Effects of L. plantarum HY7715 on the Gut Microbial Community and Riboflavin Production in a Three-Stage Semi-Continuous Simulated Gut System
Abstract
:1. Introduction
2. Materials and Methods
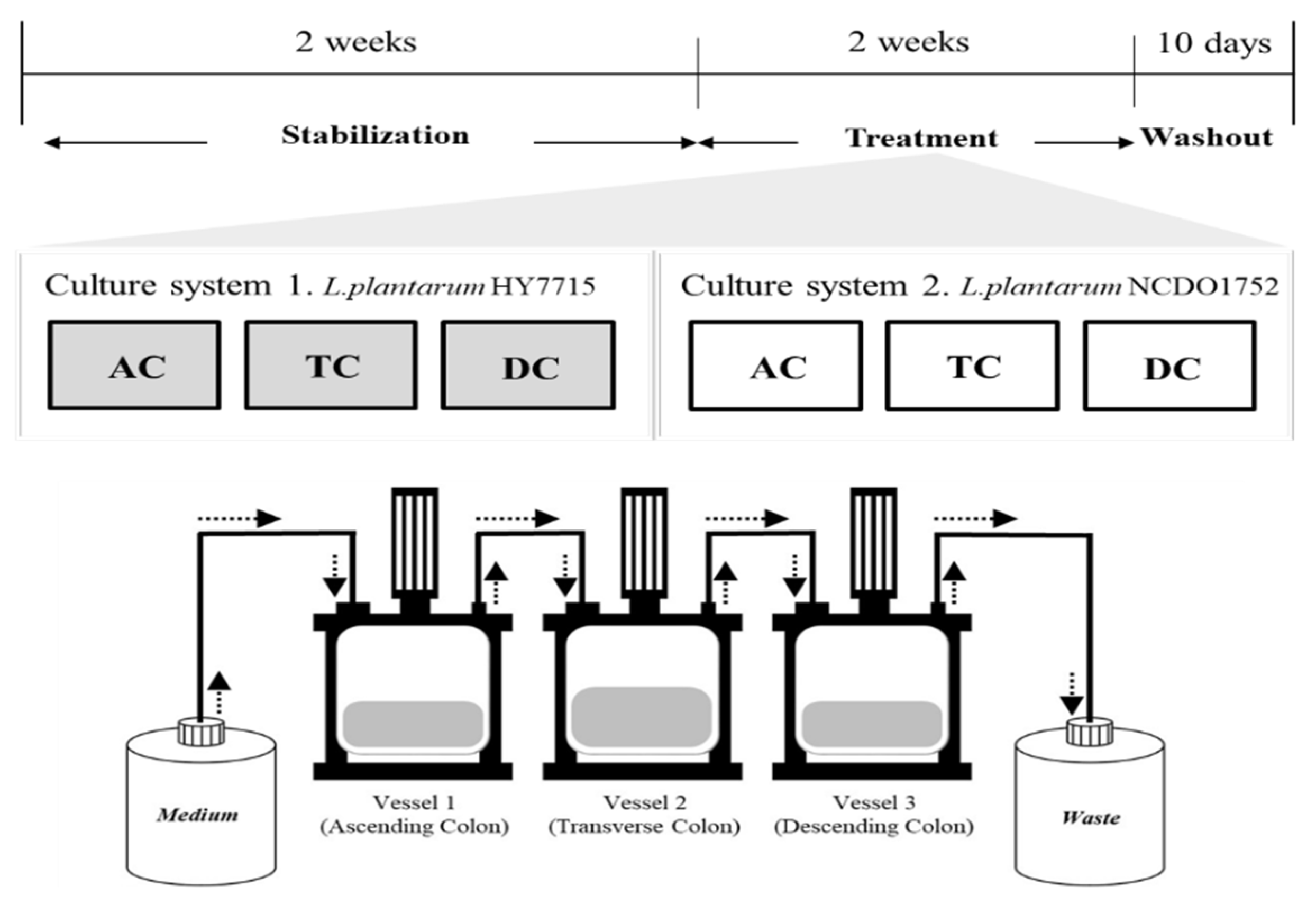
2.1. Gut Culture System Simulation
2.2. Probiotics Treatment
2.3. Riboflavin Analysis
2.4. Short-Chain Fatty Acid Measurements
2.5. Microbial Community Profiling
2.6. Statistical Analysis
3. Results
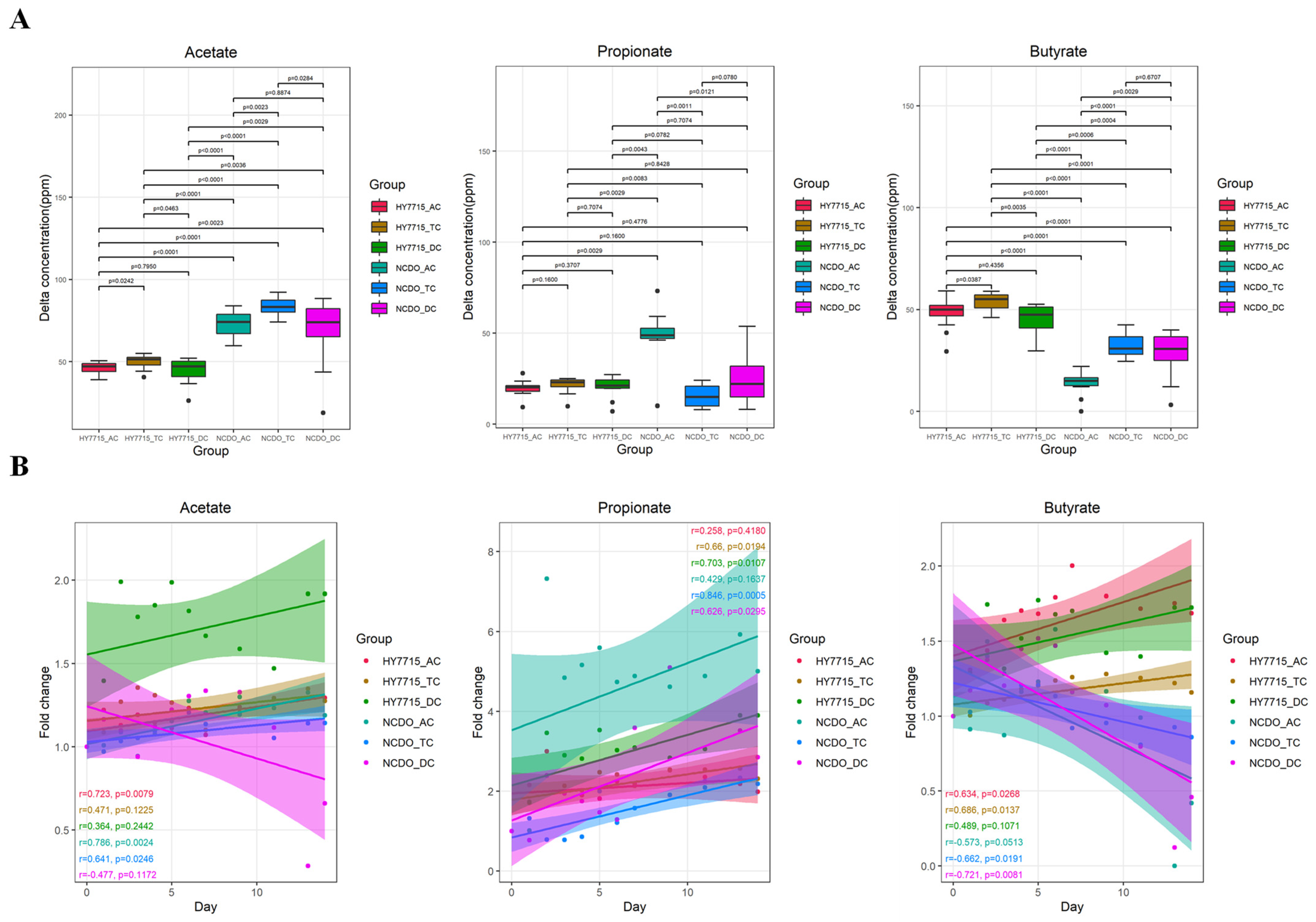
3.1. SCFA Changes in the Culture System
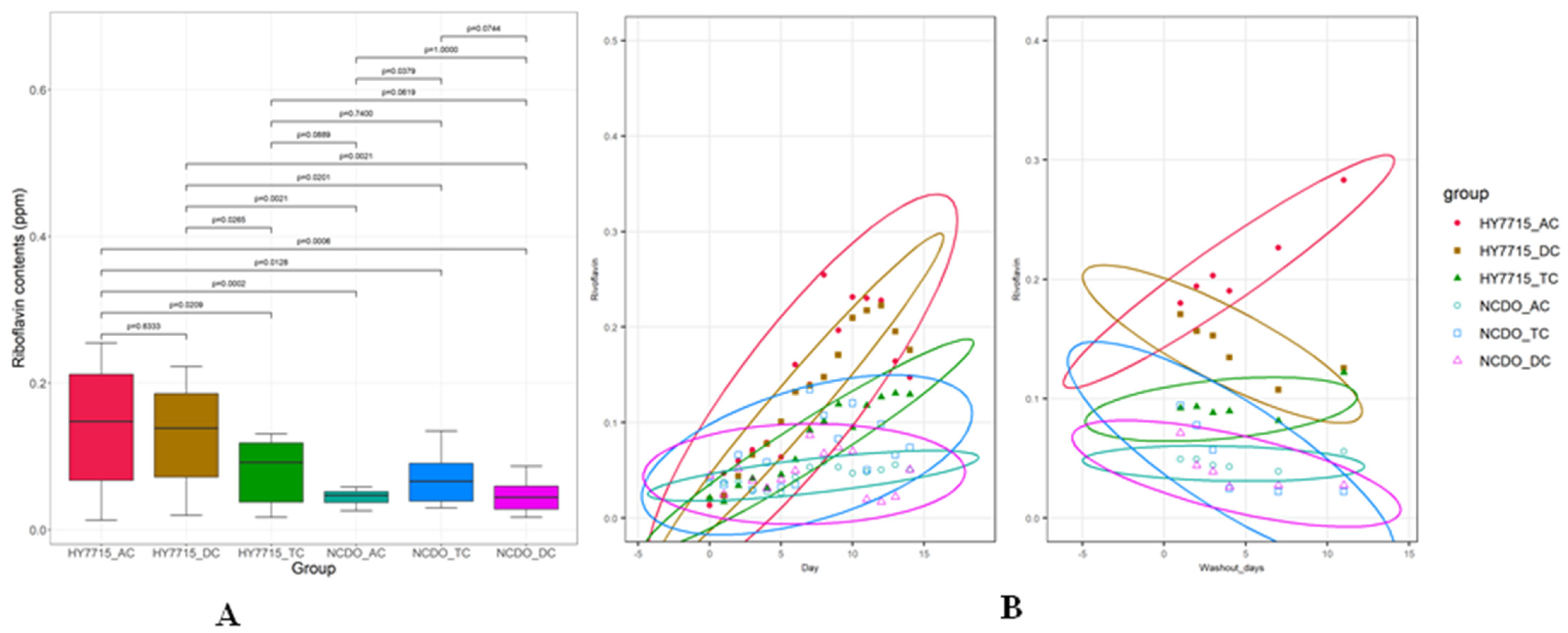
3.2. Riboflavin Production in the Culture System
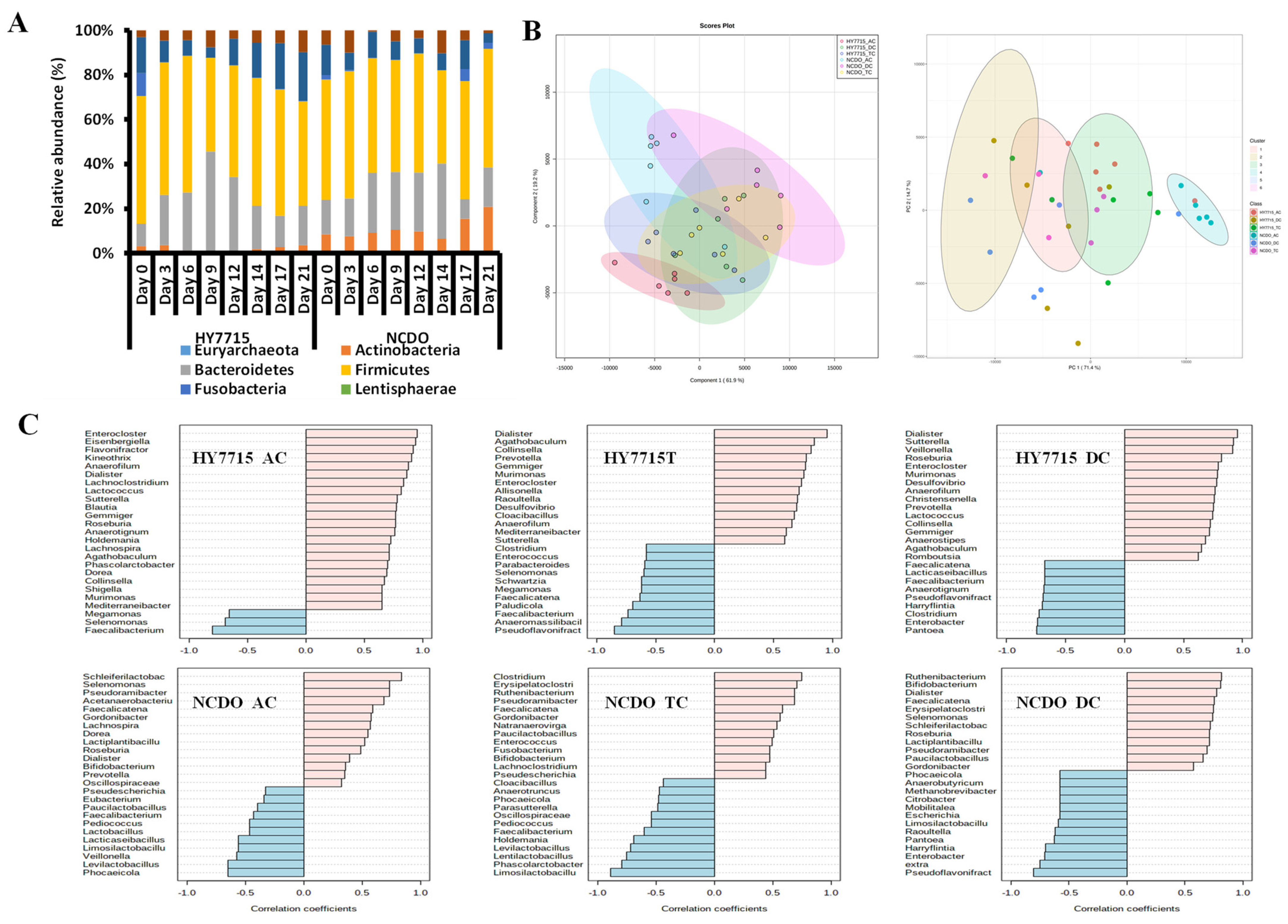
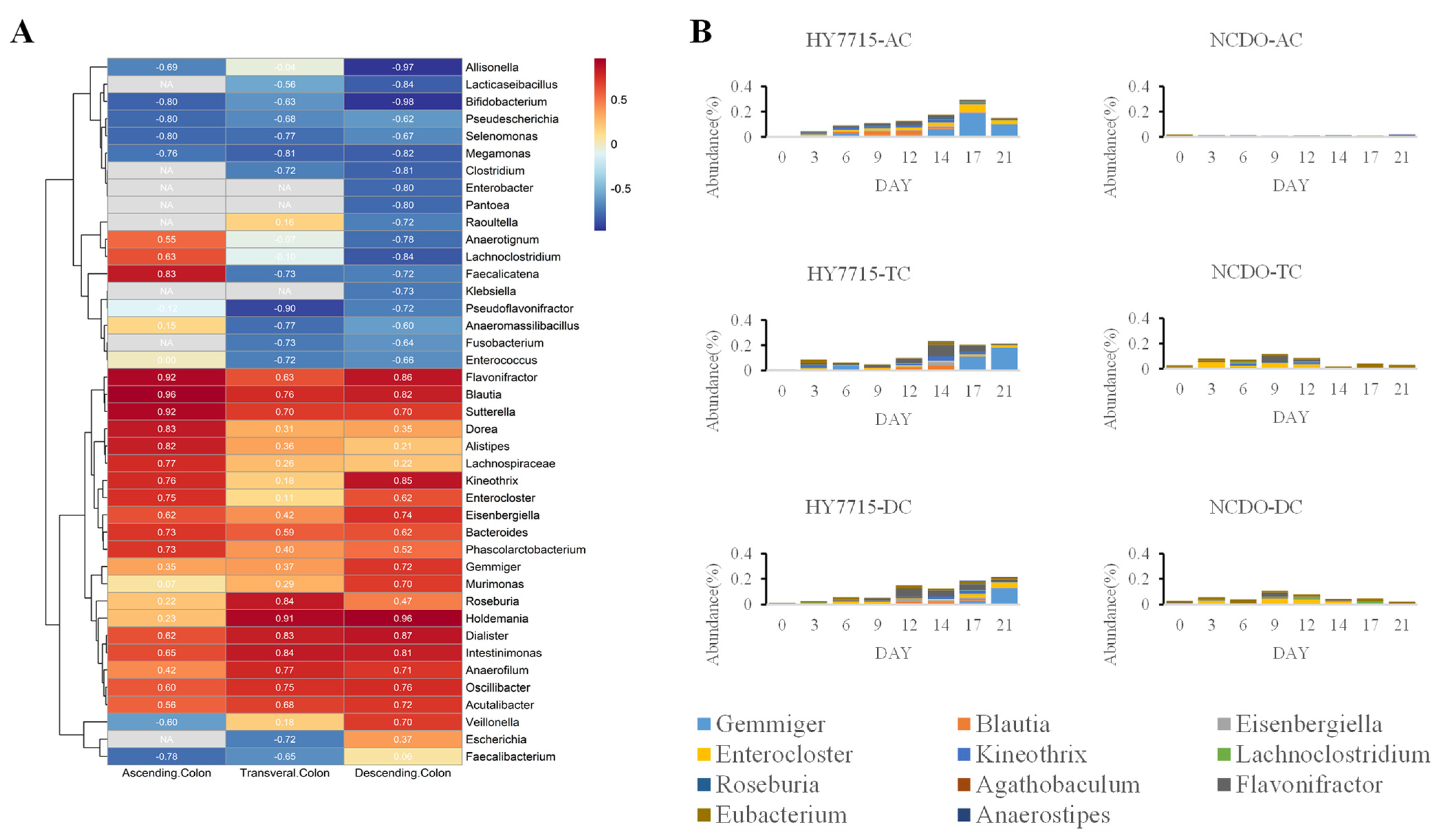
3.3. Effects of L. plantarum HY7715 on Microbial Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joint FAO/WHO Working Group. Report on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002; p. 30. [Google Scholar]
- Abraham, D.; Feher, J.; Scuderi, G.L.; Szabo, D.; Dobolyi, A.; Cservenak, M.; Juhasz, J.; Ligeti, B.; Pongor, S.; Gomez-Cabrera, M.C. Exercise and probiotics attenuate the development of Alzheimer’s disease in transgenic mice: Role of microbiome. Exp. Gerontol. 2019, 115, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Bagga, D.; Reichert, J.L.; Koschutnig, K.; Aigner, C.S.; Holzer, P.; Koskinen, K.; Moissl-Eichinger, C.; Schöpf, V. Probiotics drive gut microbiome triggering emotional brain signatures. Gut Microbes 2018, 9, 486–496. [Google Scholar] [CrossRef]
