Gut Microbiota and Short-Chain Fatty Acid Profile between Normal and Moderate Malnutrition Children in Yogyakarta, Indonesia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Population and Design
2.3. Energy and Macronutrient Analysis
2.4. Stool Sample Collection and DNA Extraction
2.5. 16S rRNA Sequencing
2.6. Sequencing Data Processing
2.7. Stool pH and SCFA Analysis
2.8. Statistical Analysis
3. Results
3.1. Participant Characteristics
3.2. The Difference in Dietary Intake between Normal and Undernutrition Children
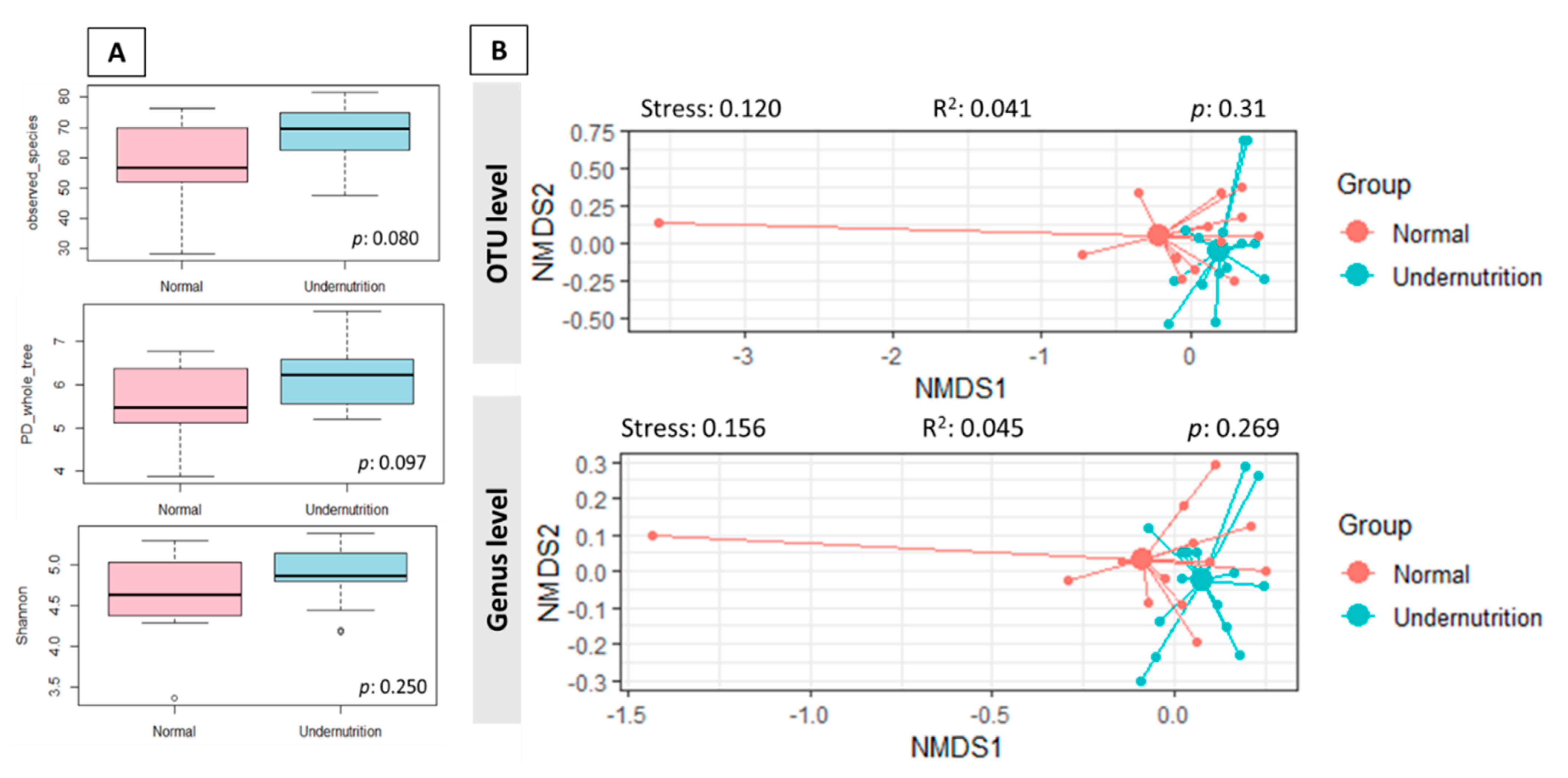
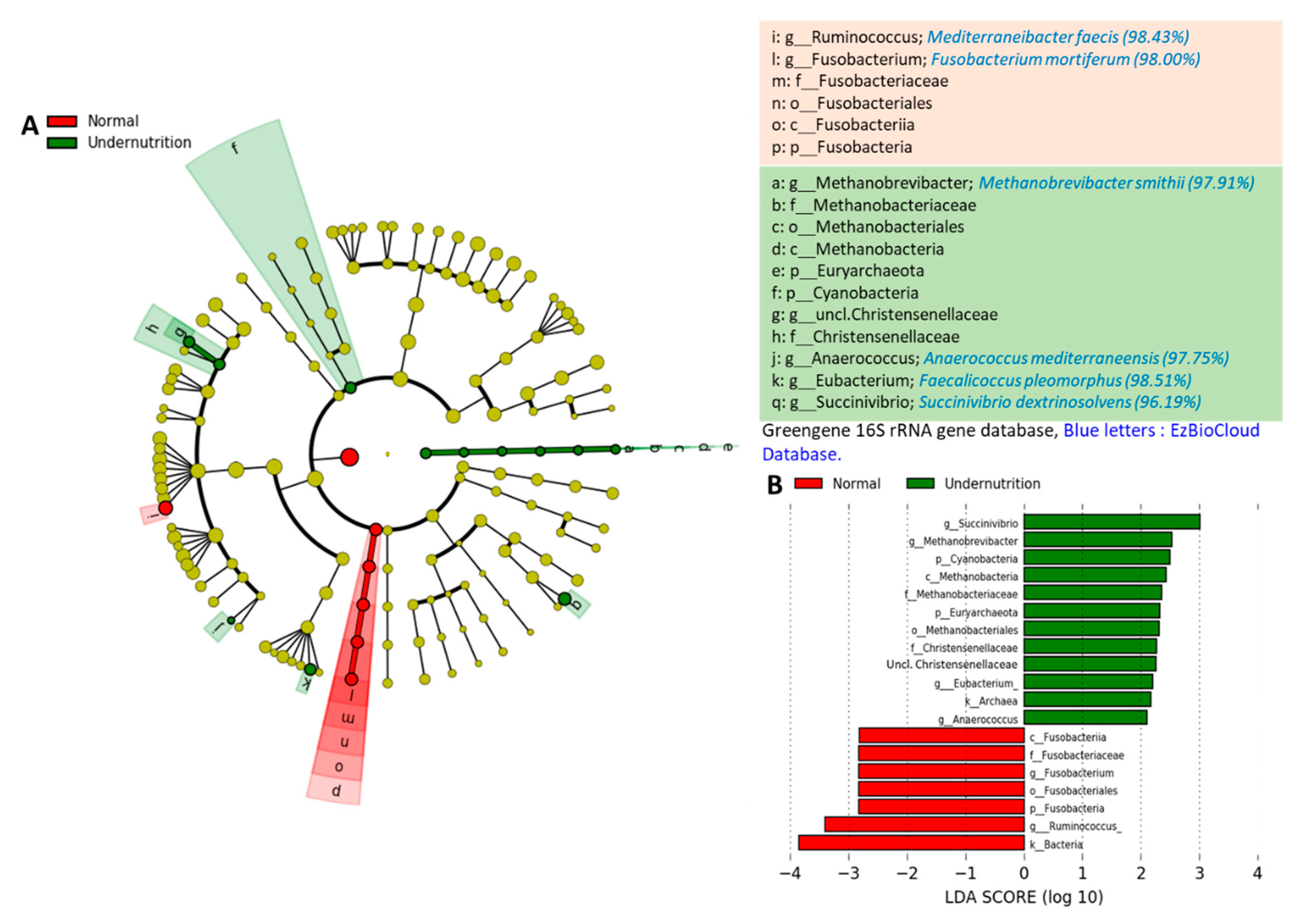
3.3. The Difference in Gut Microbiota Composition between Normal and Undernutrition Children
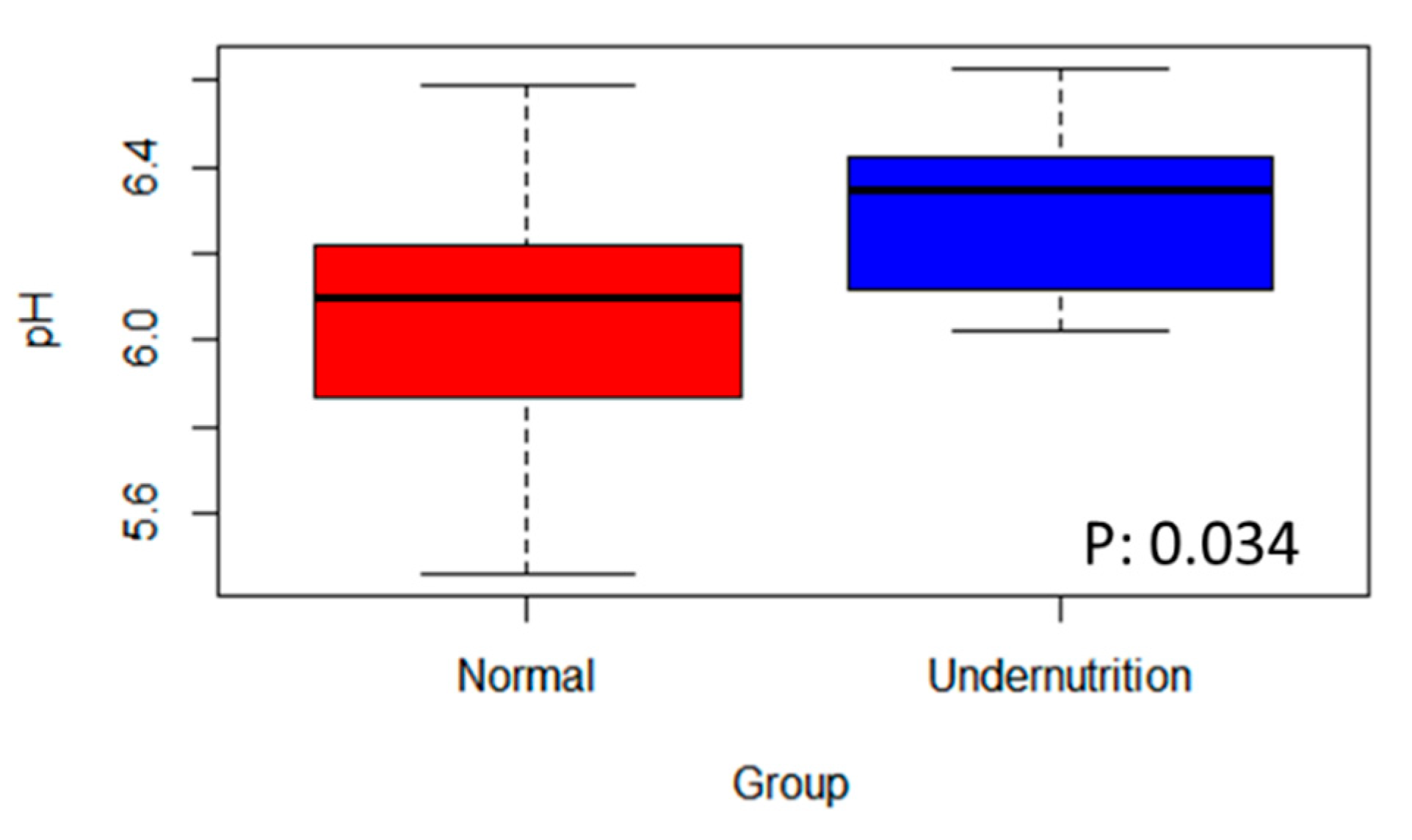
