Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens
Abstract
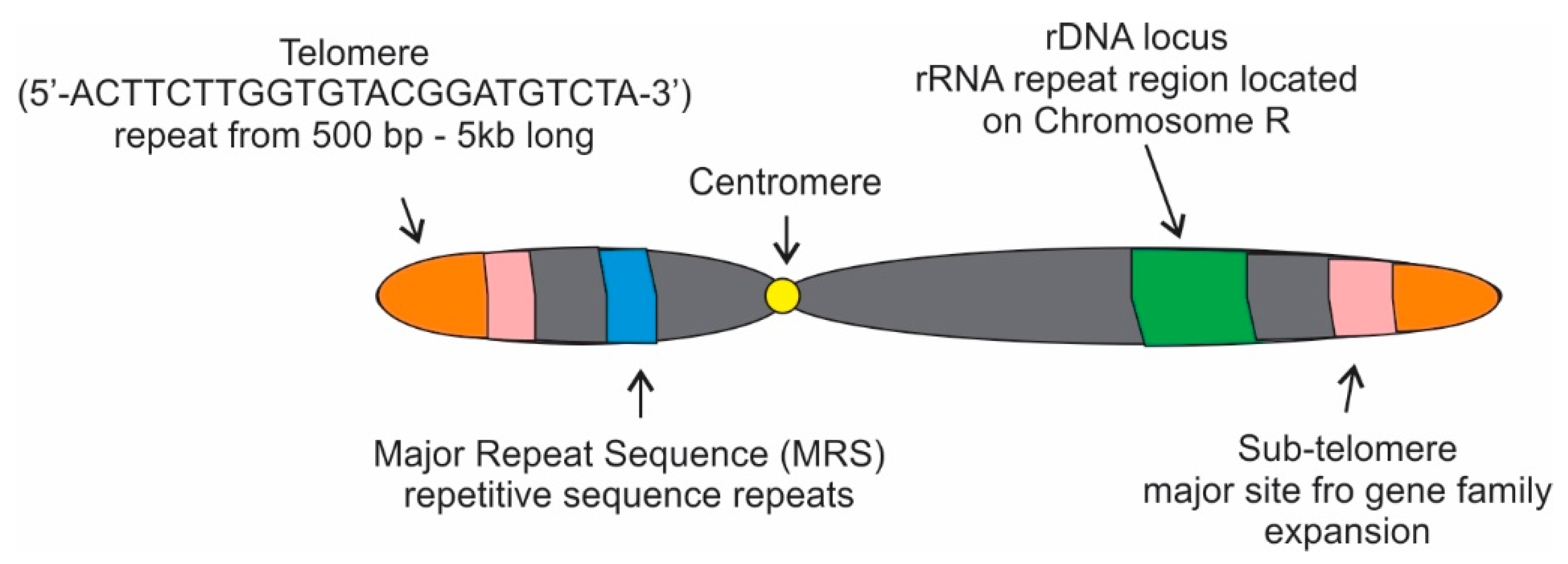
1. Introduction
2. Aspergillus fumigatus
3. Candida albicans
4. Candida glabrata
5. Pneumocystis Fungi
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Mycoses in North America. Crit. Rev. Microbiol. 2010, 36, 1–53. [Google Scholar] [CrossRef]
- O’kane, C.J.; Weild, R.; Hyland, E.M. Chromatin structure and drug resistance in candida spp. J. Fungi 2020, 6, 121. [Google Scholar] [CrossRef]
- Červenák, F.; Sepšiová, R.; Nosek, J.; Tomáška, Ľ. Step-by-Step Evolution of Telomeres: Lessons from Yeasts. Genome Biol. Evol. 2021, 13, 1–12. [Google Scholar] [CrossRef]
- Kwapisz, M.; Morillon, A. Subtelomeric Transcription and its Regulation. J. Mol. Biol. 2020, 432, 4199–4219. [Google Scholar] [CrossRef] [PubMed]
- Li, B. Telomere components as potential therapeutic targets for treating microbial pathogen infections. Front. Oncol. 2012, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shammas, M.A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Collins, K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harbor Perspect. Biol. 2011, 3, 1–9. [Google Scholar] [CrossRef]
- Saint-Leandre, B.; Levine, M.T. The telomere paradox: Stable genome preservation with rapidly evolving proteins. Trends Genet. 2020, 36, 232–242. [Google Scholar] [CrossRef]
- Mceachern, M.J.; Blackburn, E.H. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. USA 1994, 91, 3453–3457. [Google Scholar] [CrossRef] [PubMed]
- Fulnečková, J.; Ševčíková, T.; Fajkus, J.; Lukešová, A.; Lukeš, M.; Vlček, Č.; Lang, B.F.; Kim, E.; Eliáš, M.; Sýkorová, E. A broad phylogenetic survey unveils the diversity and evolution of telomeres in eukaryotes. Genome Biol. Evol. 2013, 5, 468–483. [Google Scholar] [CrossRef] [PubMed]
- Nabetani, A.; Ishikawa, F. Alternative lengthening of telomeres pathway: Recombination-mediated telomere maintenance mechanism in human cells. J. Biochem. 2011, 149, 5–14. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Diotti, R.; Loayza, D. Shelterin complex and associated factors at human telomeres. Nucleus 2011, 2, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Oestreich, S.; De Lange, T. Identification of human Rap1: Implications for telomere evolution. Cell 2000, 101, 471–483. [Google Scholar] [CrossRef]
- Kim, S.H.; Kaminker, P.; Campisi, J. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999, 23, 405–412. [Google Scholar] [CrossRef]
- Liu, D.; Safari, A.; O’Connor, M.S.; Chan, D.W.; Laegeler, A.; Qin, J.; Songyang, Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004, 6, 673–680. [Google Scholar] [CrossRef]
- Ye, J.Z.S.; Hockemeyer, D.; Krutchinsky, A.N.; Loayza, D.; Hooper, S.M.; Chait, B.T.; De Lange, T. POT1-interaction protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004, 18, 1649–1654. [Google Scholar] [CrossRef]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997, 17, 231–235. [Google Scholar] [CrossRef]
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef]
- Červenák, F.; Juríková, K.; Sepšiová, R.; Neboháčová, M.; Nosek, J.; Tomáška, L.U. Double-stranded telomeric DNA binding proteins: Diversity matters. Cell Cycle 2017, 16, 1568–1577. [Google Scholar] [CrossRef]
