Complete Genome Sequence and Function Gene Identify of Prometryne-Degrading Strain Pseudomonas sp. DY-1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Genomic DNA Sequencing and Annotation
2.3. Phylogenetic Analysis
2.4. Construction of Expression Vector
2.5. Gene Expression
2.6. Analysis of Prometryne-Degrading Capacity
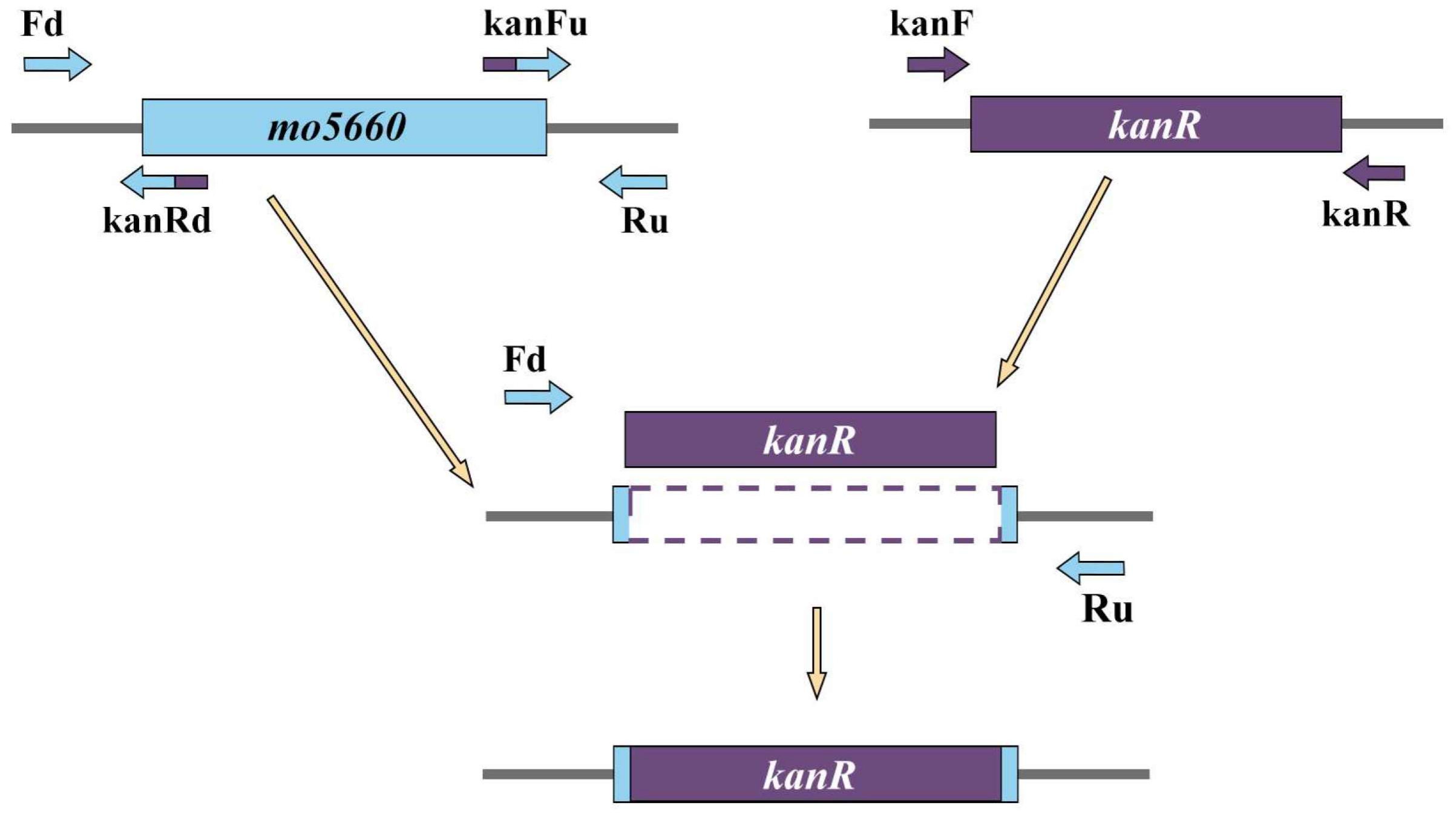
2.7. Gene Knock-Out
2.8. Bioinformatic Analysis of MO5660
3. Results
3.1. Genomic Features
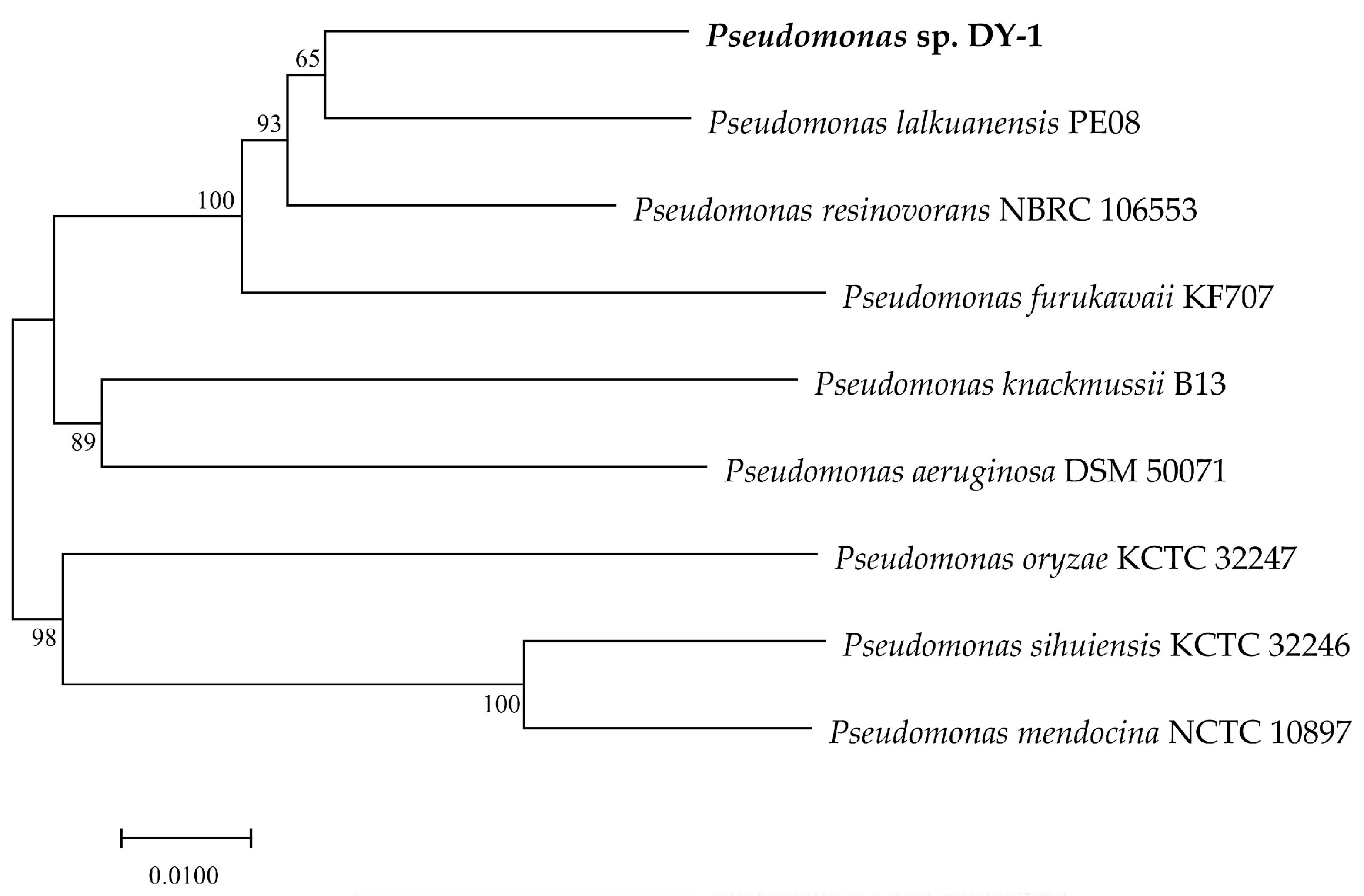
3.2. Phylogenetic and Collinearity Analysis
3.3. Database Annotation
3.3.1. VFDB Analysis
3.3.2. COG Analysis
3.3.3. KEGG Analysis
3.4. Molecular Characteristics of Pseudomonas sp. DY-1 Genome
3.4.1. Carbohydrate Metabolism
3.4.2. Nitrogen Metabolism
3.4.3. Sulfur Metabolism
3.4.4. Potential of Plant Growth Promoting
3.4.5. Environmental Stress Resistance
3.4.6. Metal/Metalloid Transport and Resistance
3.4.7. Polymer Biosynthesis
3.4.8. Xenobiotics Biodegradation and Metabolism
3.5. Sequence Analysis of MO5660 Monooxygenase
3.6. Heterologous Expression and Function Verification of MO5660
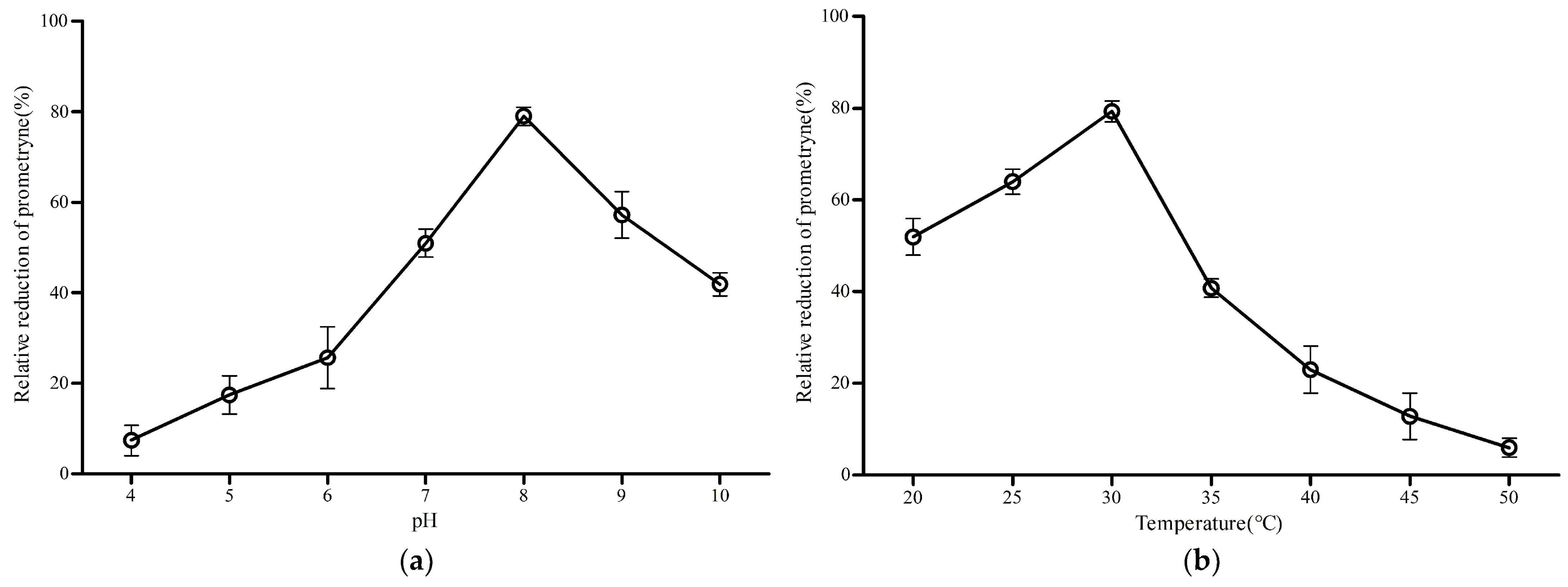
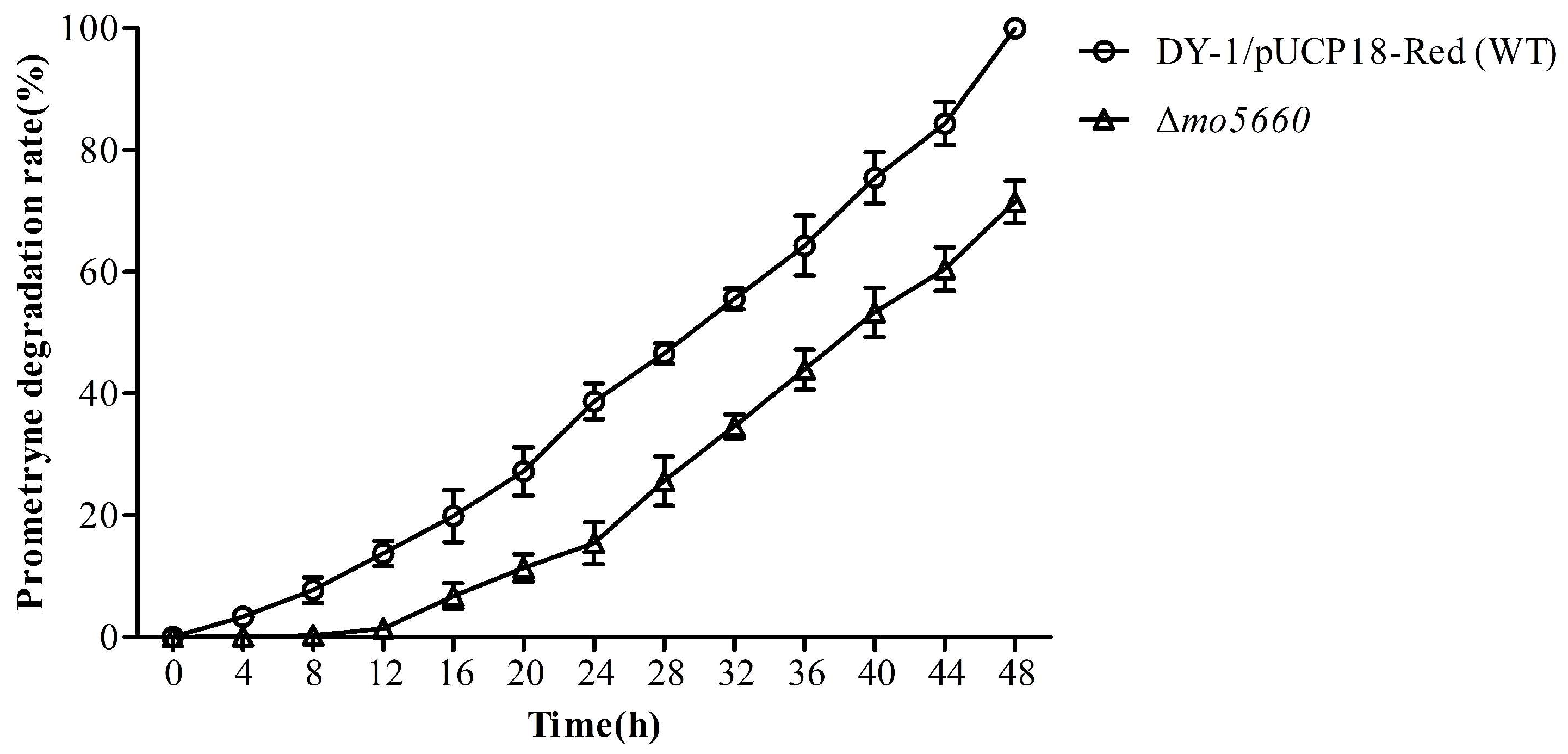
3.7. Degrading Characteristics of MO5660
3.8. Confirmation of mo5660-Defective Mutation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Boulahia, K.; Carol, P.; Planchais, S.; Abrous-Belbachir, O. Phaseolus vulgaris L. Seedlings Exposed to Prometryn Herbicide Contaminated Soil Trigger an Oxidative Stress Response. J. Agric. Food Chem. 2016, 64, 3150–3160. [Google Scholar] [CrossRef] [PubMed]
- Dikić, D.; Zidovec-Lepej, S.; Remenar, A.; Bendelja, K.; Benković, V.; Horvat-Knežević, A.; Brozović, G.; Oršolić, N. Effects of prometryne on apoptosis and necrosis in thymus, lymph node and spleen in mice. Environ. Toxicol. Pharmacol. 2009, 27, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Brack, W.; Altenburger, R.; Ensenbach, U.; Moder, M.; Segner, H.; Schuurmann, G. Bioassay-directed identification of organic toxicants in river sediment in the industrial region of bitterfeld (Germany)-A contribution to hazard assessment. Arch. Environ. Contam. Toxicol. 1999, 37, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zhou, G.; Feng, D.; Fang, W.; Chen, T.; Hu, K. Transcriptome analysis of Penaeus vannamei hepatopancreas reveals differences in toxicity mechanisms between phoxim and prometryne. Fish Shellfish. Immunol. 2020, 105, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Dsikowitzky, L.; Nguyen, T.M.; Konzer, L.; Zhao, H.; Wang, D.; Yang, F.; Schwarzbauer, J. Occurrence and origin of triazine herbicides in a tropical coastal area in China: A potential ecosystem threat. Estuar. Coast. Shelf Sci. 2020, 235, 106612. [Google Scholar] [CrossRef]
- Singh Sankhla, M.; Kumari, M.; Sharma, K.; Kushwah, R.; Kumar, R. Water Contamination through Pesticide & Their Toxic Effect on Human Health. Int. J. Res. Appl. Sci. Eng. Technol. 2018, 6, 967–970. [Google Scholar] [CrossRef]
- Quintella, C.; Mata, A.M.T.; Lima, L. Overview of bioremediation with technology assessment and emphasis on fungal bioremediation of oil contaminated soils. J. Environ. Manag. 2019, 241, 156–166. [Google Scholar] [CrossRef]
- Lovley, D.R. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat. Rev. Microbiol. 2003, 1, 35–44. [Google Scholar] [CrossRef]