3.4. The Difference in SCFA Profile between Normal and Undernutrition Children
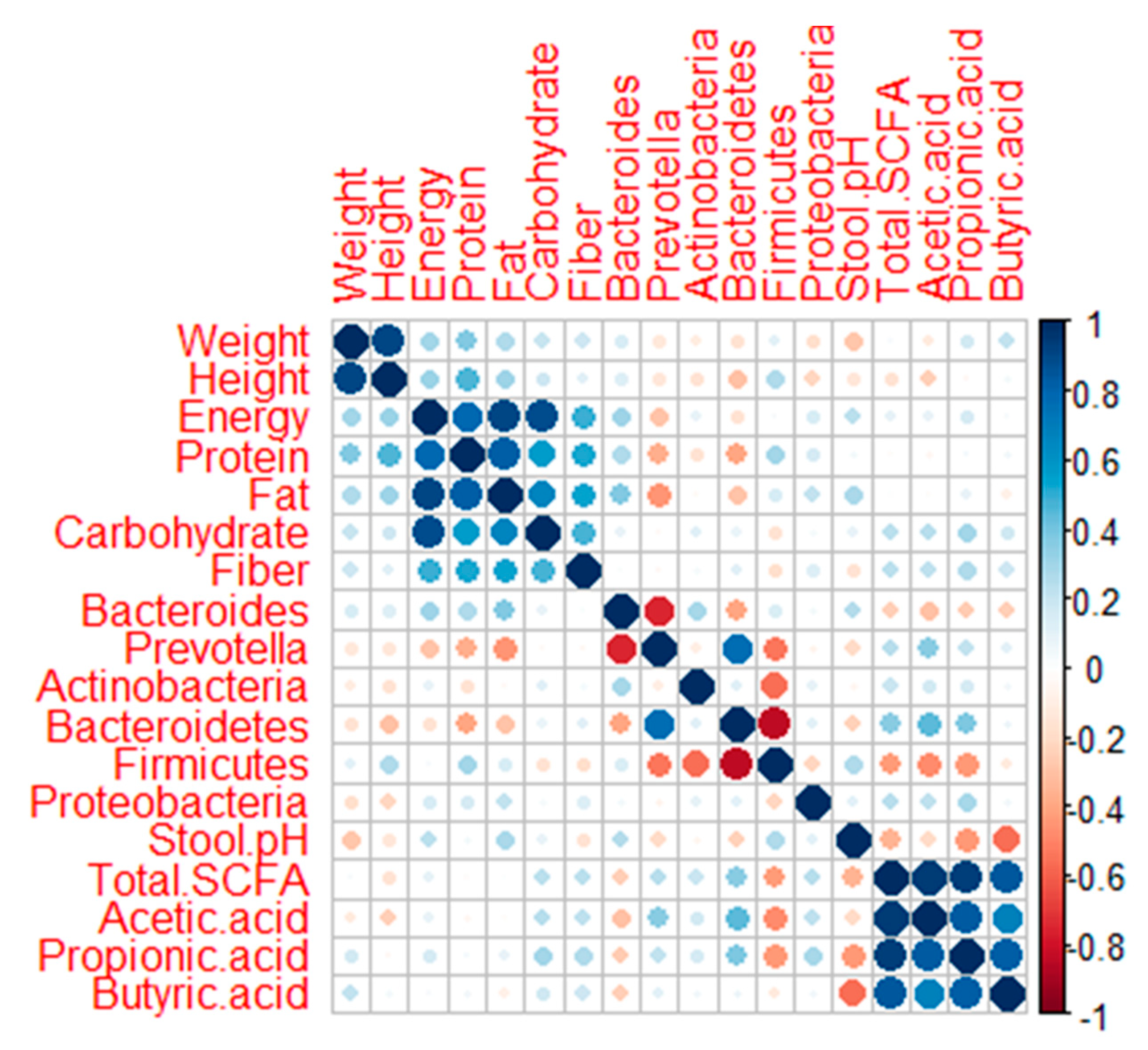
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO/UNICEF. WHO Child Growth Standards and the Identification of Severe Acute Malnutrition in Infants and Children; World Health Organization United Nations Childrens Fund: New York, NY, USA, 2009; ISBN 978-92-4-159816-3. [Google Scholar]
- Chan, M. Ten Years in Public Health; World Health Organization United Nations Childrens Fund: New York, NY, USA, 2017; ISBN 9789241512442. [Google Scholar]
- Kementerian Kesehatan Badan Penelitian dan Kesehatan HASIL UTAMA RISKESDAS 2018. 2018. Available online: http://www.depkes.go.id/resources/download/info-terkini/materi_rakorpop_2018/HasilRiskesdas2018.pdf (accessed on 26 February 2019).