- Miyake, Y.; Nakamura, M.; Nabetani, A.; Shimamura, S.; Tamura, M.; Yonehara, S.; Saito, M.; Ishikawa, F. RPA-like Mammalian Ctc1-Stn1-Ten1 Complex Binds to Single-Stranded DNA and Protects Telomeres Independently of the Pot1 Pathway. Mol. Cell 2009, 36, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Gast, A.; Choi, G.; Kulkarni, R.; Quijote, J.; Graham-Yooll, A.; Park, D.; Green, E.M. The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics 2017, 12, 93–104. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PloS Pathog. 2014, 10, e1003859. [Google Scholar] [CrossRef] [PubMed]
- Poláková, S.; Blume, C.; Zárate, J.Á.; Mentel, M.; Jørck-Ramberg, D.; Stenderup, J.; Piškur, J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 2009, 106, 2688–2693. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2020, 33, 310–350. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Denning, D.W. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- McDonagh, A.; Fedorova, N.D.; Crabtree, J.; Yu, Y.; Kim, S.; Chen, D.; Loss, O.; Cairns, T.; Goldman, G.; Armstrong-James, D.; et al. Sub-telomere directed gene expression during initiation of invasive Aspergillosis. PLoS Pathog. 2008, 4, e1000154. [Google Scholar] [CrossRef]
- Doyle, S.; Jones, G.W.; Dolan, S.K. Dysregulated gliotoxin biosynthesis attenuates the production of unrelated biosynthetic gene cluster-encoded metabolites in Aspergillus fumigatus. Fungal Biol. 2018, 122, 214–221. [Google Scholar] [CrossRef]
- Lind, A.L.; Lim, F.Y.; Soukup, A.A.; Keller, N.P.; Rokas, A. An LaeA-and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. Msphere 2018, 3, e00050-18. [Google Scholar] [CrossRef]
- Kowalski, C.H.; Morelli, K.A.; Stajich, J.E.; Nadell, C.D.; Cramer, R.A. A heterogeneously expressed gene family modulates biofilm architecture and hypoxic growth of Aspergillus fumigatus. mBio 2020. [Google Scholar] [CrossRef]
- Dos Reis, T.F.; Silva, L.P.; de Castro, P.A.; de Lima, P.B.A.; do Carmo, R.A.; Marini, M.M.; da Silveira, J.F.; Ferreira, B.H.; Rodrigues, F.; Malavazi, I.; et al. The influence of genetic stability on Aspergillus fumigatus virulence and azole resistance. G3 Genes Genom. Genet. 2018, 8, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Perrin, R.M.; Fedorova, N.D.; Bok, J.W.; Cramer, R.A., Jr.; Wortman, J.R.; Kim, H.S.; Nierman, W.C.; Keller, N.P. Transcriptional Regulation of Chemical Diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007, 3, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Bowyer, P.; Bignell, E.; Alcazar-Fuolia, L.; Cairnsb, T.; Lopez, J.F.; Zonja, B.; Pérez, S.; Barceló, D. A modified recombineering protocol for the genetic manipulation of gene clusters in Aspergillus fumigatus. PLoS ONE 2014, 9, e111875. [Google Scholar]
- Palmer, J.M.; Keller, N.P. Secondary metabolism in fungi: Does chromosomal location matter? Curr. Opin. Microbiol. 2010, 13, 431–436. [Google Scholar] [CrossRef]
- Canela, H.M.S.; Cardoso, B.; Vitali, L.H.; Coelho, H.C.; Martinez, R.; da Silva Ferreira, M.E. Prevalence, virulence factors and antifungal susceptibility of Candida spp. isolated from bloodstream infections in a tertiary care hospital in Brazil. Mycoses 2018, 61, 11–21. [Google Scholar] [CrossRef]
