Growth Inhibition by Amino Acids in Saccharomyces cerevisiae
Abstract
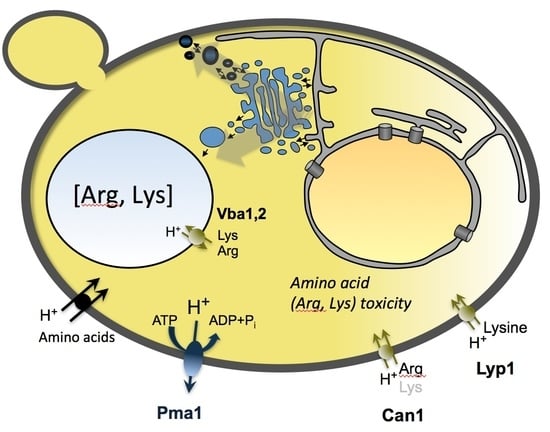
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Plasmids
2.3. Microscopy
2.4. Growth Assays
2.5. Transport Assays
2.6. Measurement of Cytosolic pH
3. Results and Discussion
3.1. All Standard Proteinogenic Amino Acids, as Well as Ornithine and Citrulline, Inhibit Growth of S. Cerevisiae
3.2. Amino Acid Sensitivity Reports on Transporter Activity and Substrate Specificity
3.3. Toxicity Is Caused by Amino Acid Accumulation, Not Proton Transport
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bender, D.A. Amino Acid Metabolism, 3rd ed.; John Wiley & Sons: Chichester, West Sussex, UK, 2012. [Google Scholar]
- Rowley, D. Inhibition of E. coli strains by amino-acids. Nature 1953, 171, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Rowley, D. Interrelationships between amino-acids in the growth of coliform organisms. J. Gen. Microbiol. 1953, 9, 37–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.L.; Vishniac, W. Growth inhibition in Thiobacillus neapolitanus by histidine, methionine, phenylalanine, and threonine. J. Bacteriol. 1970, 104, 1145–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, R.A.; Stenmark-Cox, S.; Ingram, L.O. Mis-regulation of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase does not account for growth inhibition by phenylalanine in Agmenellum quadruplicatum. J. Bacteriol. 1974, 120, 1124–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Englesberg, E.; Bass, R.; Heiser, W. Inhibition of the growth of mammalian cells in culture by amino acids and the isolation and characterization of L-phenylalanine transport. Somatic Cell Genet. 1976, 2, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Sumrada, R.; Cooper, T. Basic amino acid inhibition of growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1976, 68, 598–602. [Google Scholar] [CrossRef]
- Nishiuch, Y.; Sasaki, M.; Nakayasu, M.; Oikawa, A. Cytotoxicity of cysteine in culture media. In Vitro 1976, 12, 635–638. [Google Scholar] [CrossRef]
- Miles, D.O.; Dyer, J.K.; Wong, J.C. Influence of amino acids on the growth of Bacteroides melaninogenicus. J. Bacteriol. 1976, 127, 899–903. [Google Scholar] [CrossRef] [Green Version]
- Kaur, J.; Bachhawat, A.K. Yct1p, a novel, high-affinity, cysteine-specific transporter from the yeast Saccharomyces cerevisiae. Genetics 2007, 176, 877–890. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, D.; Kikushima, R.; Aitoku, M.; Ohtsu, I.; Nasuno, R.; Takagi, H. Exogenous addition of histidine reduces copper availability in the yeast Saccharomyces cerevisiae. Microbial Cell 2014, 1, 241–246. [Google Scholar] [CrossRef]
- Blau, N.; van Spronsen, F.J.; Levy, H.L. Phenylketonuria. Lancet 2010, 376, 1417–1427. [Google Scholar] [CrossRef]
- Leavitt, R.I.; Umbarger, H.E. Isoleucine and valine metabolism in Escherichia coli. XI. Valine inhibition of the growth of Escherichia coli strain K-12. J. Bacteriol. 1962, 83, 624–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatnagar, R.K.; Berry, A.; Hendry, A.T.; Jensen, R.A. The biochemical basis for growth inhibition by L-phenylalanine in Neisseria gonorrhoeae. Mol. Microbiol. 1989, 3, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Li, F.; Tan, K.; Liu, H.; Li, Y.; Liu, Y.; Kong, X.; Tang, Y.; Wu, G.; Yin, Y. Key mediators of intracellular amino acids signaling to mTORC1 activation. Amino Acids 2015, 47, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Powis, K.; De Virgilio, C. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2016, 2, 15049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Zhang, W.; Zhou, Y.; Li, F.; Wei, H.; Peng, J. Recent advances in understanding amino acid sensing mechanisms that regulate mTORC1. Int. J. Mol. Sci. 2016, 17, 1636. [Google Scholar] [CrossRef] [Green Version]