- Gillespie, I.M.; Philp, J.C. Bioremediation, an environmental remediation technology for the bioeconomy. Trends Biotechnol. 2013, 31, 329–332. [Google Scholar] [CrossRef]
- Kube, M.; Chernikova, T.N.; Al-Ramahi, Y.; Beloqui, A.; Lopez-Cortez, N.; Guazzaroni, M.E.; Heipieper, H.J.; Klages, S.; Kotsyurbenko, O.R.; Langer, I.; et al. Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 2013, 4, 2156. [Google Scholar] [CrossRef] [Green Version]
- Satsuma, K. Mineralization of s-triazine herbicides by a newly isolated Nocardioides species strain DN36. Appl. Microbiol. Biotechnol. 2010, 86, 1585–1592. [Google Scholar] [CrossRef]
- Aislabie, J.; Bej, A.; Ryburn, J.; Lloyd, N.; Wilkins, A. Characterization of Arthrobacter nicotinovorans HIM, an atrazine-degrading bacterium, from agricultural soil New Zealand. Fems. Microbiol. Ecol. 2005, 52, 279–286. [Google Scholar] [CrossRef]
- Strong, L.C.; Rosendahl, C.; Johnson, G.; Sadowsky, M.J.; Wackett, L.P. Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds. Appl. Environ. Microbiol. 2002, 68, 5973–5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topp, E.; Mulbry, W.M.; Zhu, H.; Nour, S.M.; Cuppels, D. Characterization of S-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl. Environ. Microbiol. 2000, 66, 3134–3141. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hua, R.; Lv, P.; Tang, J.; Wang, Y.; Cao, H.; Wu, X.; Li, Q.X. Novel hydrolytic de-methylthiolation of the s-triazine herbicide prometryn by Leucobacter sp. JW-1. Sci. Total Environ. 2017, 579, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Takagi, K.; Hiradate, S.; Iwasaki, A.; Harada, N. Biodegradation of methylthio-s-triazines by Rhodococcus sp. strain FJ1117YT, and production of the corresponding methylsulfinyl, methylsulfonyl and hydroxy analogues. Pest Manag. Sci. 2007, 63, 254–260. [Google Scholar] [CrossRef]
- Harada, N.; Takagi, K.; Fujii, K.; Iwasaki, A. Transformation of methylthio-s-triazines via sulfur oxidation by strain JUN7, a Bacillus cereus species. Soil Biol. Biochem. 2006, 38, 2952–2957. [Google Scholar] [CrossRef]
- Tabatabaei, S.; Ehsanzadeh, P.; Etesami, H.; Alikhani, H.; Glick, B. Indole-3-acetic acid (IAA) producing Pseudomonas isolates inhibit seed germination and α-amylase activity in durum wheat (Triticum turgidum L.). Span. J. Agric. Res. 2016, 14, 15. [Google Scholar] [CrossRef] [Green Version]
- Mehri, I.; Turki, Y.; Rajeb, A.; Sana, K.; Abdennasser, H. Multi-Traits of Non-Pathogenic Fluorescent Pseudomonas and Evaluation of Their Potentiel as Biocontrol Agents. Am. J. Environ. Sci. 2014, 10, 199. [Google Scholar] [CrossRef]
- Liang, D.; Ding, M.-Y.; Xiao, C.-Y.; Shen, Y.-W.; Wang, Y.-Y.; Li, H.-T.; Liu, R.-M.; Gao, J.-G. Isolation and Identification of Pseudomonas sp. Strain DY-1 from Agricultural Soil and Its Degradation Effect on Prometryne. Curr. Microbiol. 2021, 78, 1871–1881. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M. Enzyme Annotation and Metabolic Reconstruction Using KEGG. Methods Mol. Biol. 2017, 1611, 135–145. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Fillat, M.F.; Borrias, W.E.; Weisbeek, P.J. Isolation and overexpression in Escherichia coli of the flavodoxin gene from Anabaena PCC 7119. Biochem. J. 1991, 280(Pt. 1), 187–191. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Guo, H.; Zhu, H.M.; Jiang, L.; Yang, H. Effects of SOM, surfactant and pH on the sorption-desorption and mobility of prometryne in soils. Chemosphere 2008, 70, 2127–2134. [Google Scholar] [CrossRef]
