Biotransformation of Flavonoids by Newly Isolated and Characterized Lactobacillus pentosus NGI01 Strain from Kimchi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation of LAB Strains from Kimchi
2.3. Glycosyl Hydrolase Activity Assay
2.4. Identification of LAB Strains Isolated from Kimchi
2.5. Biotransformation of Hesperidin and Rutin by LAB Strains
2.6. Production of Hesperetin and Quercetin by L. pentosus NGI01 Strain
2.7. Statistical Analysis
3. Results
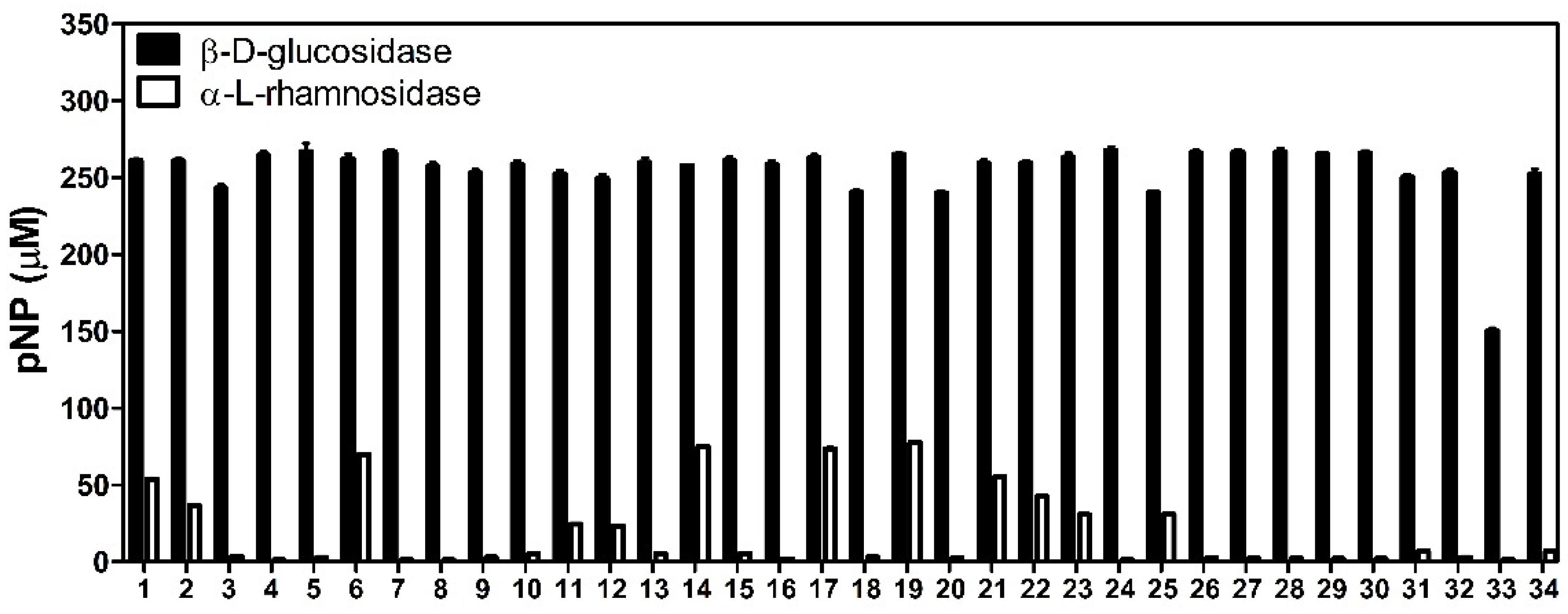
3.1. Glycosyl Hydrolase Activity of LAB Strains from Kimchi
3.2. Identification of LAB Strains from Kimchi
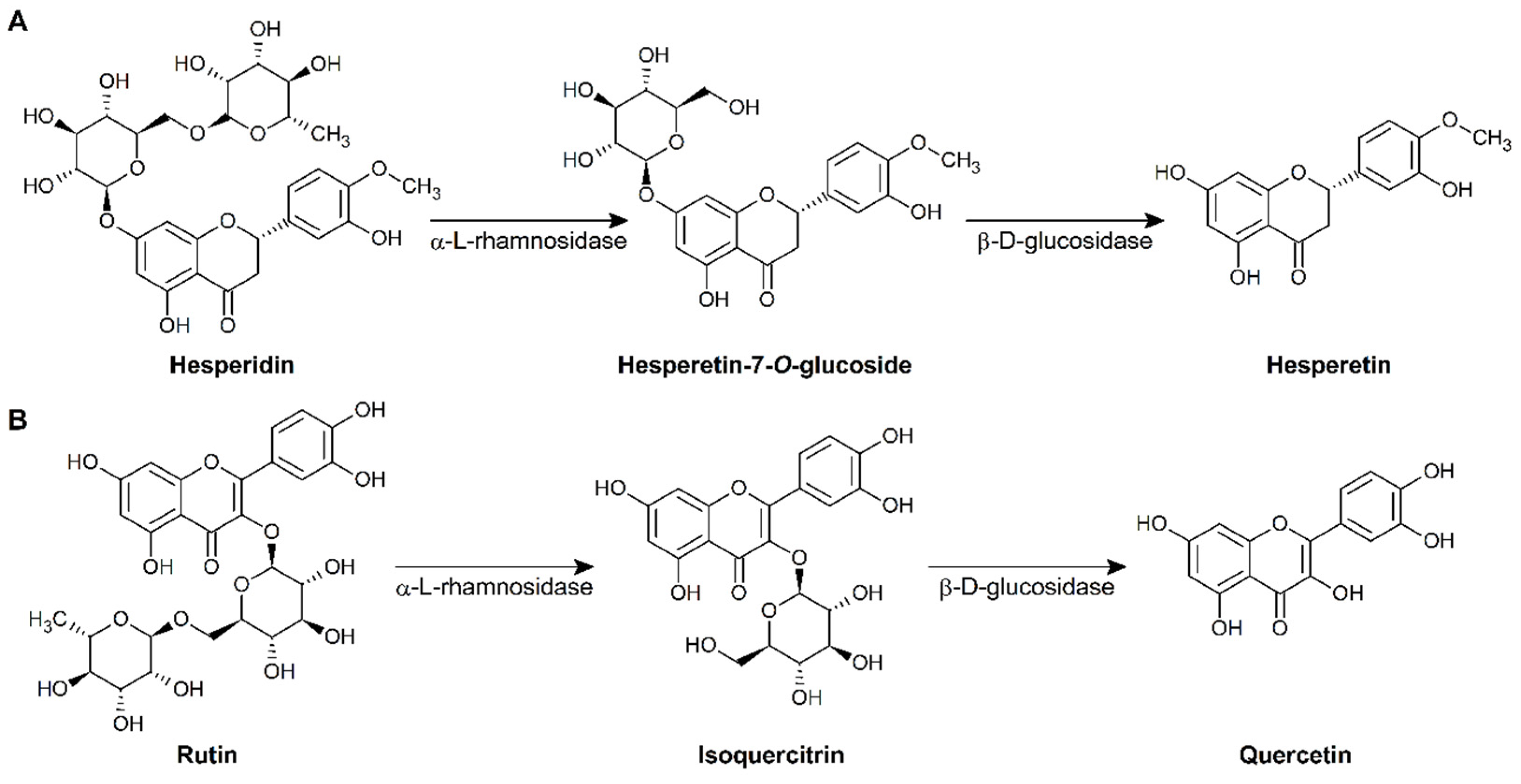
3.3. Biotransformation of Hesperidin and Rutin by the L. pentosus NGI01 Strain
3.4. Production of Hesperetin and Quercetin by L. pentosus NGI01 Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pollard, D.J.; Woodley, J.M. Biocatalysis for pharmaceutical intermediates: The future is now. Trends Biotechnol. 2007, 25, 66–73. [Google Scholar] [CrossRef]
- Porter, J.L.; Rusli, R.A.; Ollis, D.L. Directed Evolution of Enzymes for Industrial Biocatalysis. ChemBioChem 2016, 17, 197–203. [Google Scholar] [CrossRef] [PubMed]
- El Gharras, H. Polyphenols: Food sources, properties, and applications—A review. Int. J. Food Sci. Technol. 2009, 44, 2512–2518. [Google Scholar] [CrossRef]
- Gupta, C.; Prakash, D. Phytonutrients as therapeutic agents. J. Complement. Integr. Med. 2014, 11, 151–169. [Google Scholar] [CrossRef]
- Tripoli, E.; Guardia, M.L.; Giammanco, S.; Majo, D.D.; Giammanco, M. Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 2007, 104, 466–479. [Google Scholar] [CrossRef]
- Day, A.J.; DuPont, M.S.; Ridley, S.; Rhodes, M.; Rhodes, M.J.C.; Morgan, M.R.A.; Williamson, G. Deglycosylation of flavonoid and iso-flavonoid glycosides by human small intestine and liver β-glucosidase activity. FEBS Lett. 1998, 436, 71–75. [Google Scholar] [CrossRef] [Green Version]
- Hollman, P.C.H. Absorption, Bioavailability, and Metabolism of Flavonoids. Pharm. Biol. 2004, 42, 74–83. [Google Scholar] [CrossRef]