- Dinas Kesehatan DIY. Profil Kesehatan, D.I Yogyakarta tahun 2018. 2019. Available online: http://www.dinkes.jogjaprov.go.id/download/download/27 (accessed on 5 November 2020).
- Blössner, M.; de Onis, M. Malnutrition: Quantifying the health impact at national and local levels. Environ. Burd. Dis. Ser. 2005, 12, 43. [Google Scholar]
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; de Onis, M.; Ezzati, M.; Mcgregor, S.G.; Katz, J.; Martorell, R.; et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. The Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Scepanovic, P.; Hodel, F.; Mondot, S.; Partula, V.; Byrd, A.; Hammer, C.; Alanio, C.; Bergstedt, J.; Patin, E.; Touvier, M.; et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Filippo, C.; Cavalieri, D.; di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Kisuse, J.; Ongkham, O.L.; Nakphaichit, M.; Therdtatha, P.; Momoda, R.; Tanaka, M.; Fukuda, S.; Popluechai, S.; Kespechara, K.; Sonomoto, K.; et al. Urban Diets Linked to Gut Microbiome and Metabolome Alterations in Children: A Comparative Cross-Sectional Study in Thailand. Front. Microbiol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Nakayama, J.; Yamamoto, A.; Conde, L.A.P.; Higashi, K.; Sonomoto, K.; Tan, J.; Lee, Y.K. Impact of westernized diet on gut microbiota in children on Leyte island. Front. Microbiol. 2017, 8, 197. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Alou, M.T.; Lagier, J.C.; Raoult, D. Diet influence on the gut microbiota and dysbiosis related to nutritional disorders. Hum. Microbiome J. 2016, 1, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Gordon, J.I.; Dewey, K.G.; Mills, D.A.; Medzhitov, R.M. The human gut microbiota and undernutrition. Sci. Transl. Med. 2012, 4, 2–7. [Google Scholar] [CrossRef] [Green Version]
- Monira, S.; Nakamura, S.; Gotoh, K.; Izutsu, K.; Watanabe, H.; Alam, N.H.; Endtz, H.P.; Cravioto, A.; Ali, S.I.; Nakaya, T.; et al. Gut microbiota of healthy and malnourished children in Bangladesh. Front. Microbiol. 2011, 2, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; Destefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.I.; Yatsunenko, T.; Manary, M.J.; Trehan, I.; Mkakosya, R.; Cheng, J.; Kau, A.L.; Rich, S.S.; Concannon, P.; Mychaleckyj, J.C.; et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013, 339, 548–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alou, M.T.; Million, M.; Traore, S.I.; Mouelhi, D.; Khelaifia, S.; Bachar, D.; Caputo, A.; Delerce, J.; Brah, S.; Alhousseini, D.; et al. Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front. Microbiol. 2017, 8, 899. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Watanabe, K.; Jiang, J.; Matsuda, K.; Chao, S.H.; Haryono, P.; La-Ongkham, O.; Sarwoko, M.A.; Sujaya, I.N.; Zhao, L.; et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015, 5, 8397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, J. Pyrosequence-Based 16S rRNA Profiling of Gastro-Intestinal Microbiota. Biosci. Microflora 2010, 29, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Hayati, A.W.; Hardiansyah; Jalal, F.; Madanijah, S.; Briawan, D. Determinan Stunting Anak Baduta: Analisa Data Riskesdas 2010. In Pemantapan Ketahanan Pangan dan Perbaikan Gizi Masyarakat Berbasis Kemandirian dan Kearifan Lokal, Proceedings of the Widyakarya Nasional Pangan dan Gizi X; Auditorium LIPI, Indonesia, 20–21 November 2012; LIPI Press: Jakarta, Indonesia, 2012. [Google Scholar]