- Conway, P.L.; Gorbach, S.L.; Goldin, B.R. Survival of Lactic Acid Bacteria in the Human Stomach and Adhesion to Intestinal Cells. J. Dairy Sci. 1987, 70, 1–12. [Google Scholar] [CrossRef]
- Kaur, H.; Nagamoto-Combs, K.; Golovko, S.; Golovko, M.Y.; Klug, M.G.; Combs, C.K. Probiotics ameliorate intestinal pathophysiology in a mouse model of Alzheimer’s disease. Neurobiol. Aging 2020, 92, 114–134. [Google Scholar] [CrossRef] [PubMed]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health benefits of probiotics: A review. Int. Sch. Res. Not. 2013, 2013, 481651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, probiotics and neurodegenerative diseases: Deciphering the gut brain axis. Cell. Mol. Life Sci. 2017, 74, 3769–3787. [Google Scholar] [CrossRef]
- Gupta, R.; Jeevaratnam, K.; Fatima, A. Lactic Acid Bacteria: Probiotic Characteristic, Selection Criteria, and its Role in Human Health (A Review). Int. J. Emerg. Technol. Innov. Res. 2018, 5, 411–424. [Google Scholar]
- Bezkorovainy, A. Probiotics: Determinants of survival and growth in the gut. Am. J. Clin. Nutr. 2001, 73, 399s–405s. [Google Scholar] [CrossRef]
- Both, E.; Gyorgy, E.; Kibedi-Szabo, C.Z.; Tamas, E.; Abraham, B.; Miklossy, I.; Lanyi, S. Acid and bile tolerance, adhesion to epithelial cells of probiotic microorganisms. UPB Bul. Stiintific Ser. B Chem. Mater. Sci. 2010, 72, 37–44. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Bae, J.; Kim, M.J.; Kwon, H.; Park, G.; Kim, S.