- Thilakarathna, S.H.; Rupasinghe, H.P.V. Flavonoid Bioavailability and Attempts for Bioavailability Enhancement. Nutrients 2013, 5, 3367–3387. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, G.B.; Smagghe, G.; Grootaert, C.; Zotti, M.; Raes, K.; Van Camp, J. Flavonoid interactions during digestion, absorption, distribution, and metabolism: A sequential structure-activity/property relationship-based approach in the study of bioa-vailability and bioactivity. Drug Metab. Rev. 2015, 47, 175–190. [Google Scholar] [CrossRef]
- Hollman, P.C.H.; Bijsman, M.N.C.P.; van Gameren, Y.; Cnossen, E.P.J.; de Vries, J.H.M.; Katan, M.B. The sugar moiety is a major deter-minant of the absorption of dietary flavonoid glycosides in man. Free Radic. Res. 1999, 31, 569–573. [Google Scholar] [CrossRef]
- Nielsen, I.L.; Chee, W.S.; Poulsen, L.; Offord-Cavin, E.; Rasmussen, S.E.; Frederiksen, H.; Enslen, M.; Barron, D.; Horcajada, M.N.; Wil-liamson, G. Bioavailability is improved by enzymatic modification of the citrus flavonoid hesperidin in humans: A ran-domized, double-blind, crossover trial. J. Nutr. 2006, 136, 404–408. [Google Scholar] [CrossRef] [Green Version]
- Amaretti, A.; Raimondi, S.; Leonardi, A.; Quartieri, A.; Rossi, M. Hydrolysis of the rutinose-conjugates flavonoids rutin and hes-peridin by the gut microbiota and bifidobacteria. Nutrients 2015, 7, 2788–2800. [Google Scholar] [CrossRef] [Green Version]
- Mueller, M.; Zartl, B.; Schleritzko, A.; Stenzl, M.; Viernstein, H.; Unger, F.M. Rhamnosidase activity of selected probiotics and their ability to hydrolyse flavonoid rhamnoglucosides. Bioprocess Biosyst. Eng. 2017, 41, 221–228. [Google Scholar] [CrossRef] [Green Version]
- Beekwilder, J.; Marcozzi, D.; Vecchi, S.; De Vos, R.; Janssen, P.; Francke, C.; Vlieg, J.V.H.; Hall, R.D. Characterization of Rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl. Environ. Microbiol. 2009, 75, 3447–3454. [Google Scholar] [CrossRef] [Green Version]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Yadav, P.K.; Yadav, S.; Yadav, K.D.S. α-L-Rhamnosidase: A review. Process Biochem. 2010, 45, 1226–1235. [Google Scholar] [CrossRef]
- Busto, M.; Meza, V.; Ortega, N.; Perez-Mateos, M. Immobilization of naringinase from Aspergillus niger CECT 2088 in poly(vinyl alcohol) cryogels for the debittering of juices. Food Chem. 2007, 104, 1177–1182. [Google Scholar] [CrossRef]