- Laplante, M.; Sabatini, D.M. MTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.-Y.; Sohn, B.H.; Johnson, R.L.; Kang, M.-H.; Kim, S.B.; Shim, J.-J.; Mangala, L.S.; Kim, J.H.; Yoo, J.E.; Rodriguez-Aguayo, C.; et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2016, 63, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Cormerais, Y.; Giuliano, S.; LeFloch, R.; Front, B.; Durivault, J.; Tambutté, E.; Massard, P.-A.; de la Ballina, L.R.; Endou, H.; Wempe, M.F.; et al. Genetic disruption of the multifunctional CD98/LAT1 complex demonstrates the key role of essential amino acid transport in the control of mTORC1 and tumor growth. Cancer Res. 2016, 76, 4481–4492. [Google Scholar] [CrossRef] [Green Version]
- Krall, A.S.; Xu, S.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef] [Green Version]
- Sanayama, Y.; Matsumoto, A.; Shimojo, N.; Kohno, Y.; Nakaya, H. Phenylalanine sensitive K562-D cells for the analysis of the biochemical impact of excess amino acid. Sci. Rep. 2014, 4, 6941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonner, C.A.; Rodrigues, A.M.; Miller, J.A.; Jensen, R.A. Amino acids are general growth inhibitors of Nicotiana silvestris in tissue culture. Physiol. Plant. 1992, 84, 319–328. [Google Scholar] [CrossRef]
- Bonner, C.A.; Williams, D.S.; Aldrich, H.C.; Jensen, R.A. Antagonism by L-glutamine of toxicity and growth inhibition caused by other amino acids in suspension cultures of Nicotiana silvestris. Plant Sci. 1996, 113, 43–58. [Google Scholar] [CrossRef]
- Bonner, C.A.; Jensen, R.A. Recognition of specific patterns of amino acid inhibition of growth in higher plants, uncomplicated by glutamine-reversible ‘general amino acid inhibition’. Plant Sci. 1997, 130, 133–143. [Google Scholar] [CrossRef]
- Durán, R.V.; Oppliger, W.; Robitaille, A.M.; Heiserich, L.; Skendaj, R.; Gottlieb, E.; Hall, M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 2012, 47, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef] [Green Version]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.-X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.-L. Differential regulation of mTORC1 by leucine and glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef] [Green Version]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [Green Version]
- Gournas, C.; Prévost, M.; Krammer, E.-M.; André, B. Function and regulation of fungal amino acid transporters: Insights from predicted structure. In Yeast Membrane Transport; Ramos, J., Sychrová, H., Kschischo, M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 892, pp. 69–106. [Google Scholar]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Ljungdahl, P.O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 2009, 37, 242–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.H.; MacGurn, J.A.; Chu, T.; Stefan, C.J.; Emr, S.D. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 2008, 135, 714–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikko, E.; Pelham, H.R.B. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 2009, 10, 1856–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatakeyama, R.; Kamiya, M.; Takahara, T.; Maeda, T. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 2010, 30, 5598–5607. [Google Scholar] [CrossRef] [Green Version]
- Keener, J.M.; Babst, M. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic 2013, 14, 412–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaddar, K.; Merhi, A.; Saliba, E.; Krammer, E.M.; Prevost, M.; André, B. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol. Cell. Biol. 2014, 34, 4447–4463. [Google Scholar] [CrossRef] [Green Version]