- Velly, H.; Britton, R.A.; Preidis, G.A. Mechanisms of cross-talk between the diet, the intestinal microbiome, and the undernourished host. Gut Microbes 2017, 8, 98–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Mohammed, M.; Ghosh, T.; Kanungo, S.; Nair, G.; Mande, S.S. Metagenome of the gut of a malnourished child. Gut Pathog. 2011, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, E.O.M.; López, M.G.O.; Silvestre, M.D.L.Á.G.; González, B.P.; Menjivar, M. Altered gut microbiota and compositional changes in Firmicutes and Proteobacteria in mexican undernourished and obese children. Front. Microbiol. 2018, 9, 2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutoyo, D.A.; Atmaka, D.R.; Sidabutar, L.M.G.B. Dietary Factors Affecting Firmicutes and Bacteroidetes Ratio in Solving Obesity Problem: A Literature Review. Media Gizi Indones. 2020, 15, 94. [Google Scholar] [CrossRef]
- Bervoets, L.; Vankerckhoven, V.; Vael, C.; Desager, K.N.; Goossens, H.; van Hoorenbeeck, K.; van Noten, C.; Hens, N.; Kortleven, I. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, R.K.; Ilhan, Z.E.; Kang, D.W.; di Baise, J.K. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 2012, 27, 201–214. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Preidis, G.A.; Ajami, N.J.; Wong, M.C.; Bessard, B.C.; Conner, M.E.; Petrosino, J.F. Composition and function of the undernourished neonatal mouse intestinal microbiome. J. Nutr. Biochem. 2015, 26, 1050–1057. [Google Scholar] [CrossRef]
- Khine, W.W.T.; Rahayu, E.S.; See, T.Y.; Kuah, S.; Salminen, S.; Nakayama, J.; Lee, Y.K. Indonesian children fecal microbiome from birth until weaning was different from microbiomes of their mothers. Gut Microbes 2020, 12, 1761240. [Google Scholar] [CrossRef]
- Rahayu, E.S.; Utami, T.; Mariyatun, M.; Hasan, P.N.; Kamil, R.Z.; Setyawan, R.H.; Pamungkaningtyas, F.H.; Harahap, I.A.; Wiryohanjoyo, D.V.; Pramesi, P.C.; et al. Gut Microbiota Profile in Healthy Indonesians. World J. Gastroenterol. 2019, 9327, 1478–1491. [Google Scholar] [CrossRef]
- Gao, X.; Jia, R.; Xie, L.; Kuang, L.; Feng, L.; Wan, C. Obesity in school-aged children and its correlation with Gut E.coli and Bifidobacteria: A case-control study. BMC Pediatr. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Correlation between body mass index and faecal microbiota from children. Clin. Microbiol. Infect. 2016, 22, 258.e1–258.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Eilam, O.; Zarecki, R.; Oberhardt, M.; Ursell, L.K.; Kupiec, M.; Knight, R.; Gophna, U.; Ruppin, E. Glycan Degradation (GlyDeR) Analysis Predicts Mammalian Gut Microbiota Abundance and Host Diet-Specific Adaptations. mBio 2014, 5, e01526-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Faden, H.S.; Zhu, L. The Response of the Gut Microbiota to Dietary Changes in the First Two Years of Life. Front. Pharmacol. 2020, 11, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pols, T.W.H.; Noriega, L.G.; Nomura, M.; Auwerx, J.; Schoonjans, K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 2011, 54, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E.; et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014, 8, 2218–2230. [Google Scholar] [CrossRef]
- Kvissberg, M.A.; Dalvi, P.S.; Kerac, M.; Voskuijl, W.; Berkley, J.A.; Priebe, M.G.; Bandsma, R.H.J. Carbohydrate malabsorption in acutely malnourished children and infants: A systematic review. Nutr. Rev. 2016, 74, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated Fecal pH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century. mSphere 2018, 3, e00041-18. [Google Scholar] [CrossRef] [Green Version]