- Spagna, G.; Barbagallo, R.N.; Martino, A.; Pifferi, P.G. A simple method for purifying glycosidases: α-l-rhamnopyranosidase from Aspergillus niger to increase the aroma of Moscato wine. Enzym. Microb. Technol. 2000, 27, 522–530. [Google Scholar] [CrossRef]
- Németh, K.; Plumb, G.W.; Berrin, J.G.; Juge, N.; Jacob, R.; Naim, H.Y.; Williamson, G.; Swallow, D.M.; Kroon, P.A. Deglycosylation by small intestinal epithelial cell beta-glucosidases is a critical step in the absorption and metabolism of dietary flavonoid glycosides in humans. Eur. J. Nutr. 2003, 42, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria: Their applications in foods. J. Bacteriol. Mycol. Open Access. 2018, 6, 89–94. [Google Scholar]
- Patra, J.K.K.; Das, G.; Paramithiotis, S.; Shin, H.-S. Kimchi and Other Widely Consumed Traditional Fermented Foods of Korea: A Review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.J.; Lee, J.-E.; Lee, C.-H. Importance of lactic acid bacteria in Asian fermented foods. Microb. Cell Factories 2011, 10, S5. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.-B.; Kim, K.-S.; Rhee, J.-S. Hydrolysis of soybean isoflavone glucosides by lactic acid bacteria. Biotechnol. Lett. 2002, 24, 2113–2116. [Google Scholar] [CrossRef]
- Lee, N.-K.; Paik, H.-D. Bioconversion Using Lactic Acid Bacteria: Ginsenosides, GABA, and Phenolic Compounds. J. Microbiol. Biotechnol. 2017, 27, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Michlmayr, H.; Kneifel, W. β-Glucosidase activities of lactic acid bacteria: Mechanisms, impact on fermented food and human health. FEMS Microbiol. Lett. 2014, 352, 1–10. [Google Scholar] [CrossRef] [Green Version]
- McMahon, H.; Zoecklein, B.W.; Fugelsang, K.; Jasinski, Y. Quantification of glycosidase activities in selected yeasts and lactic acid bacteria. J. Ind. Microbiol. Biotechnol. 1999, 23, 198–203. [Google Scholar] [CrossRef]
- Bowers, G.N.; McComb, R.B.; Christensen, R.C.; Schaffer, R. High-Purity 4-Nitrophenol: Purification, Characterization, and Speci-fications for Use as a Spectrophotometric Reference Material. Clin. Chem. 1980, 26, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Liu, X.Q.; Du, X.P.; Wu, L.; Jiang, Z.D.; Ni, H.; Li, Q.B.; Chen, F. Preparation of isoquercitrin by biotransformation of rutin using α-L-rhamnosidase from Aspergillus niger JMU-TS528 and HSCCC purification. Prep. Biochem. Biotechnol. 2019, 50, 1–9. [Google Scholar] [CrossRef]