- MacGurn, J.A.; Hsu, P.-C.; Emr, S.D. Ubiquitin and membrane protein turnover: From cradle to grave. Annu. Rev. Biochem. 2012, 81, 231–259. [Google Scholar] [CrossRef]
- Melnykov, A.V. New mechanisms that regulate Saccharomyces cerevisiae short peptide transporter achieve balanced intracellular amino acid concentrations. Yeast 2016, 33, 21–31. [Google Scholar] [CrossRef]
- Risinger, A.L.; Cain, N.E.; Chen, E.J.; Kaiser, C.A. Activity-dependent reversible inactivation of the general amino acid permease. Mol. Biol. Cell 2006, 17, 4411–4419. [Google Scholar] [CrossRef] [Green Version]
- Schreve, J.L.; Sin, J.K.; Garrett, J.M. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J. Bacteriol. 1998, 180, 2556–2559. [Google Scholar] [CrossRef] [Green Version]
- Iraqui, I.; Vissers, S.; Bernard, F.; de Craene, J.O.; Boles, E.; Urrestarazu, A.; André, B. Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 1999, 19, 989–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Düring-Olsen, L.; Regenberg, B.; Gjermansen, C.; Kielland-Brandt, M.C.; Hansen, J. Cysteine uptake by Saccharomyces cerevisiae is accomplished by multiple permeases. Curr. Genet. 1999, 35, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, C.; Neve, E.P.A.; Ljungdahl, P.O. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 2004, 21, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, D.A.; Chianelli, M.S.; Stella, C.A. L-Phenylalanine transport in Saccharomyces cerevisiae: Participation of GAP1, BAP2, and AGP1. J. Amino Acids 2014, 2014, 283962. [Google Scholar] [CrossRef]
- Grauslund, M.; Didion, T.; Kielland-Brandt, M.C.; Andersen, H.A. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1995, 1269, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Schreve, J.; Garrett, J.M. The branched-chain amino acid permease gene of Saccharomyces cerevisiae, BAP2, encodes the high-affinity leucine permease (S1). Yeast 1997, 13, 435–439. [Google Scholar] [CrossRef]
- Usami, Y.; Uemura, S.; Mochizuki, T.; Morita, A.; Shishido, F.; Inokuchi, J.-I.; Abe, F. Functional mapping and implications of substrate specificity of the yeast high-affinity leucine permease Bap2. Biochim. Biophys. Acta 2014, 1838, 1719–1729. [Google Scholar] [CrossRef] [Green Version]
- Grenson, M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. II. Evidence for a specific lysine-transporting system. Biochim. Biophys. Acta 1966, 127, 339–346. [Google Scholar] [CrossRef]
- Ghaddar, K.; Krammer, E.-M.; Mihajlovic, N.; Brohée, S.; André, B.; Prévost, M. Converting the yeast arginine Can1 permease to a lysine permease. J. Biol. Chem. 2014, 289, 7232–7246. [Google Scholar] [CrossRef] [Green Version]
- Regenberg, B.; Holmberg, S.; Olsen, L.D.; Kielland-Brandt, M.C. Dip5p mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate in Saccharomyces cerevisiae. Curr. Genet. 1998, 33, 171–177. [Google Scholar] [CrossRef]
- Zhu, X.; Garrett, J.; Schreve, J.; Michaeli, T. GNP1, the high-affinity glutamine permease of S. cerevisiae. Curr. Genet. 1996, 30, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Sychrová, H.; Chevallier, M.R. Cloning and sequencing of the Saccharomyces cerevisiae gene LYP1 coding for a lysine-specific permease. Yeast 1993, 9, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Sychrová, H.; Matĕjčková, A.; Kotyk, A. Kinetic properties of yeast lysine permeases coded by genes on multi-copy vectors. FEMS Microbiol. Lett. 1993, 113, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P.F.; Brandriss, M.C. Proline transport in Saccharomyces cerevisiae. J. Bacteriol. 1981, 148, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jauniaux, J.C.; Vandenbol, M.; Vissers, S.; Broman, K.; Grenson, M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur. J. Biochem. 1987, 164, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Vandenbol, M.; Jauniaux, J.C.; Grenson, M. Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: Similarities between CAN1, HIP1 and PUT4 permeases. Gene 1989, 83, 153–159. [Google Scholar] [CrossRef]