- Vonaesch, P.; Morien, E.; Andrianonimiadana, L.; Sanke, H.; Mbecko, J.R.; Huus, K.E.; Naharimanananirina, T.; Gondje, B.P.; Nigatoloum, S.N.; Vondo, S.S.; et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc. Natl. Acad. Sci. USA 2018, 115, E8489–E8498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osuka, A.; Shimizu, K.; Ogura, H.; Tasaki, O.; Hamasaki, T.; Asahara, T.; Nomoto, K.; Morotomi, M.; Kuwagata, Y.; Shimazu, T. Prognostic impact of fecal pH in critically ill patients. Crit. Care 2012, 16, R119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Covián, D.R.; Madiedo, P.R.; Margolles, A.; Gueimonde, M.; Gavilán, C.G.d.l.R.; Salazar, N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; O’Riordan, M.X.D. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv. Appl. Microbiol. 2013, 85, 93–118. [Google Scholar] [CrossRef] [Green Version]
- Monira, S.; Hoq, M.M.; Chowdhury, A.K.A.; Suau, A.; Magne, F.; Endtz, H.P.H.; Alam, M.; Rahman, M.; Pochart, P.; Desjeux, J.F.; et al. Short-chain fatty acids and commensal microbiota in the faeces of severely malnourished children with cholera rehydrated with three different carbohydrates. Eur. J. Clin. Nutr. 2010, 64, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Litvak, Y.; Byndloss, M.X.; Bäumler, A.J. Colonocyte metabolism shapes the gut microbiota Single sentence summary. Science 2018, 362, 6418. [Google Scholar] [CrossRef] [Green Version]
- Cobas, A.E.P.; Artacho, A.; Knecht, H.; Ferrús, M.L.; Friedrichs, A.; Ott, S.J.; Moya, A.; Latorre, A.; Gosalbes, M.J. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS ONE 2013, 8, e80201. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. 2014, 20, 16062–16078. [Google Scholar] [CrossRef]
- Diop, K.; Bretelle, F.; Fournier, P.E.; Fenollar, F. ‘Anaerococcus mediterraneensis’ sp. nov., a new species isolated from human female genital tract. New Microbes New Infect. 2017, 17, 75–76. [Google Scholar] [CrossRef] [PubMed]
- Dione, N.; Bellali, S.; Yasir, M.; Azhar, E.I.; Bibi, F.; Beye, M.; Armstrong, N.; Cadoret, F.; Jiman-Fatani, A.A.; Helmy, N.; et al. Anaerococcus jeddahensis sp. nov., a New Bacterial Species Isolated From Healthy Nomadic Bedouin Woman From Saudi Arabia. Curr. Microbiol. 2018, 75, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- de Maesschalck, C.; van Immerseel, F.; Eeckhaut, V.; de Baere, S.D.; Cnockaert, M.; Croubels, S.; Haesebrouck, F.; Ducatelle, R.; Vandamme, P. Faecalicoccus acidiformans gen. nov., Sp. nov., Isolated from the chicken caecum, And reclassification of streptococcus pleomorphus (barnes et al. 1977), Eubacterium biforme (eggerth 1935) and eubacterium cylindroides (cato et al. 1974) as faecalicoccus p. Int. J. Syst. Evol. Microbiol. 2014, 64, 3877–3884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Southern, P.M. Bacteremia due to Succinivibrio dextrinosolvens: Report of a case. Am. J. Clin. Pathol. 1975, 64, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J.D. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Togo, A.H.; Diop, A.; Bittar, F.; Maraninchi, M.; Valero, R.; Armstrong, N.; Dubourg, G.; Labas, N.; Richez, M.; Delerce, J.; et al. Description of Mediterraneibacter massiliensis, gen. nov., sp. nov., a new genus isolated from the gut microbiota of an obese patient and reclassification of Ruminococcus faecis, Ruminococcus lactaris, Ruminococcus torques, Ruminococcus gnavus and Clostridium glycyrrhizinilyticum as Mediterraneibacter faecis comb. nov., Mediterraneibacter lactaris comb. nov., Mediterraneibacter torques cob. nov., Mediterraneibacter gnavus comb. nov. and Mediterraneibacter glycyrrhizinilyticus comb. nov. Antonie Leeuwenhoek 2018, 111, 2107–2128. [Google Scholar] [CrossRef] [PubMed]