- Naglik, J.R.; Richardson, J.P.; Moyes, D.L. Candida albicans pathogenicity and epithelial immunity. PLoS Pathog. 2014, 10, e1004257. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Legrand, M.; Jaitly, P.; Feri, A.; d’Enfert, C.; Sanyal, K. Candida albicans: An emerging yeast model to study eukaryotic genome plasticity. Trends Genet. 2019, 35, 292–307. [Google Scholar] [CrossRef]
- Dunn, M.J.; Anderson, M.Z. To repeat or not to repeat: Repetitive sequences regulate genome stability in Candida albicans. Genes 2019, 10, 866. [Google Scholar] [CrossRef]
- Haran, J.; Boyle, H.; Hokamp, K.; Yeomans, T.; Liu, Z.; Church, M.; Fleming, A.B.; Anderson, M.Z.; Berman, J.; Myers, L.C.; et al. Telomeric ORFs (TLO s) in Candida spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits. PLoS Genet. 2014, 10, e1004658. [Google Scholar] [CrossRef]
- McEachern, M.J.; Hicks, J.B. Unusually large telomeric repeats in the yeast Candida albicans. Mol. Cell. Biol. 1993, 13, 551–560. [Google Scholar] [CrossRef]
- Underwood, A.P.; Louis, E.J.; Borts, R.H.; Stringer, J.R.; Wakefield, A.E. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol. Microbiol. 1996, 19, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.; McEachern, M.J.; Dandjinou, A.T.; Tzfati, Y.; Orr, E.; Blackburn, E.H.; Lue, N.F. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. USA 2007, 104, 11682–11687. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Ong, E.B.B. Genomic Instability and Cellular Senescence: Lessons from the Budding Yeast. Front. Cell Dev. Biol. 2020, 8, 619126. [Google Scholar] [CrossRef]
- Yu, E.Y.; Wang, F.; Lei, M.; Lue, N.F. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 2008, 15, 985. [Google Scholar] [PubMed]
- Yu, E.Y.; Yen, W.F.; Steinberg-Neifach, O.; Lue, N.F. Rap1 in Candida albicans: An unusual structural organization and a critical function in suppressing telomere recombination. Mol. Cell. Biol. 2010, 30, 1254–1268. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.Z.; Baller, J.A.; Dulmage, K.; Wigen, L.; Berman, J. The three clades of the telomere-associated TLO gene family of Candida albicans have different splicing, localization, and expression features. Eukaryot. Cell 2012, 11, 1268–1275. [Google Scholar] [CrossRef]
- Anderson, M.Z.; Wigen, L.J.; Burrack, L.S.; Berman, J. Real-time evolution of a subtelomeric gene family in Candida albicans. Genetics 2015, 200, 907–919. [Google Scholar] [CrossRef]
- Fidel, P.L.; Vazquez, J.A.; Sobel, J.D. Candida glabrata: Review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 1999, 12, 80–96. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive Candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A.; Surveillance, A. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Rodrigues, M.E.; Silva, S.; Henriques, M. Candida glabrata biofilms: How far have we come? J. Fungi 2017, 3, 11. [Google Scholar] [CrossRef]
- Horn, D.L.; Neofytos, D.; Anaissie, E.J.; Fishman, J.A.; Steinbach, W.J.; Olyaei, A.J.; Mar, K.A.; Pfaller, M.A.; Chang, C.-H.; Webster, K.M. Epidemiology and outcomes of candidemia in 2019 patients: Data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 2009, 48, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Trick, W.E.; Fridkin, S.K.; Edwards, J.R.; Hajjeh, R.A.; Gaynes, R.P. National Nosocomial Infections Surveillance System Hospitals. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989–1999. Clin. Infect. Dis. 2002, 35, 627–630. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Hollis, R.J.; Boyken, L.; Tendolkar, S.; Kroeger, J.; Diekema, D.J. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location in the United States in 2001 to 2007. J. Clin. Microbiol. 2009, 47, 3185–3190. [Google Scholar] [CrossRef] [PubMed]