-J.; Choe, Y.H.; Kim, J.; Park, S.-H.; Choe, B.-H. Delayed establishment of gut microbiota in infants delivered by cesarean section. Front. Microbiol. 2020, 11, 2099. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The effects of vegetarian and vegan diets on gut microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savage, N. The complex relationship between drugs and the microbiome. Nature 2020, 577, S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catania, F.; Baedke, J.; Fábregas-Tejeda, A.; Nieves Delgado, A.; Vitali, V.; Long, L.A.N. Global climate change, diet, and the complex relationship between human host and microbiome: Towards an integrated picture. BioEssays 2021, 43, 2100049. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, R. Humanizing the gut microbiota of mice: Opportunities and challenges. Lab. Anim. 2019, 53, 244–251. [Google Scholar] [CrossRef]
- Hugenholtz, F.; de Vos, W.M. Mouse models for human intestinal microbiota research: A critical evaluation. Cell. Mol. Life Sci. 2018, 75, 149–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venema, K.; Van den Abbeele, P. Experimental models of the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Marsh, P.D. The role of continuous culture in modelling the human microflora. J. Chem. Technol. Biotechnol. 1995, 64, 1–9. [Google Scholar] [CrossRef]
- Eid, H.M.; Wright, M.L.; Anil Kumar, N.; Qawasmeh, A.; Hassan, S.T.; Mocan, A.; Nabavi, S.M.; Rastrelli, L.; Atanasov, A.G.; Haddad, P.S. Significance of microbiota in obesity and metabolic diseases and the modulatory potential by medicinal plant and food ingredients. Front. Pharmacol. 2017, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.N.; Zihler, A.; Chassard, C.; Lacroix, C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012, 30, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Rodriguez, M.; McDonald, J.A.; Hyde, R.; Allen-Vercoe, E.; Claud, E.C.; Sheth, P.M.; Petrof, E.O. Using bioreactors to study the effects of drugs on the human microbiota. Methods 2018, 149, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Joo, W.; Ling, L.; Choi, H.S.; Han, N.S. In vitro digestion and fermentation of sialyllactoses by infant gut microflora. J. Funct. Foods 2016, 21, 497–506. [Google Scholar] [CrossRef]
- Moon, J.S.; Li, L.; Bang, J.; Han, N.S. Application of in vitro gut fermentation models to food components: A review. Food Sci. Biotechnol. 2016, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Lee, E.H.; Yoon, H.S.; Lee, J.H.; Kim, J.Y.; Kang, K.; Park, J.-S.; Jin, J.B.; Ko, G.; Pan, C.-H. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem. 2018, 263, 216–224. [Google Scholar] [CrossRef]
- Mosegaard, S.; Dipace, G.; Bross, P.; Carlsen, J.; Gregersen, N.; Olsen, R.K.J. Riboflavin deficiency-implications for general human health and inborn errors of metabolism. Int. J. Mol. Sci. 2020, 21, 3847. [Google Scholar] [CrossRef] [PubMed]