- Abed, J.; Maalouf, N.; Manson, A.L.; Earl, A.M.; Parhi, L.; Emgård, J.E.M.; Klutstein, M.; Tayeb, S.; Almogy, G.; Atlan, K.A.; et al. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front. Cell. Infect. Microbiol. 2020, 10, 400. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, J.; Yao, H.; Hu, H. Fusobacterium and colorectal cancer. Front. Oncol. 2018, 8, 371. [Google Scholar] [CrossRef]
- Portrait, V.; Cottenceau, G.; Pons, A.M. A Fusobacterium mortiferum strain produces a bacteriocin-like substance(s) inhibiting Salmonella enteritidis. Lett. Appl. Microbiol. 2000, 31, 115–117. [Google Scholar] [CrossRef] [Green Version]
- Prout, J.; Glymph, R. Anaerobic septicemia secondary to Fusobacterium mortiferum. J. Natl. Med. Assoc. 1986, 78, 334–337. [Google Scholar]
- Belzer, C.; De Vos, W.M. Microbes insidefrom diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, M.; van Baarlen, P.; Hooiveld, G.; Norin, E.; Müller, M.; de Vos, W.M. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011, 2, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Normal (n:13) | Undernutrition (n:15) | p | |
|---|---|---|---|
| Male | 8 (61.54%) | 7 (46.66%) | |
| Female | 5 (38.46 %) | 8 (53.33%) | |
| Age (Mo) | 41.15 ± 14.34 | 40.73 ± 11.00 | 0.931 |
| Weight (kg) | 14.80 ± 3.98 | 10.70 ± 1.54 | 0.000 |
| Height (cm) | 95.94 ± 8.98 | 87.30 ± 6.49 | 0.007 |
| Unit | Normal | Undernutrition | p | RDA | %RDA | ||
|---|---|---|---|---|---|---|---|
| Normal | Undernutrition | ||||||
| Energy | kcal | 933.99 ± 137.86 | 684.31 ± 225.95 | 0.002 | 1350 | 69.18 | 50.69 |
| Protein | g | 38.80 ± 7.65 | 27.25 ± 7.96 | 0.000 | 20 | 194.00 | 136.27 |
| Fat | g | 37.09 ± 9.85 | 24.57 ± 9.36 | 0.002 | 45 | 82.43 | 54.61 |
| Carbohydrate | g | 111.74 ± 18.00 | 88.16 ± 31.37 | 0.016 | 215 | 51.97 | 41.00 |
| Dietary Fiber | g | 5.72 ± 1.93 | 3.35 ± 1.53 | 0.010 | 19 | 30.12 | 17.61 |
| Organic Acid | Mmol/g Feces (Mean ± SD) | p | |
|---|---|---|---|
| Normal | Undernutrition | ||
| Total organic acid | 33.54 ± 13.86 | 27.01 ± 16.74 | 0.112 |
| Acetic acid | 18.21 ± 8.14 | 16.75 ± 11.95 | 0.279 |
| Propionic acid | 7.43±3.61 | 5.08±3.74 | 0.025 |
| Butyric acid | 5.14±2.46 | 3.35±1.73 | 0.033 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamil, R.Z.; Murdiati, A.; Juffrie, M.; Nakayama, J.; Rahayu, E.S. Gut Microbiota and Short-Chain Fatty Acid Profile between Normal and Moderate Malnutrition Children in Yogyakarta, Indonesia. Microorganisms 2021, 9, 127. https://doi.org/10.3390/microorganisms9010127
Kamil RZ, Murdiati A, Juffrie M, Nakayama J, Rahayu ES. Gut Microbiota and Short-Chain Fatty Acid Profile between Normal and Moderate Malnutrition Children in Yogyakarta, Indonesia. Microorganisms. 2021; 9(1):127. https://doi.org/10.3390/microorganisms9010127
Chicago/Turabian StyleKamil, Rafli Zulfa, Agnes Murdiati, Mohammad Juffrie, Jiro Nakayama, and Endang Sutriswati Rahayu. 2021. "Gut Microbiota and Short-Chain Fatty Acid Profile between Normal and Moderate Malnutrition Children in Yogyakarta, Indonesia" Microorganisms 9, no. 1: 127. https://doi.org/10.3390/microorganisms9010127