- Mota, S.; Alves, R.; Carneiro, C.; Silva, S.; Brown, A.J.; Istel, F.; Kuchler, K.; Sampaio, P.; Casal, M.; Henriques, M.; et al. Candida glabrata susceptibility to antifungals and phagocytosis is modulated by acetate. Front. Microbiol. 2015, 6, 919. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Fatahinia, M.; Halvaeizadeh, M.; Mahmoudabadi, A.Z.; AboualiGalehdari, E.; Kiasat, N. In vitro antifungal susceptibilities of six antifungal drugs against clinical Candida glabrata isolates according to EUCAST. Curr. Med. Mycol. 2020, 6, 1–6. [Google Scholar] [CrossRef]
- Iraqui, I.; Garcia-Sanchez, S.; Aubert, S.; Dromer, F.; Ghigo, J.M.; D’Enfert, C.; Janbon, G.; Enfert, C.; Janbon, G. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol. Microbiol. 2005, 55, 1259–1271. [Google Scholar] [CrossRef]
- Kachouri-Lafond, R.; Dujon, B.; Gilson, E.; Westhof, E.; Fairhead, C.; Teixeira, M.T. Large telomerase RNA, telomere length heterogeneity and escape from senescence in Candida glabrata. FEBS Lett. 2009, 583, 3605–3610. [Google Scholar] [CrossRef]
- Cormack, B.P.; Ghori, N.; Falkow, S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef]
- Gallegos-García, V.; Pan, S.J.; Juárez-Cepeda, J.; Ramírez-Zavaleta, C.Y.; Martin-del-Campo, M.B.; Martínez-Jiménez, V.; Castaño, I.; Cormack, B.; de Las Peñas, A. A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in candida glabrata. Genetics 2012, 190, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- De Las Peñas, A.; Pan, S.J.; Castaño, I.; Alder, J.; Cregg, R.; Cormack, B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003, 17, 2245–2258. [Google Scholar] [CrossRef]
- Castaño, I.; Pan, S.J.; Zupancic, M.; Hennequin, C.; Dujon, B.; Cormack, B.P. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2005, 55, 1246–1258. [Google Scholar] [CrossRef]
- Shubin, C.B.; Greider, C.W. The role of Rif1 in telomere length regulation is separable from its role in origin firing. Elife 2020, 9, e58066. [Google Scholar] [CrossRef]
- Rosas-Hernández, L.L.; Juárez-Reyes, A.; Arroyo-helguera, O.E.; De Las Peñas, A.; Pan, S.J.; Cormack, B.P.; Castaño, I.; Rosas-herna, L.L.; Jua, A.; Arroyo-helguera, O.E.; et al. yKu70/yKu80 and Rif1 regulate silencing differentially at telomeres in Candida glabrata. Eukaryot. Cell 2008, 7, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregela, S.; Lafentaine, I.; De Montigny, J.; Marck, C.; Talla, E.; Goffard, N.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, S.C.; Smith, M.C.A.; Muston, P.; Holland, S.L.; Avery, S.V. Heterogeneous expression of the virulence-related adhesin epa1 between individual cells and strains of the pathogen Candida glabrata. Eukaryot. Cell 2012, 11, 141–150. [Google Scholar] [CrossRef]