- Langston, J.; Sheehy, N.; Xu, F. Substrate specificity of Aspergillus oryzae family 3 β-glucosidase. Biochim. Biophys. Acta Proteins Proteom. 2006, 1764, 972–978. [Google Scholar] [CrossRef]
- Volkov, P.V.; Rozhkova, A.M.; Zorov, I.N.; Sinitsyn, A.P. Cloning, purification and study of recombinant GH3 family β-glucosidase from Penicillium verruculosum. Biochimie 2020, 168, 231–240. [Google Scholar] [CrossRef]

| Strain | Isolation Source | Species | Accession Number |
|---|---|---|---|
| NGI01 | Cucumber kimchi | L. pentosus | MT898558 |
| NGI02 | Cucumber kimchi | L. senmaizukei | MT898562 |
| NGI06 | Cabbage kimchi | L. pentosus | MT898563 |
| NGI14 | Cabbage kimchi | L. plantarum | MT898568 |
| NGI17 | Cabbage kimchi | L. pentosus | MT919355 |
| NGI19 | Cabbage kimchi | L. pentosus | MT898576 |
| NGI21 | Cabbage kimchi | L. paraplantarum | MT898577 |
| NGI22 | Cabbage kimchi | L. pentosus | MT898642 |
| NGI23 | Cabbage kimchi | L. pentosus | MT898646 |
| NGI25 | Cabbage kimchi | L. curvatus | MT898647 |
| Strain | Product Yield (%) | |
|---|---|---|
| Hesperidin → Hesperetin | Rutin → Quercetin | |
| NGI01 | 30.3 ± 0.1 | 3.9 ± 0.2 |
| NGI02 | 26.3 ± 2.3 | 0 |
| NGI06 | 28.3 ± 0.8 | 0 |
| NGI14 | 20.4 ± 0.1 | 0 |
| NGI17 | 26.7 ± 0.1 | 0 |
| NGI19 | 24.9 ± 0.2 | 0 |
| NGI21 | 22.7 ± 0.1 | 0 |
| NGI22 | 23.1 ± 0.1 | 0 |
| NGI23 | 12.5 ± 0.1 | 0 |
| NGI25 | 15.5 ± 0.1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, C.-M.; Kim, G.-M.; Cha, G.-S. Biotransformation of Flavonoids by Newly Isolated and Characterized Lactobacillus pentosus NGI01 Strain from Kimchi. Microorganisms 2021, 9, 1075. https://doi.org/10.3390/microorganisms9051075
Park C-M, Kim G-M, Cha G-S. Biotransformation of Flavonoids by Newly Isolated and Characterized Lactobacillus pentosus NGI01 Strain from Kimchi. Microorganisms. 2021; 9(5):1075. https://doi.org/10.3390/microorganisms9051075
Chicago/Turabian StylePark, Chan-Mi, Gyoung-Min Kim, and Gun-Su Cha. 2021. "Biotransformation of Flavonoids by Newly Isolated and Characterized Lactobacillus pentosus NGI01 Strain from Kimchi" Microorganisms 9, no. 5: 1075. https://doi.org/10.3390/microorganisms9051075






