Riboswitch theo/metE as a Transcription Regulation Tool for Xanthomonas citri subsp. citri
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Theophylline Stock Solution
2.3. Molecular Biology Procedures
2.4. Compound Susceptibility and Cell Viability Analyses
2.5. Fluorescence Microscopy
2.6. Real-Time Reverse Transcription PCR (qRT-PCR)
3. Results
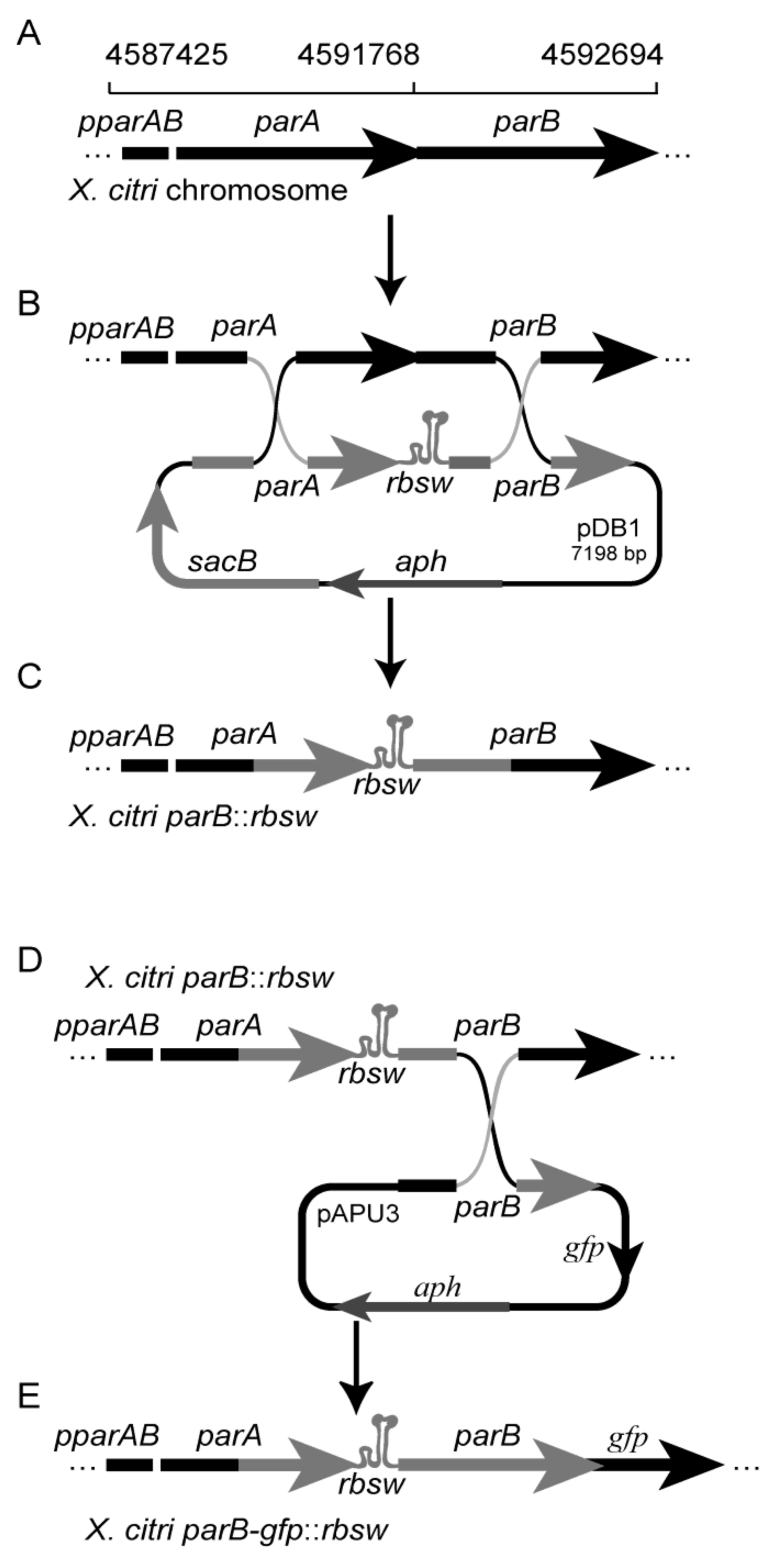
3.1. pNPTS138: A Tool for Allele Exchange in Gram-Negative Bacteria
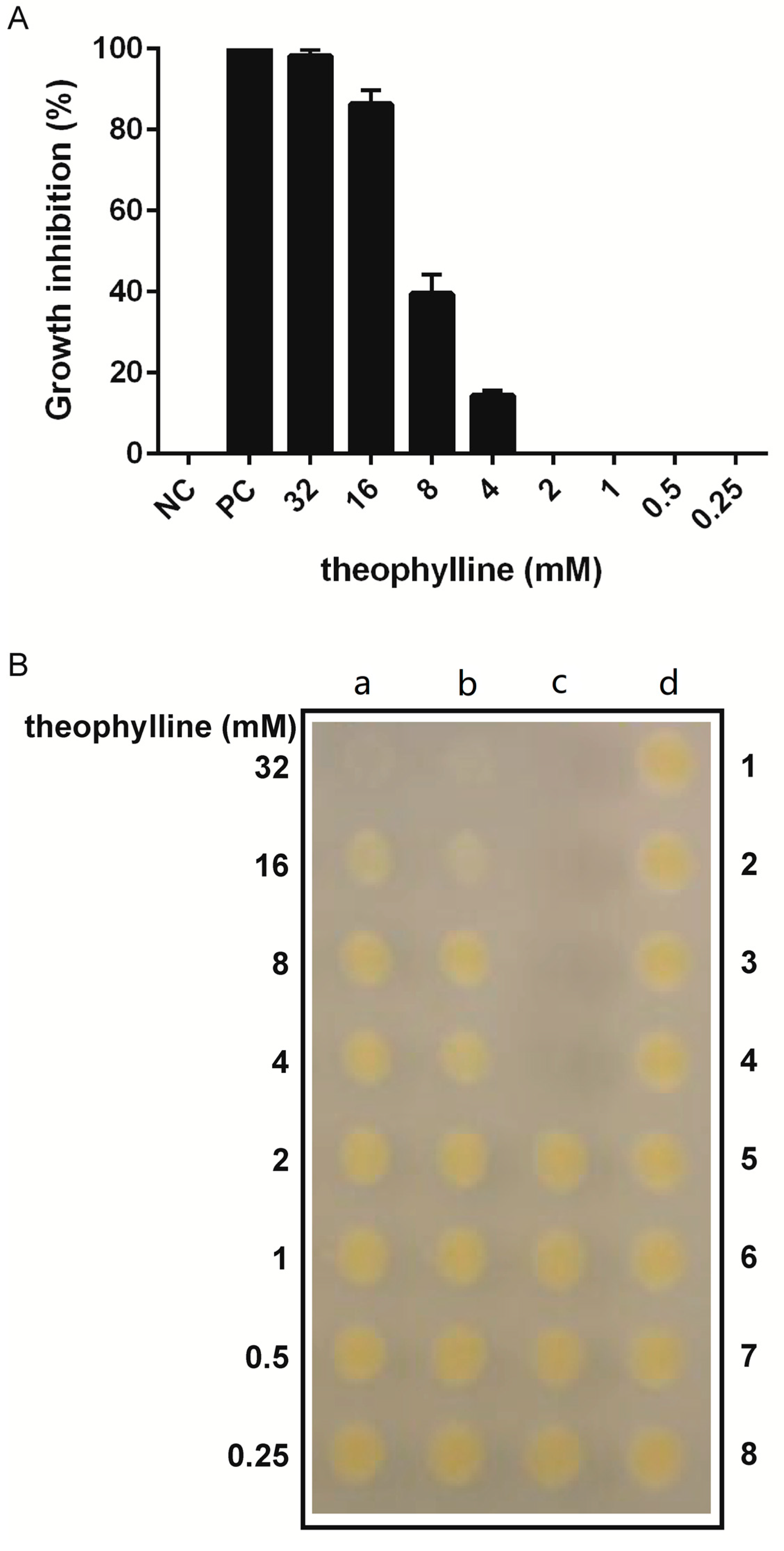
3.2. Effect of Theophylline Concentration on X. citri Growth
3.3. Theophylline Influences the Expression of parB
3.4. Chromosome Segregation Is Not Affected by the Downregulation of parB Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schaad, N.W.; Postnikova, E.; Lacy, G.; Sechler, A.; Agarkova, I.; Stromberg, P.E.; Stromberg, V.K.; Vidaver, A.K. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 2006, 29, 690–695. [Google Scholar] [CrossRef] [Green Version]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [Green Version]
- Gottwald, T.R.; Graham, J.H.; Schubert, T.S. Citrus canker: The pathogen and its impact. Online Plant Health Prog. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Hutchison, C.A., 3rd; Merryman, C.; Sun, L.; Assad-Garcia, N.; Richter, R.A.; Smith, H.O.; Glass, J.I. Polar Effects of Transposon Insertion into a Minimal Bacterial Genome. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutkenhaus, J.F.; Wolf-Watz, H.; Donachie, W.D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J. Bacteriol. 1980, 142, 615–620. [Google Scholar] [CrossRef] [Green Version]
- Lacerda, L.A.; Cavalca, L.B.; Martins, P.M.M.; Govone, J.S.; Bacci, M., Jr.; Ferreira, H. Protein depletion using the arabinose promoter in Xanthomonas citri subsp. citri. Plasmid 2017, 90, 44–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livny, J.; Yamaichi, Y.; Waldor, M.K. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J. Bacteriol. 2007, 189, 8693–8703. [Google Scholar] [CrossRef] [Green Version]