- Domergue, R.; Castaño, I.; De Las Peñas, A.; Zupancic, M.; Lockatell, V.; Hebel, J.R.; Johnson, D.; Cormack, B.P. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 2005, 308, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Orta-Zavalza, E.; Guerrero-Serrano, G.; Gutiérrez-Escobedo, G.; Cañas-Villamar, I.; Juárez-Cepeda, J.; Castaño, I.; De Las Peñas, A. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol. Microbiol. 2013, 88, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Reyes, A.; Ramírez-Zavaleta, C.Y.; Medina-Sánchez, L.; de Las Peñas, A.; Castaño, I. A protosilencer of subtelomeric gene expression in Candida glabrata with unique properties. Genetics 2012, 190, 101–111. [Google Scholar] [CrossRef]
- López-Fuentes, E.; Hernández-Hernández, G.; Castanedo, L.; Gutiérrez-Escobedo, G.; Oktaba, K.; De las Peñas, A.; Castaño, I. Chromatin loop formation induced by a subtelomeric protosilencer represses EPA genes in Candida glabrata. Genetics 2018, 210, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Zavaleta, C.Y.; Salas-Delgado, G.E.; de Las Peñas, A.; Castaño, I. Subtelomeric silencing of the MTL3 locus of Candida glabrata requires yKu70, yKu80, and Rif1 proteins. Eukaryot. Cell 2010, 9, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Jong, H.S.; Myung, J.C.; Jeong, W.S.; Jung, S.I.; Cho, D.; Seung, J.K.; Soo, H.K.; Myung, G.S.; Soon, P.S.; Dong, W.R. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 2007, 45, 2385–2391. [Google Scholar]
- Skalski, J.H.; Kottom, T.J.; Limper, A.H. Pathobiology of Pneumocystis pneumonia: Life cycle, cell wall and cell signal transduction. FEMS Yeast Res. 2015, 15, 1–12. [Google Scholar] [CrossRef]
- Cissé, O.H.; Pagni, M.; Hauser, P.M. Comparative genomics suggests that the human pathogenic fungus pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol. Evol. 2014, 6, 1938–1948. [Google Scholar] [CrossRef]
- Ambrose, H.E.; Keely, S.P.; Aliouat, E.M.; Dei-Cas, E.; Wakefield, A.E.; Miller, R.F.; Stringer, J.R. Expression and complexity of the PRT1 multigene family of Pneumocystis carinii. Microbiology 2004, 150, 293–300. [Google Scholar] [CrossRef]
- Keely, S.P.; Renauld, H.; Wakefield, A.E.; Cushion, M.T.; Smulian, A.G.; Fosker, N.; Fraser, A.; Harris, D.; Murphy, L.; Price, C.; et al. Gene arrays at Pneumocystis carinii telomeres. Genetics 2005, 170, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, Z.; Huang, D.W.; Kutty, G.; Ishihara, M.; Wang, H.; Abouelleil, A.; Bishop, L.; Davey, E.; Deng, R.; et al. Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat. Commun. 2016, 7, 10740. [Google Scholar] [CrossRef]
- Schmid-Siegert, E.; Richard, S.; Luraschi, A.; Mühlethaler, K.; Pagni, M.; Hauser, P.M. Mechanisms of surface antigenic variation in the human pathogenic fungus Pneumocystis jirovecii. bioRxiv 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Keely, S.P.; Stringer, J.R. Complexity of the MSG gene family of Pneumocystis carinii. BMC Genom. 2009, 10, 367. [Google Scholar] [CrossRef] [PubMed]
- Stringer, J.R.; Cushion, M.T., II. The genome of Pneumocystis carinii. FEMS Immunol. Med. Microbiol. 1998, 22, 15–26. [Google Scholar] [CrossRef]
- Zhang, J.; Cushion, M.T.; Stringer, J.R. Molecular characterization of a novel repetitive element from Pneumocystis carinii from rats. J. Clin. Microbiol. 1993, 31, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M. Telomeres and genomic evolution. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160437. [Google Scholar] [CrossRef] [PubMed]