- Schmidt, A.; Hall, M.N.; Koller, A. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol. Cell. Biol. 1994, 14, 6597–6606. [Google Scholar] [CrossRef] [Green Version]
- Regenberg, B.; Düring-Olsen, L.; Kielland-Brandt, M.C.; Holmberg, S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 1999, 36, 317–328. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Bianchi, F.; Klooster, J.S.; Ruiz, S.J.; Luck, K.; Pols, T.; Urbatsch, I.L.; Poolman, B. Asymmetry in inward- and outward-affinity constant of transport explain unidirectional lysine flux in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 31443. [Google Scholar] [CrossRef]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Nguyen, A.W.; Daugherty, P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Chee, M.K.; Haase, S.B. New and redesigned pRS plasmids shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3: Genes, Genomes, Genetics 2012, 2, 515–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orij, R.; Postmus, J.; Beek Ter, A.; Brul, S.; Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Bitinaite, J.; Nichols, N.M. DNA cloning and engineering by uracil excision. Curr. Protoc. Mol. Biol. 2009, 86, 3–21. [Google Scholar] [CrossRef]
- Nørholm, M.H.H. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 2010, 10, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Warringer, J.; Blomberg, A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 2003, 20, 53–67. [Google Scholar] [CrossRef]
- O’Donnell, A.F.; Apffel, A.; Gardner, R.G.; Cyert, M.S. α-Arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 2010, 21, 3552–3566. [Google Scholar] [CrossRef] [Green Version]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Ro, D.-K.; Ouellet, M.; Paradise, E.M.; Burd, H.; Eng, D.; Paddon, C.J.; Newman, J.D.; Keasling, J.D. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008, 8, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Österberg, M.; Kim, H.; Warringer, J.; Melén, K.; Blomberg, A.; von Heijne, G. Phenotypic effects of membrane protein overexpression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2006, 103, 11148–11153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.A.; Clark, K.M.; Grayhack, E.J.; Dumont, M.E. Characteristics affecting expression and solubilization of yeast membrane proteins. J. Mol. Biol. 2007, 365, 621–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courchesne, W.E.; Magasanik, B. Ammonia regulation of amino acid permeases in Saccharomyces cerevisiae. Mol. Cell. Biol. 1983, 3, 672–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.J.; Kaiser, C.A. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 14837–14842. [Google Scholar] [CrossRef] [Green Version]
- Jauniaux, J.C.; Grenson, M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 1990, 190, 39–44. [Google Scholar] [CrossRef]
- Kanda, N.; Abe, F. Structural and functional implications of the yeast high-affinity tryptophan permease Tat2. Biochemistry 2013, 52, 4296–4307. [Google Scholar] [CrossRef]
- Grenson, M.; Mousset, M.; Wiame, J.M.; Bechet, J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 1966, 127, 325–338. [Google Scholar] [CrossRef]