- Britton, H.T.S.; Robinson, R.A. CXCVIII—Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1931, 1456–1462. [Google Scholar] [CrossRef]
- Lesic, B.; Rahme, L.G. Use of the lambda red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol. 2008, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, H.P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 1991, 97, 109–121. [Google Scholar] [CrossRef]
- Choi, K.H.; Kumar, A.; Schweizer, H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 2006, 64, 391–397. [Google Scholar] [CrossRef]
- UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiederschain, G.Y. The proteomics protocols handbook. Biochemistry 2006, 71, 696. [Google Scholar] [CrossRef]
- Mitaku, S.; Hirokawa, T.; Tsuji, T. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 2002, 18, 608–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, T.; Ramos, J.L.; Rodríguez-Herva, J.J.; Fuhrer, T.; Sauer, U.; Duque, E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: Genomic and flux analysis. J. Bacteriol. 2007, 189, 5142–5152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daddaoua, A.; Krell, T.; Ramos, J.L. Regulation of glucose metabolism in Pseudomonas: The phosphorylative branch and entner-doudoroff enzymes are regulated by a repressor containing a sugar isomerase domain. J. Biol. Chem. 2009, 284, 21360–21368. [Google Scholar] [CrossRef] [Green Version]
- Chavarría, M.; Nikel, P.I.; Pérez-Pantoja, D.; de Lorenzo, V. The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 2013, 15, 1772–1785. [Google Scholar] [CrossRef]
- Kim, J.; Jeon, C.O.; Park, W. Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology 2008, 154, 3905–3916. [Google Scholar] [CrossRef] [Green Version]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; de Lorenzo, V. Pseudomonas putida KT2440 Strain Metabolizes Glucose through a Cycle Formed by Enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and Pentose Phosphate Pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef] [Green Version]
- Rediers, H.; Vanderleyden, J.; De Mot, R. Nitrate respiration in Pseudomonas stutzeri A15 and its involvement in rice and wheat root colonization. Microbiol. Res. 2009, 164, 461–468. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biessy, A.; Novinscak, A.; Blom, J.; Léger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirleau, P.; Philippot, L.; Corberand, T.; Lemanceau, P. Involvement of nitrate reductase and pyoverdine in competitiveness of Pseudomonas fluorescens strain C7R12 in soil. Appl. Environ. Microbiol. 2001, 67, 2627–2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bünemann, E.K.; Condron, L.M. Phosphorus and Sulphur Cycling in Terrestrial Ecosystems. In Nutrient Cycling in Terrestrial Ecosystems; Marschner, P., Rengel, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 65–92. [Google Scholar]
- Kredich, N. Biosynthesis of Cysteine. EcoSal Plus 2008. [Google Scholar] [CrossRef]
- Scherer, H.W. Sulphur in crop production—invited paper. Eur. J. Agron. 2001, 14, 81–111. [Google Scholar] [CrossRef]
- Mirleau, P.; Wogelius, R.; Smith, A.; Kertesz, M.A. Importance of organosulfur utilization for survival of Pseudomonas putida in soil and rhizosphere. Appl. Environ. Microbiol. 2005, 71, 6571–6577. [Google Scholar] [CrossRef] [Green Version]