- Hauser, P.M. Is the unique camouflage strategy of Pneumocystis associated with its particular niche within host lungs? PLoS Pathog. 2019, 15, e1007480. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Z.; Huang, D.W.; Cissé, O.H.; Rothenburger, J.L.; Latinne, A.; Bishop, L.; Blair, R.; Brenchley, J.M.; Chabé, M.; et al. Diversity and Complexity of the Large Surface Protein Family in the Compacted Genomes of Multiple Pneumocystis Species. mBio 2020, 11, e02878-19. [Google Scholar] [CrossRef]
- Wada, M.; Nakamura, Y. Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 1996, 3, 55–64. [Google Scholar] [CrossRef]
- Shay, J.W. Telomeres and aging. Curr. Opin. Cell Biol. 2018, 52, 1–7. [Google Scholar] [CrossRef]
- Wyatt, N.A.; Richards, J.K.; Brueggeman, R.S.; Friesen, T.L. A comparative genomic analysis of the barley pathogen Pyrenophora teres f. teres identifies subtelomeric regions as drivers of virulence. Mol. Plant Microbe Interact. 2020, 33, 173–188. [Google Scholar] [CrossRef]
- Gunisova, S.; Elboher, E.; Nosek, J.; Gorkovoy, V.; Brown, Y.; Lucier, J.F.; Laterreur, N.; Wellinger, R.J.; Tzfati, Y.; Tomaska, L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. Rna 2009, 15, 546–559. [Google Scholar] [CrossRef]
- Koering, C.E.; Fourel, G.; Binet-Brasselet, E.; Laroche, T.; Klein, F.; Gilson, E. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 2000, 28, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Preti, M.; Ribeyre, C.; Pascali, C.; Bosio, M.C.; Cortelazzi, B.; Rougemont, J.; Guarnera, E.; Naef, F.; Shore, D.; Dieci, G. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell. 2010, 38, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Buscaino, A. Chromatin-Mediated Regulation of Genome Plasticity in Human Fungal Pathogens. Genes 2019, 10, 855. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.Z.; Hollin, T.; Lenz, T.; Le Roch, K.G. Three-dimensional chromatin in infectious disease—A role for gene regulation and pathogenicity? PLoS Pathog. 2021, 17, e1009207. [Google Scholar] [CrossRef]

| Fungal Species | Telomere Repeat Sequence | Number of bp Repeats |
|---|---|---|
| Aspergillus fumigatus | 5′-TTAGGG-3′ | 6 |
| Candida albicans | 5′-ACTTCTTGGTGTACGGATGTCTA-3′ | 23 |
| Candida glabrata | 5′-CTGGGTGCTGTGGGGT-3′ | 16 |
| Pneumocystis jirovecii | 5′-TTAGGG-3′ | 6 |
| Gene/Genes in Subtelomeres | Protein/Metabolite | Function | References |
|---|---|---|---|
| Fumitremorgin biosynthesis supercluster | Pseurotin | Neuritogenic, nematicidal quinone | [28,29] |
| LaeA gene | LaeA methyltransferase | Global transcriptional regulator of secondary metabolite biosynthesis; virulence factor | [30] |
| bafA | Biofilm Architecture Factor A | Modulates hyphae formation and ability to survive low oxygen conditions increasing pathogenicity | [31] |
| atmA and atrA | Ataxia-telangiectasia mutated kinases | Chromosomal polymorphism | [32] |
| Gene/Genes | Protein/Metabolite | Function | References |
|---|---|---|---|
| TER1 | Telomerase RNA component | Involved in production and maintenance of telomeres to prevent cell senescence | [40,44] |
| EST2 | Telomerase reverse transcriptase | Involved in production and maintenance of telomeres to prevent cell senescence | [40,44] |
