_016690 Glutamate dehydrogenase | 0.33➘ |
| AWN76_017295 methylmalonyl-CoA epimerase | ~ |
| AWN76_018450 3-phosphoserine/phosphohydroxythreonine transaminase | 0.42➘ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liew, K.J.; Bruce, N.C.; Sani, R.K.; Chong, C.S.; Yaakop, A.S.; Shamsir, M.S.; Goh, K.M. Global Transcriptomic Responses of Roseithermus sacchariphilus Strain RA in Media Supplemented with Beechwood Xylan. Microorganisms 2020, 8, 976. https://doi.org/10.3390/microorganisms8070976
Liew KJ, Bruce NC, Sani RK, Chong CS, Yaakop AS, Shamsir MS, Goh KM. Global Transcriptomic Responses of Roseithermus sacchariphilus Strain RA in Media Supplemented with Beechwood Xylan. Microorganisms. 2020; 8(7):976. https://doi.org/10.3390/microorganisms8070976
Chicago/Turabian StyleLiew, Kok Jun, Neil C. Bruce, Rajesh Kumar Sani, Chun Shiong Chong, Amira Suriaty Yaakop, Mohd Shahir Shamsir, and Kian Mau Goh. 2020. "Global Transcriptomic Responses of Roseithermus sacchariphilus Strain RA in Media Supplemented with Beechwood Xylan" Microorganisms 8, no. 7: 976. https://doi.org/10.3390/microorganisms8070976