- Toyosawa, T.; Suzuki, M.; Kodama, K.; Araki, S. Effects of intravenous infusion of highly purified vitamin B2 on lipopolysaccharide-induced shock and bacterial infection in mice. Eur. J. Pharmacol. 2004, 492, 273–280. [Google Scholar] [CrossRef]
- Toyosawa, T.; Suzuki, M.; Kodama, K.; Araki, S. Potentiation by amino acid of the therapeutic effect of highly purified vitamin B2 in mice with lipopolysaccharide-induced shock. Eur. J. Pharmacol. 2004, 493, 177–182. [Google Scholar] [CrossRef]
- Levit, R.; de Giori, G.S.; de LeBlanc, A.d.M.; LeBlanc, J.G. Protective effect of the riboflavin-overproducing strain Lactobacillus plantarum CRL2130 on intestinal mucositis in mice. Nutrition 2018, 54, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.; Tomar, S.K. Invitro study of riboflavin producing lactobacilli as potential probiotic. LWT-Food Sci. Technol. 2016, 68, 570–578. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Choi, E.-J.; Lee, J.-H.; Yoo, M.-S.; Heo, K.; Shim, J.-J.; Lee, J.-L. Probiotic Potential of a Novel Vitamin B2-Overproducing Lactobacillus plantarum Strain, HY7715, Isolated from Kimchi. Appl. Sci. 2021, 11, 5765. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Factories 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, L.; Li, P.; Wang, D.; Wang, T.; Hao, D.; Qu, X. Alterations in intestinal microbiota diversity, composition, and function in patients with sarcopenia. Sci. Rep. 2021, 11, 4628. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.; Park, S.D.; Shim, J.J.; Lee, J.L. Lactobacillus plantarum HY7715 Ameliorates Sarcopenia by Improving Skeletal Muscle Mass and Function in Aged Balb/c Mice. Int. J. Mol. Sci. 2021, 22, 10023. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.S.; Sabui, S.; Heskett, C.W.; Said, H.M. Sodium butyrate enhances intestinal riboflavin uptake via induction of expression of riboflavin transporter-3 (RFVT3). Dig. Dis. Sci. 2019, 64, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Mayangsari, Y.; Suzuki, T. Resveratrol enhances intestinal barrier function by ameliorating barrier disruption in Caco-2 cell monolayers. J. Funct. Foods 2018, 51, 39–46. [Google Scholar] [CrossRef]
- Teixeira, T.F.; Collado, M.C.; Ferreira, C.L.; Bressan, J.; Maria do Carmo, G.P. Potential mechanisms for the emerging link between obesity and increased intestinal permeability. Nutr. Res. 2012, 32, 637–647. [Google Scholar] [CrossRef]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Khalid, N.; Arif, S.; Kobayashi, I.; Nakajima, M. Chapter 14—Lab-on-a-chip techniques for high-throughput proteomics and drug discovery. In Microfluidics for Pharmaceutical Applications; Santos, H.A., Liu, D., Zhang, H., Eds.; William Andrew Publishing: Norwich, NY, USA, 2019; pp. 371–422. [Google Scholar]