- Wilson, D.M.; Kusch, M.; Flagg-Newton, J.L.; Wilson, T.H. Control of lactose transport in Escherichia coli. FEBS Lett. 1980, 117, K37–K44. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.M.; Putzrath, R.M.; Wilson, T.H. Inhibition of growth of Escherichia coli by lactose and other galactosides. Biochim. Biophys. Acta 1981, 649, 377–384. [Google Scholar] [CrossRef]
- Ohsumi, Y.; Anraku, Y. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 1981, 256, 2079–2082. [Google Scholar] [PubMed]
- Sato, T.; Ohsumi, Y.; Anraku, Y. Substrate specificities of active transport systems for amino acids in vacuolar-membrane vesicles of Saccharomyces cerevisiae. Evidence of seven independent proton/amino acid antiport systems. J. Biol. Chem. 1984, 259, 11505–11508. [Google Scholar] [PubMed]
- Sherman, F. Getting started with yeast. Meth. Enzymol. 2002, 350, 3–41. [Google Scholar]
- Dechant, R.; Binda, M.; Lee, S.S.; Pelet, S.; Winderickx, J.; Peter, M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 2010, 29, 2515–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarsio, M.; Zheng, H.; Smardon, A.M.; Martínez-Muñoz, G.A.; Kane, P.M. Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 2011, 286, 28089–28096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orij, R.; Urbanus, M.L.; Vizeacoumar, F.J.; Giaever, G.; Boone, C.; Nislow, C.; Brul, S.; Smits, G.J. Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae. Genome Biol. 2012, 13, R80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dechant, R.; Saad, S.; Ibáñez, A.J.; Peter, M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol. Cell 2014, 55, 409–421. [Google Scholar] [CrossRef] [Green Version]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef] [Green Version]
- Pickart, C.M.; Rose, I.A. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 1985, 260, 1573–1581. [Google Scholar]
- Wenzel, D.M.; Lissounov, A.; Brzovic, P.S.; Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 2011, 474, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Stoll, K.E.; Brzovic, P.S.; Davis, T.N.; Klevit, R.E. The essential Ubc4/Ubc5 function in yeast is HECT E3-dependent, and RING E3-dependent pathways require only monoubiquitin transfer by Ubc4. J. Biol. Chem. 2011, 286, 15165–15170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gournas, C.; Gkionis, S.; Carquin, M.; Twyffels, L.; Tyteca, D.; André, B. Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains. Proc. Natl. Acad. Sci. USA 2018, 115, E3145–E3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, F.; Van't Klooster, J.S.; Ruiz, S.J.; Poolman, B. Regulation of amino acid transport in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2019, 83, 720. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Syga, Ł.; Moiset, G.; Spakman, D.; Schavemaker, P.E.; Punter, C.M.; Seinen, A.-B.; van Oijen, A.M.; Robinson, A.; Poolman, B. Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nat. Commun. 2018, 9, 501. [Google Scholar] [CrossRef] [PubMed]





| Name | Transport Substrates | Reference(s) * |
|---|---|---|
| Agp1 | His, Asp, Glu, Ser, Thr, Asn (0.29 mM), Gln (0.79 mM), Cys, Gly, Pro, Ala, Val, Ile (0.6 mM), Leu (0.16 mM), Met, Phe (0.6 mM), Tyr, Trp | [42,43,44,45,46] |
| Bap2 | Cys, Ala, Val, Ile, Leu (37 µM), Met, Phe, Tyr, Trp | [46,47,48,49] |
| Can1 | His, Arg (10–20 µM), Lys (150–250 µM), Orn | [50,51] |
| Dip5 | Glu (48 µM), Asp (56 µM), Ser, Asn, Gln, Gly, Ala | [52] |
| Gnp1 | Ser, Thr, Asn, Gln (0.59 mM), Cys, Pro, Leu, Met | [44,45,53] |
| Lyp1 | Lys (10–25 µM), Met | [50,51,54,55] |
| Put4 | Gly, Pro, Ala | [45,56,57,58] |
| Tat2 | Gly, Ala, Phe, Tyr, Trp, Cys | [44,59] |
| Name | Description | Source |
|---|---|---|
| pRSII425 | LEU2, 2μ (multicopy) | [65] |
| pRSII426 | URA3, 2μ (multicopy) | [65] |
| pFB022 | pRS426 (URA3, 2μ) derivative with Lyp1YPet under ADH1 promoter | [62] |