- Fogel, M.A.; Waldor, M.K. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006, 20, 3269–3282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ptacin, J.L.; Lee, S.F.; Garner, E.C.; Toro, E.; Eckart, M.; Comolli, L.R.; Moerner, W.E.; Shapiro, L. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell. Biol. 2010, 12, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, A.; Bikard, D. CRISPR Tools To Control Gene Expression in Bacteria. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.C.; Halper, S.M.; Vezeau, G.E.; Cetnar, D.P.; Hossain, A.; Clauer, P.R.; Salis, H.M. Simultaneous repression of multiple bacterial genes using nonrepetitive extra-long sgRNA arrays. Nat. Biotechnol. 2019, 37, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.M.M.; da Silva Xavier, A.; Takita, M.A.; Alfemas-Zerbini, P.; de Souza, A.A. CRISPR-Cas systems in the plant pathogen Xanthomonas spp. and their impact on genome plasticity. BioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Jeong, K.; Munoz-Bodnar, A.; Arias Rojas, N.; Poulin, L.; Rodriguez, R.L.; Gagnevin, L.; Verniere, C.; Pruvost, O.; Koebnik, R. CRISPR elements provide a new framework for the genealogy of the citrus canker pathogen Xanthomonas citri pv. citri. BMC Genom. 2019, 20, 917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043. [Google Scholar] [CrossRef] [Green Version]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef]
- Ceres, P.; Garst, A.D.; Marcano-Velazquez, J.G.; Batey, R.T. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth. Biol. 2013, 2, 463–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherlock, M.E.; Sudarsan, N.; Stav, S.; Breaker, R.R. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. eLife 2018, 7. [Google Scholar] [CrossRef]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [Green Version]
- Collier, J.; Shapiro, L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 2009, 191, 5706–5716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrokhi, A.; Liu, H.; Szatmari, G. Characterization of the Chromosome Dimer Resolution Site in Caulobacter crescentus. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Krysciak, D.; Petersen, K.; Utpatel, C.; Knapp, A.; Schmeisser, C.; Daniel, R.; Voget, S.; Jaeger, K.E.; Streit, W.R. Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated Burkholderia glumae PG1 strain. Appl. Environ. Microbiol. 2015, 81, 7993–8007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schredl, A.T.; Perez Mora, Y.G.; Herrera, A.; Cuajungco, M.P.; Murray, S.R. The Caulobacter crescentus ctrA P1 promoter is essential for the coordination of cell cycle events that prevent the overinitiation of DNA replication. Microbiology 2012, 158, 2492–2503. [Google Scholar] [CrossRef] [Green Version]