- Hummerjohann, J.; Küttel, E.; Quadroni, M.; Ragaller, J.; Leisinger, T.; Kertes, M.A. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: Role of cysteine biosynthetic intermediates. Microbiology 1998, 144(Pt. 5), 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Kertesz, M.A.; Mirleau, P. The role of soil microbes in plant sulphur nutrition. J. Exp. Bot. 2004, 55, 1939–1945. [Google Scholar] [CrossRef] [Green Version]
- Sirko, A.; Zatyka, M.; Sadowy, E.; Hulanicka, D. Sulfate and thiosulfate transport in Escherichia coli K-12: Evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 1995, 177, 4134–4136. [Google Scholar] [CrossRef] [Green Version]
- van der Ploeg, J.R.; Weiss, M.A.; Saller, E.; Nashimoto, H.; Saito, N.; Kertesz, M.A.; Leisinger, T. Identification of sulfate starvation-regulated genes in Escherichia coli: A gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 1996, 178, 5438–5446. [Google Scholar] [CrossRef] [Green Version]
- Kertesz, M.A.; Schmidt-Larbig, K.; Wüest, T. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 1464–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davison, J.; Brunel, F.; Phanopoulos, A.; Prozzi, D.; Terpstra, P. Cloning and sequencing of Pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene 1992, 114, 19–24. [Google Scholar] [CrossRef]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef] [Green Version]
- Dakora, F.D.; Matiru, V.N.; Kanu, A.S. Rhizosphere ecology of lumichrome and riboflavin, two bacterial signal molecules eliciting developmental changes in plants. Front. Plant Sci. 2015, 6, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. Fems. Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [Green Version]
- Seneviratne, G.; Weerasekara, M.L.M.A.W.; Seneviratne, K.A.C.N.; Zavahir, J.S.; Kecskés, M.L.; Kennedy, I.R. Importance of Biofilm Formation in Plant Growth Promoting Rhizobacterial Action. In Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 81–95. [Google Scholar]
- Audrain, B.; Farag, M.A.; Ryu, C.-M.; Ghigo, J.-M. Role of bacterial volatile compounds in bacterial biology. Fems Microbiol. Rev. 2015, 39, 222–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; Norman, D.; El-Sayed, A.S. Chapter Ten—Soluble and Volatile Metabolites of Plant Growth-Promoting Rhizobacteria (PGPRs): Role and Practical Applications in Inhibiting Pathogens and Activating Induced Systemic Resistance (ISR). In Advances in Botanical Research; Bais, H., Sherrier, J., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 75, pp. 241–284. [Google Scholar]
- Robin, A.; Vansuyt, G.; Hinsinger, P.; Meyer, J.M.; Briat, J.F.; Lemanceau, P. Chapter 4 Iron Dynamics in the Rhizosphere: Consequences for Plant Health and Nutrition. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2008; Volume 99, pp. 183–225. [Google Scholar]
- Thomashow, L.; Bakker, P.A.H.M. Microbial Control of Root-Pathogenic Fungi and Oomycetes. In Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture; Lugtenberg, B., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 165–173. [Google Scholar]
- Wandersman, C.; Delepelaire, P. Bacterial iron sources: From siderophores to hemophores. Annu. Rev. Microbiol. 2004, 58, 611–647. [Google Scholar] [CrossRef] [PubMed]