- Pham, V.; Mohajeri, M. The application of in vitro human intestinal models on the screening and development of pre-and probiotics. Benef. Microbes 2018, 9, 725–742. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, K.; Spee, B.; Costa, P.; Sachs, N.; Clevers, H.; Malda, J. Converging biofabrication and organoid technologies: The next frontier in hepatic and intestinal tissue engineering? Biofabrication 2017, 9, 013001. [Google Scholar] [CrossRef] [PubMed]
- Chilton, C.; Freeman, J.; Crowther, G.; Todhunter, S.; Wilcox, M. Effectiveness of a short (4 day) course of oritavancin in the treatment of simulated Clostridium difficile infection using a human gut model. J. Antimicrob. Chemother. 2012, 67, 2434–2437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, D.-K.; Yoo, M.-S.; Heo, K.; Shim, J.-J.; Lee, J.-L. Effects of L. plantarum HY7715 on the Gut Microbial Community and Riboflavin Production in a Three-Stage Semi-Continuous Simulated Gut System. Microorganisms 2021, 9, 2478. https://doi.org/10.3390/microorganisms9122478
Hong D-K, Yoo M-S, Heo K, Shim J-J, Lee J-L. Effects of L. plantarum HY7715 on the Gut Microbial Community and Riboflavin Production in a Three-Stage Semi-Continuous Simulated Gut System. Microorganisms. 2021; 9(12):2478. https://doi.org/10.3390/microorganisms9122478
Chicago/Turabian StyleHong, Dong-Ki, Myeong-Seok Yoo, Keon Heo, Jae-Jung Shim, and Jung-Lyoul Lee. 2021. "Effects of L. plantarum HY7715 on the Gut Microbial Community and Riboflavin Production in a Three-Stage Semi-Continuous Simulated Gut System" Microorganisms 9, no. 12: 2478. https://doi.org/10.3390/microorganisms9122478