- Skerker, J.M.; Prasol, M.S.; Perchuk, B.S.; Biondi, E.G.; Laub, M.T. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: A system-level analysis. PLoS Biol. 2005, 3, e334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- da Silva, A.C.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Ucci, A.P.; Martins, P.M.; Lau, I.F.; Bacci, M., Jr.; Belasque, J., Jr.; Ferreira, H. Asymmetric chromosome segregation in Xanthomonas citri ssp. citri. MicrobiologyOpen 2014, 3, 29–41. [Google Scholar] [CrossRef]
- Ferreira, H.; Barrientos, F.J.A.; Baldini, R.L.; Rosato, Y.B. Electrotransformation in three pathovars of Xanthomonas campestris. Appl. Microbiol. Biotechnol. 1995, 43, 651–655. [Google Scholar] [CrossRef]
- Silva, I.C.; Regasini, L.O.; Petronio, M.S.; Silva, D.H.; Bolzani, V.S.; Belasque, J.; Sacramento, L.V.; Ferreira, H. Antibacterial Activity of Alkyl Gallates against Xanthomonas citri subsp. citri. J. Bacteriol. 2013, 195, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Martins, P.M.; Lau, I.F.; Bacci, M.; Belasque, J.; do Amaral, A.M.; Taboga, S.R.; Ferreira, H. Subcellular localization of proteins labeled with GFP in Xanthomonas citri ssp. citri: Targeting the division septum. FEMS Microbiol. Lett. 2010, 310, 76–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, T.R.; Laia, M.L.; Ferro, J.A.; Ferro, M.I. Selection and validation of reference genes for gene expression studies by reverse transcription quantitative PCR in Xanthomonas citri subsp. citri during infection of Citrus sinensis. Biotechnol. Lett. 2011, 33, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzoni, A.S.G.; Dantas, G.C.; Bergsma, T.; Ferreira, H.; Scheffers, D.J. Xanthomonas citri MinC Oscillates from Pole to Pole to Ensure Proper Cell Division and Shape. Front. Microbiol. 2017, 8, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recorbet, G.; Robert, C.; Givaudan, A.; Kudla, B.; Normand, P.; Faurie, G. Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Appl. Environ. Microbiol. 1993, 59, 1361–1366. [Google Scholar] [CrossRef] [Green Version]
- Suess, B.; Fink, B.; Berens, C.; Stentz, R.; Hillen, W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004, 32, 1610–1614. [Google Scholar] [CrossRef] [Green Version]