| EST3 | Subunit of telomerase regulatory complex | Telomerase stimulatory function | [45,46] |
| RAP1 | Repressor activator protein 1 | Regulator of telomere length and structure; works in conjunction with Ku70, Stn1, and Ten1 | [47] |
| TLO genes | Highly variable Telomere associated metabolites | Includes Med2 and other subunit components of mediator complex and transcriptional responses; Involved in adaptability, hyphal growth, and oxidative stress responses | [41,48] |
| Gene/Genes | Protein/Metabolite | Function | References |
|---|---|---|---|
| EPA1–7 | Large adhesin proteins (glycosylphosphatidylinositol-anchored cell wall protein) | Adherence; clusters of subtelomeric EPA genes are involved in colonization and biofilm formation | [62,63] |
| SIR 1–4 | Histone deacetylase and silencing proteins | Involved in transcriptional repression, gene silencing, and adhesion | [64] |
| RAP1 | Repressor activator protein 1 | Regulator of telomere length and structure; Involved in gene silencing | [64] |
| RIF1–2 | Rif1 and Rif2 regulatory Proteins | Regulators of telomere length and structure; Involved in gene silencing | [66] |
| HDF1–2 | yKu70/80 | Telomere maintenance; non-homologous end join repair | [67] |
| Gene/Genes | Protein/Metabolite | Function | References |
|---|---|---|---|
| MSG | Major Surface Glycoproteins | Components of cell wall and involved in antigenic variation evading host immune responses | [81,83] |
| MSR | MSG-related antigens | Part of the MSG superfamily involved in isoform switching and immune evasion with antigenic variation | [77] |
| PRT1 | Cell surface protease | Involved with MSR in immune evasion with antigenic variation | [77] |
| Family | No. of MSG Genes | Characteristics | ||||||
|---|---|---|---|---|---|---|---|---|
| P. murina | P. jirovecii | Average Gene (kb)/Protein (kDa) Size | No. of Introns | 5′-end Leader Sequence | No. of Domains | Expression Mode | ||
| MSG-A1c | 22 | 86 | 3.2/120 | 0 | CRJEd | 9 | Mutually exclusive | |
| MSG-A2c | 14 | 0 | 2.9/218 | 1 | Highly conserved | 7–9 | Independently | |
| MSG-A3c | 6 | 33 | 3.1/117 | 1–2 | Highly variable | 9 | Independently | |
| MSG-B | 0 | 21 | 1.4/55 | 1–2 | Highly variable | 2–3 | Independently | |
| MSG-C | 6 | 2 | 1.7/60 | 1 | Highly conserved | 3 | Independently | |
| MSG-D | 1 | 20 | 2.9/111 | 1 | Highly variable | 3–6 | Independently | |
| MSG-E | 7 | 5 | 1.2/49 | 1 | Highly variable | 1 | Independently | |
| Unclassified | 8 | 12 | 0.9/37 | 0–8 | Highly variable | 0–1 | Unknown | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diotti, R.; Esposito, M.; Shen, C.H. Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens. Microorganisms 2021, 9, 1405. https://doi.org/10.3390/microorganisms9071405
Diotti R, Esposito M, Shen CH. Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens. Microorganisms. 2021; 9(7):1405. https://doi.org/10.3390/microorganisms9071405
Chicago/Turabian StyleDiotti, Raffaella, Michelle Esposito, and Chang Hui Shen. 2021. "Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens" Microorganisms 9, no. 7: 1405. https://doi.org/10.3390/microorganisms9071405
APA StyleDiotti, R., Esposito, M., & Shen, C. H. (2021). Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens. Microorganisms, 9(7), 1405. https://doi.org/10.3390/microorganisms9071405