| pFB023 | pFB022 derivative containing Lyp1(62-590)YPet | [62] |
| pSR053 | pFB023 derivative containing Lyp1(62-590) | This study |
| pSR045 | pFB022 derivative containing Agp1YPet | This study |
| pSR046 | pFB022 derivative containing Bap2YPet | This study |
| pSR047 | pFB022 derivative containing Dip5YPet | This study |
| pSR048 | pFB022 derivative containing Gnp1YPet | This study |
| pSR049 | pFB022 derivative containing Put4YPet | This study |
| pSR050 | pFB022 derivative containing Tat2YPet | This study |
| pSR051 | pFB022 derivative containing Can1YPet | This study |
| pSR057 | pRSII425 derivative containing Lyp1(62-590) under ADH1 promoter | This study |
| pYES2-PACT1-pHluorin | pYES2 (URA3, 2μ) derivative with pHluorin under ACT1 promoter | [66] |
| Name | Sequence (5′ to 3′) | Description 1 |
|---|---|---|
| 5273 | GGAGGGGAAAATTTATATTTTCAAGGTTC | pFB022 (F) |
| 5954 | CATTTTGGGATCCACTAGTTCTAG | pFB022 (R) |
| 5480 | TCTAGAACTAGTGGATCCCAAAATGTCGTCGTCGAAGTCTC | AGP1 (F) |
| 5481 | TCTAGAACTAGTGGATCCCAAAATGCTATCTTCAGAAGATTTTGGATC | BAP2 (F) |
| 5561 | TCTAGAACTAGTGGATCCCAAAATGGGAACAAATTCAAAAGAAG | CAN1 (F) |
| 5482 | TCTAGAACTAGTGGATCCCAAAATGAAGATGCCTCTAAAGAAGATG | DIP5 (F) |
| 5483 | TCTAGAACTAGTGGATCCCAAAATGACGCTTGGTAATAGACGC | GNP1 (F) |
| 5484 | TCTAGAACTAGTGGATCCCAAAATGGTAAATATACTGCCCTTCC | PUT4 (F) |
| 5485 | TCTAGAACTAGTGGATCCCAAAATGACCGAAGACTTTATTTCTTCTG | TAT2 (F) |
| 5486 | ACCTTGAAAATATAAATTTTCCCCTCCACACCAGAAGGCAACGAC | AGP1 (R) |
| 5487 | ACCTTGAAAATATAAATTTTCCCCTCCACACCAGAAATGATAAGCTTTTCTC | BAP2 (R) |
| 5562 | ACCTTGAAAATATAAATTTTCCCCTCCTGCTACAACATTCCAAAATTTG | CAN1 (R) |
| 5488 | ACCTTGAAAATATAAATTTTCCCCTCCGAAGATATTACCCAAAAATTTTTCATAG | DIP5 (R) |
| 5489 | ACCTTGAAAATATAAATTTTCCCCTCCACACCAGAAATCAAGAACTCTTTTC | GNP1 (R) |
| 5490 | ACCTTGAAAATATAAATTTTCCCCTCCCAACAAGGCGTCCAAGAAC | PUT4 (R) |
| 5491 | ACCTTGAAAATATAAATTTTCCCCTCCACACCAGAAATGGAACTGTCTC | TAT2 (R) |
| 4258 | ACCACCACCAUCATCATCATCATTAACTGCAGGAATTC | pFB023-A (F) |
| 3631 | AGCACTACCCUTTAGCTGTTCTATATGCTGCC | pFB023-A (R) |
| 3630 | AGGGTAGTGCUGAAGGAAGCATACGATACCC | pFB023-B (F) |
| 5159 | ATTTTGGGAUCCACTAGTTCTAGAGCGGCCAGCTTGGAGTTGATTG | pFB023-B (R) |
| 5087 | ATCCCAAAAUGCATGGGTCATTGCAAGGTGG | LYP1(62-590) (F) |
| 5089 | ATGGTGGTGGUGTCCCCCTCCTTCGATTTCTCTTCTGTCGGAATC | LYP1(62-590) (R) |
| Amino Acids (3-letter, 1-letter code) | |
| Alanine (Ala, A) | 0.853 |
| Arginine (Arg, R) | 0.361 |
| Asparagine (Asn, N) | 0.575 |
| Aspartic acid (Asp, D) | 0.571 |
| Cysteine (Cys, C) | 0.627 |
| Glutamine (Gln, Q) | 0.520 |
| Glutamate (Glu, E) | 0.517 |
| Glycine (Gly, G) | 1.012 |
| Histidine (His, H) | 0.490 |
| Isoleucine (Ile, I) | 0.579 |
| Leucine (Leu, L) | 2.897 |
| Lysine (Lys, K) | 0.520 |
| Methionine (Met, M) | 0.509 |
| Phenylalanine (Phe, F) | 0.460 |
| Proline (Pro, P) | 0.460 |
| Serine (Ser, S) | 0.723 |
| Threonine (Thr, T) | 0.638 |
| Tryptophan (Trp, W) | 0.372 |
| Tyrosine (Tyr, Y) | 0.419 |
| Valine (Val, V) | 0.649 |
| Other | |
| Adenine | 0.133 |
| myo-Inositol | 0.422 |
| 4-Aminobenzoic acid | 0.058 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz, S.J.; van ’t Klooster, J.S.; Bianchi, F.; Poolman, B. Growth Inhibition by Amino Acids in Saccharomyces cerevisiae. Microorganisms 2021, 9, 7. https://doi.org/10.3390/microorganisms9010007
Ruiz SJ, van ’t Klooster JS, Bianchi F, Poolman B. Growth Inhibition by Amino Acids in Saccharomyces cerevisiae. Microorganisms. 2021; 9(1):7. https://doi.org/10.3390/microorganisms9010007
Chicago/Turabian StyleRuiz, Stephanie J., Joury S. van ’t Klooster, Frans Bianchi, and Bert Poolman. 2021. "Growth Inhibition by Amino Acids in Saccharomyces cerevisiae" Microorganisms 9, no. 1: 7. https://doi.org/10.3390/microorganisms9010007