- Kamiura, R.; Toya, Y.; Matsuda, F.; Shimizu, H. Theophylline-inducible riboswitch accurately regulates protein expression at low level in Escherichia coli. Biotechnol. Lett. 2019, 41, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Glaser, P.; Sharpe, M.E.; Raether, B.; Perego, M.; Ohlsen, K.; Errington, J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes. Dev. 1997, 11, 1160–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, H.; Ferreira, H.; Errington, J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 2006, 61, 1352–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wachsmuth, M.; Findeiss, S.; Weissheimer, N.; Stadler, P.F.; Morl, M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 2013, 41, 2541–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strains | Characteristics | References |
|---|---|---|
| X. citri 306 | Xanthomonas citri subsp. citri strain 306 (wild type strain), ApR | IBSBF 1594 [29] |
| E. coli DH10B | Cloning strain | Invitrogen, Thermo Fisher Scientific, Waltham, USA |
| X. citri parB::rbsw | parA-rbsw-parB; ApR | This work |
| X. citri parB-gfp::rbsw | parA-rbsw-parB-gfpmut1; ApR and KmR | This work |
| Plasmids | ||
| pNPTS138 | ColE1-like ori, sacB, aph | This work (GenBank MK533795) |
| pDB1 | pNPTS138 carrying the riboswitch theo/metE located between parA (4590983.4591768) * and parB (4591768.4592311) * = parA-rbsw-parB | This work |
| pAPU3 | xylR, pxyl, parB-gfpmut1, bla, aph | [30] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bueno, D.; Pedrolli, D.B.; Martins, P.M.M.; Bocchini, D.A.; Moraes, K.C.M.; Facincani, A.P.; Ferro, J.A.; Varani, A.M.; Pena, M.M.; Ferreira, H. Riboswitch theo/metE as a Transcription Regulation Tool for Xanthomonas citri subsp. citri. Microorganisms 2021, 9, 329. https://doi.org/10.3390/microorganisms9020329
Bueno D, Pedrolli DB, Martins PMM, Bocchini DA, Moraes KCM, Facincani AP, Ferro JA, Varani AM, Pena MM, Ferreira H. Riboswitch theo/metE as a Transcription Regulation Tool for Xanthomonas citri subsp. citri. Microorganisms. 2021; 9(2):329. https://doi.org/10.3390/microorganisms9020329
Chicago/Turabian StyleBueno, Danilo, Danielle B. Pedrolli, Paula M. M. Martins, Daniela A. Bocchini, Karen C. M. Moraes, Agda P. Facincani, Jesus A. Ferro, Alessandro M. Varani, Michelle M. Pena, and Henrique Ferreira. 2021. "Riboswitch theo/metE as a Transcription Regulation Tool for Xanthomonas citri subsp. citri" Microorganisms 9, no. 2: 329. https://doi.org/10.3390/microorganisms9020329