- Thieringer, H.A.; Jones, P.G.; Inouye, M. Cold shock and adaptation. Bioessays 1998, 20, 49–57. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B.; Yu, Y.; Shu, Y.L.; Rong, J.C.; Zhang, Y.J.; Zhao, D.L.; Chen, X.L.; Zhang, X.Y.; Chen, B.; et al. Comparative genomics of the marine bacterial genus Glaciecola reveals the high degree of genomic diversity and genomic characteristic for cold adaptation. Environ. Microbiol. 2014, 16, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.F.; Xu, Z.S.; Guo, J.K.; Wang, Y.X.; Abernathy, B.; Fu, J.D.; Chen, X.; Zhou, Y.B.; Chen, M.; Ye, X.G.; et al. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 2017, 7, 44050. [Google Scholar] [CrossRef]
- Messner, K.R.; Imlay, J.A. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J. Biol. Chem. 1999, 274, 10119–10128. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.N.; Dieckmann, G.S. Antarctic Sea ice—A habitat for extremophiles. Science 2002, 295, 641–644. [Google Scholar] [CrossRef] [Green Version]
- Seaver, L.C.; Imlay, J.A. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 2001, 183, 7173–7181. [Google Scholar] [CrossRef] [Green Version]
- Padan, E.; Venturi, M.; Gerchman, Y.; Dover, N. Na(+)/H(+) antiporters. Biochim. Et. Biophys. Acta 2001, 1505, 144–157. [Google Scholar] [CrossRef] [Green Version]
- Takami, H.; Takaki, Y.; Uchiyama, I. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 2002, 30, 3927–3935. [Google Scholar] [CrossRef]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. Mmbr. 1998, 62, 504–544. [Google Scholar] [CrossRef] [Green Version]
- Kosono, S.; Kitada, M.; Kudo, T. A novel type of Na+/H+ antiporter: Its unique characteristics and function. In Progress in Biotechnology; Endo, I., Kudo, T., Osada, H., Shibata, T., Yamaguchi, I., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; Volume 22, pp. 75–84. [Google Scholar]
- Bucarey, S.A.; Penn, K.; Paul, L.; Fenical, W.; Jensen, P.R. Genetic complementation of the obligate marine actinobacterium Salinispora tropica with the large mechanosensitive channel gene mscL rescues cells from osmotic downshock. Appl. Environ. Microbiol. 2012, 78, 4175–4182. [Google Scholar] [CrossRef]
- Huertas, M.J.; Luque-Almagro, V.M.; Martínez-Luque, M.; Blasco, R.; Moreno-Vivián, C.; Castillo, F.; Roldán, M.D. Cyanide metabolism of Pseudomonas pseudoalcaligenes CECT5344: Role of siderophores. Biochem. Soc. Trans. 2006, 34, 152–155. [Google Scholar] [CrossRef]
- Igeño, M.I.; Becerra, G.; Guijo, M.I.; Merchán, F.; Blasco, R. Metabolic adaptation of Pseudomonas pseudoalcaligenes CECT5344 to cyanide: Role of malate-quinone oxidoreductases, aconitase and fumarase isoenzymes. Biochem. Soc. Trans. 2011, 39, 1849–1853. [Google Scholar] [CrossRef] [Green Version]
- Estepa, J.; Luque-Almagro, V.M.; Manso, I.; Escribano, M.P.; Martínez-Luque, M.; Castillo, F.; Moreno-Vivián, C.; Roldán, M.D. The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT5344 involved in assimilation of nitriles is essential for growth on cyanide. Environ. Microbiol. Rep. 2012, 4, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Luque-Almagro, V.M.; Merchán, F.; Blasco, R.; Igeño, M.I.; Martínez-Luque, M.; Moreno-Vivián, C.; Castillo, F.; Roldán, M.D. Cyanide degradation by Pseudomonas pseudoalcaligenes CECT5344 involves a malate:quinone oxidoreductase and an associated cyanide-insensitive electron transfer chain. Microbiology 2011, 157, 739–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Płociniczak, T.; Kukla, M.; Wątroba, R.; Piotrowska-Seget, Z. The effect of soil bioaugmentation with strains of Pseudomonas on Cd, Zn and Cu uptake by Sinapis alba L. Chemosphere 2013, 91, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Y.; Huang, W.J.; Sinclair, R.B.; Powers, L. The structure of the zinc sites of Escherichia coli DNA-dependent RNA polymerase. J. Biol. Chem. 1992, 267, 25560–25567. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Park, D.; Yun, Y.-S.; Jo, J.H.; Moon, J. Biosorption Process for Treatment of Electroplating Wastewater Containing Cr(VI): Laboratory-Scale Feasibility Test. Ind. Eng. Chem. Res. 2006, 45, 5059–5065. [Google Scholar] [CrossRef]
- Xie, P.; Hao, X.; Herzberg, M.; Luo, Y.; Nies, D.H.; Wei, G. Genomic analyses of metal resistance genes in three plant growth promoting bacteria of legume plants in Northwest mine tailings, China. J. Environ. Sci. 2015, 27, 179–187. [Google Scholar] [CrossRef]
- Cruz, M.V.; Freitas, F.; Paiva, A.; Mano, F.; Dionísio, M.; Ramos, A.M.; Reis, M.A. Valorization of fatty acids-containing wastes and byproducts into short- and medium-chain length polyhydroxyalkanoates. New Biotechnol. 2016, 33, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ashby, R.D.; Needleman, D.S.; Lee, K.T.; Solaiman, D.K. Cloning, sequencing, and characterization of lipase genes from a polyhydroxyalkanoate (PHA)-synthesizing Pseudomonas resinovorans. Appl. Microbiol. Biotechnol. 2012, 96, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Huisman, G.W.; Wonink, E.; Meima, R.; Kazemier, B.; Terpstra, P.; Witholt, B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J. Biol. Chem. 1991, 266, 2191–2198. [Google Scholar] [CrossRef]
- Tarazona, N.A.; Hernández-Arriaga, A.M.; Kniewel, R.; Prieto, M.A. Phasin interactome reveals the interplay of PhaF with the polyhydroxyalkanoate transcriptional regulatory protein PhaD in Pseudomonas putida. Environ. Microbiol. 2020, 22, 3922–3936. [Google Scholar] [CrossRef]
- Jiménez, J.I.; Miñambres, B.; García, J.L.; Díaz, E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 2002, 4, 824–841. [Google Scholar] [CrossRef]
- Galán, B.; Díaz, E.; Prieto, M.A.; García, J.L. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: A prototype of a new Flavin:NAD(P)H reductase subfamily. J. Bacteriol. 2000, 182, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Díaz, E.; Ferrández, A.; Prieto, M.A.; García, J.L. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. Mmbr. 2001, 65, 523–569. [Google Scholar] [CrossRef] [Green Version]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef] [Green Version]
- Kindl, H.; Schiefer, S. Aldoximes as intermediates in the biosynthesis of tyrosol and tyrosol derivatives. Phytochemistry 1971, 10, 1795–1802. [Google Scholar] [CrossRef]
- Yun-Shu, F.; Pei-Chung, C.; Fu-Shyan, W.; Wan-Yi, H.; Chi-Mei, L. Nitrile hydratase from Mesorhizobium sp. F28 and its potential for nitrile biotransformation. Process. Biochem. 2008, 43, 1391–1397. [Google Scholar] [CrossRef]
- Kato, Y.; Ooi, R.; Asano, Y. Distribution of aldoxime dehydratase in microorganisms. Appl. Environ. Microbiol. 2000, 66, 2290–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolz, A. Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol. 2001, 56, 69–80. [Google Scholar] [CrossRef]
- Nakanishi, M.; Yatome, C.; Ishida, N.; Kitade, Y. Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J. Biol. Chem. 2001, 276, 46394–46399. [Google Scholar] [CrossRef] [Green Version]
- Robinson, A.; Edwards, K.J.; Carr, P.D.; Barton, J.D.; Ewart, G.D.; Ollis, D.L. Structure of the C123S mutant of dienelactone hydrolase (DLH) bound with the PMS moiety of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF). Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 1376–1384. [Google Scholar] [CrossRef] [PubMed]
- Pathak, D.; Ashley, G.; Ollis, D. Thiol protease-like active site found in the enzyme dienelactone hydrolase: Localization using biochemical, genetic, and structural tools. Proteins 1991, 9, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Táncsics, A.; Benedek, T.; Szoboszlay, S.; Veres, P.G.; Farkas, M.; Máthé, I.; Márialigeti, K.; Kukolya, J.; Lányi, S.; Kriszt, B. The detection and phylogenetic analysis of the alkane 1-monooxygenase gene of members of the genus Rhodococcus. Syst. Appl. Microbiol. 2015, 38, 1–7. [Google Scholar] [CrossRef]
- Yang, R.; Liu, G.; Chen, T.; Li, S.; An, L.; Zhang, G.; Li, G.; Chang, S.; Zhang, W.; Chen, X.; et al. Characterization of the genome of a Nocardia strain isolated from soils in the Qinghai-Tibetan Plateau that specifically degrades crude oil and of this biodegradation. Genomics 2019, 111, 356–366. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Kamerbeek, N.M.; Heidekamp, A.J.; Fortin, R.; Janssen, D.B. The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase. J. Biol. Chem. 2004, 279, 3354–3360. [Google Scholar] [CrossRef] [Green Version]
- Fraaije, M.W.; Kamerbeek, N.M.; van Berkel, W.J.; Janssen, D.B. Identification of a Baeyer-Villiger monooxygenase sequence motif. Febs. Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Ferroni, F.; Smit, M.; Opperman, D. Functional divergence between closely related Baeyer-Villiger monooxygenases from Aspergillus flavus. J. Mol. Catal. B Enzym. 2014, 107, 47–54. [Google Scholar] [CrossRef]
- Balderrama Carmona, A.P.; Silva, N.; Alvarez, L.; Bante, N.; Morán Palacio, E.F. Consequences of Herbicide Use in Rural Environments and Their Effect on Agricultural Workers. In Sustainability Concept in Developing Countries; BoD—Books on Demand: Norderstedt, Germany, 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, X.; Jiang, Y.; Wu, Y.; Chen, J.; Hu, F.; Li, H. Combined effects of bacterial-feeding nematodes and prometryne on the soil microbial activity. J. Hazard. Mater. 2011, 192, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.Y.; Zhang, J.B.; Gao, J.G.; Li, H.T.; Liang, D.; Liu, R.M. Isolation and characterization of atrazine-degrading strain Shewanella sp. YJY4 from cornfield soil. Lett. Appl. Microbiol. 2016, 63, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, L.; Ma, F.; Bai, S.; Yang, J.; Qi, S. Pseudomonas sp. ZXY-1, a newly isolated and highly efficient atrazine-degrading bacterium, and optimization of biodegradation using response surface methodology. J. Environ. Sci. 2017, 54, 152–159. [Google Scholar] [CrossRef]
- Zhang, B.; Ni, Y.; Liu, J.; Yan, T.; Zhu, X.; Li, Q.X.; Hua, R.; Pan, D.; Wu, X. Bead-immobilized Pseudomonas stutzeri Y2 prolongs functions to degrade s-triazine herbicides in industrial wastewater and maize fields. Sci. Total Environ. 2020, 731, 139183. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Poindexter, J.S. Oligotrophy. In Advances in Microbial Ecology; Alexander, M., Ed.; Springer: Boston, MA, USA, 1981; pp. 63–89. [Google Scholar]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Weller, D.M. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Husen, E.; Wahyudi, A.T.; Suwanto, A. Soybean Response to 1-Aminocyclopropane-1-Carboxylate Deaminase-Producing Pseudomonas under Field Soil Conditions. Am. J. Agric. Biol. Sci. 2011, 6, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Zhou, M.; Zhao, Q.; Wang, F.; Gao, J.; Sheng, H.; An, L. Complete genome sequence of Sphingomonas sp. Cra20, a drought resistant and plant growth promoting rhizobacteria. Genomics 2020, 112, 3648–3657. [Google Scholar] [CrossRef] [PubMed]
- Yoshikane, Y.; Yokochi, N.; Ohnishi, K.; Yagi, T. Coenzyme precursor-assisted cooperative overexpression of an active pyridoxine 4-oxidase from Microbacterium luteolum. Protein Expr. Purif. 2004, 34, 243–248. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W. Coenzyme precursor-assisted expression of a cholesterol oxidase from Brevibacterium sp. in Escherichia coli. Biotechnol. Lett. 2007, 29, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Bisagni, S. Baeyer-Villiger Monooxygenases from a Dietzia sp.—Enzyme Discovery, Characterization and Engineering. Ph.D. Thesis, Division of Biotechnology, Lund University, Lund, Sweden, October 2014. [Google Scholar]
- Zhang, Y.; Liu, F.; Xu, N.; Wu, Y.; Zheng, Y.-C.; Zhao, Q.; Lin, G.; Yu, H.; Xu, J.-H. The discovery of two native Baeyer-Villiger monooxygenases for asymmetric synthesis of bulky chiral sulfoxides. Appl. Environ. Microbiol. 2018, 84, e00638-18. [Google Scholar] [CrossRef] [Green Version]
- Fürst, M.J.; Savino, S.; Dudek, H.M.; Gómez Castellanos, J.R.; Gutiérrez de Souza, C.; Rovida, S.; Fraaije, M.W.; Mattevi, A. Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila: A Structurally Distinct Biocatalyst for Bulky Substrates. J. Am. Chem. Soc. 2017, 139, 627–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fordwour, O.B.; Luka, G.; Hoorfar, M.; Wolthers, K.R. Kinetic characterization of acetone monooxygenase from Gordonia sp. strain TY-5. Amb. Express 2018, 8, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahnert, A.; Mirleau, P.; Wait, R.; Kertesz, M.A. The LysR-type regulator SftR is involved in soil survival and sulphate ester metabolism in Pseudomonas putida. Environ. Microbiol. 2002, 4, 225–237. [Google Scholar] [CrossRef] [PubMed]

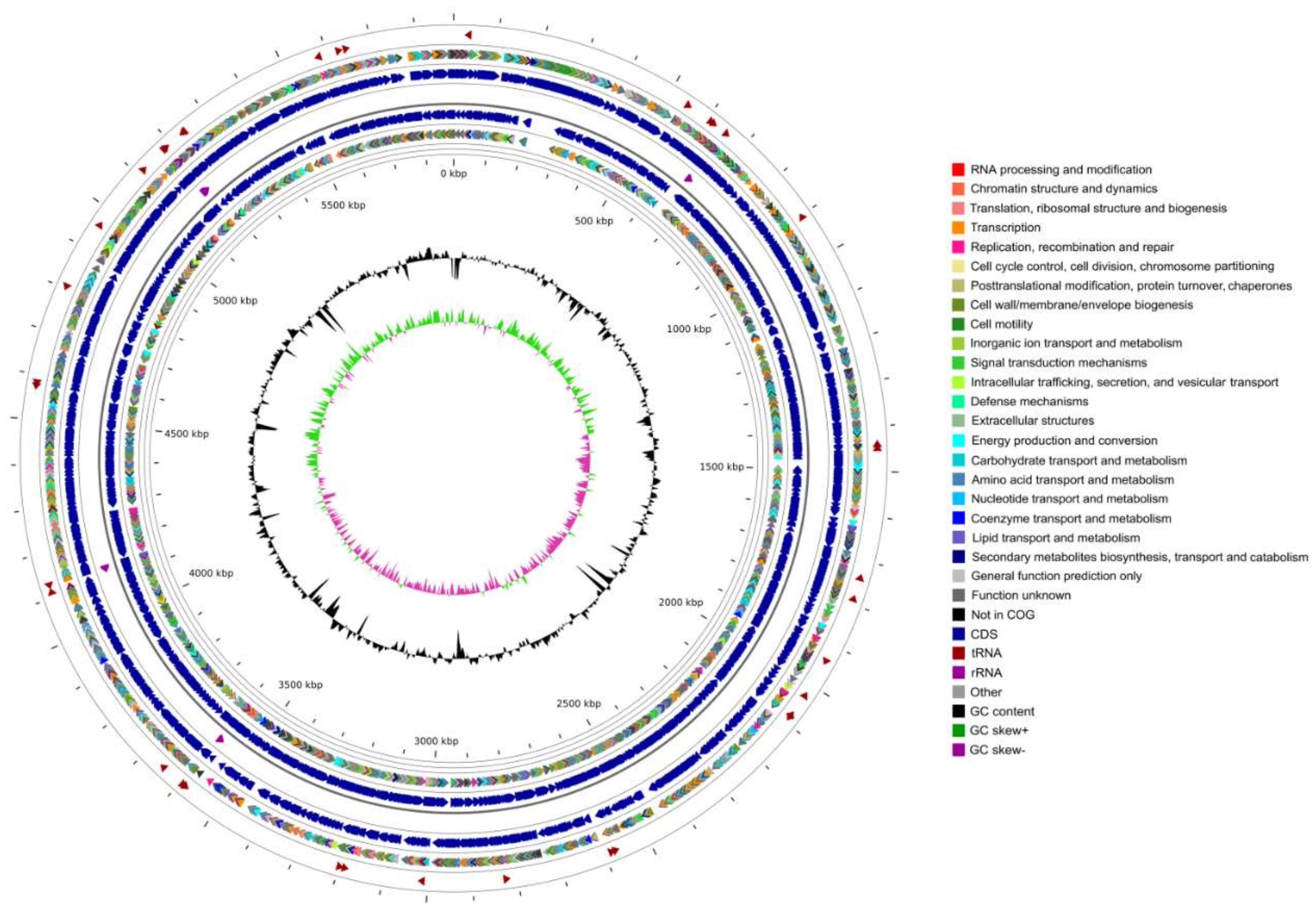



| Features | Chromosome | Plasmid |
|---|---|---|
| Genome size (bp) | 5,886,398 | 26,350 |
| GC content (mol %) | 62.96 | 56.80 |
| Total genes | 5501 | 42 |
| Total CDS | 5410 | 42 |
| Number of rRNAs (5S, 16S, 23S) | 16 (6,5,5) | 0 |
| Number of tRNAs | 71 | 0 |
| Number of ncRNAs | 4 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, D.; Xiao, C.; Song, F.; Li, H.; Liu, R.; Gao, J. Complete Genome Sequence and Function Gene Identify of Prometryne-Degrading Strain Pseudomonas sp. DY-1. Microorganisms 2021, 9, 1261. https://doi.org/10.3390/microorganisms9061261
Liang D, Xiao C, Song F, Li H, Liu R, Gao J. Complete Genome Sequence and Function Gene Identify of Prometryne-Degrading Strain Pseudomonas sp. DY-1. Microorganisms. 2021; 9(6):1261. https://doi.org/10.3390/microorganisms9061261
Chicago/Turabian StyleLiang, Dong, Changyixin Xiao, Fuping Song, Haitao Li, Rongmei Liu, and Jiguo Gao. 2021. "Complete Genome Sequence and Function Gene Identify of Prometryne-Degrading Strain Pseudomonas sp. DY-1" Microorganisms 9, no. 6: 1261. https://doi.org/10.3390/microorganisms9061261







