Stimulation of Nicotiana tabacum L. In Vitro Shoot Growth by Endophytic Bacillus cereus Group Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Cultivable Endophytic Bacteria
2.2. Identification of Bacterial Isolates and Bacterial Genome Analysis
2.3. Bacterial Genome Sequencing, Annotation and Comparative Analysis
2.4. Tobacco In Vitro Shoot Co-Cultivation with Endophytic Bacteria
2.5. Analysis of Endophytic Bacteria Density in Tobacco Shoot Tissues
3. Results
3.1. Isolation and Identification of Cultivable Endophytic Bacteria
3.2. Endophytic Bacteria Co-Cultivation Effect on Tobacco Shoot Biomass Accumulation
3.3. Survival of Endophytic Bacteria Isolates in Tobacco Shoot Tissues
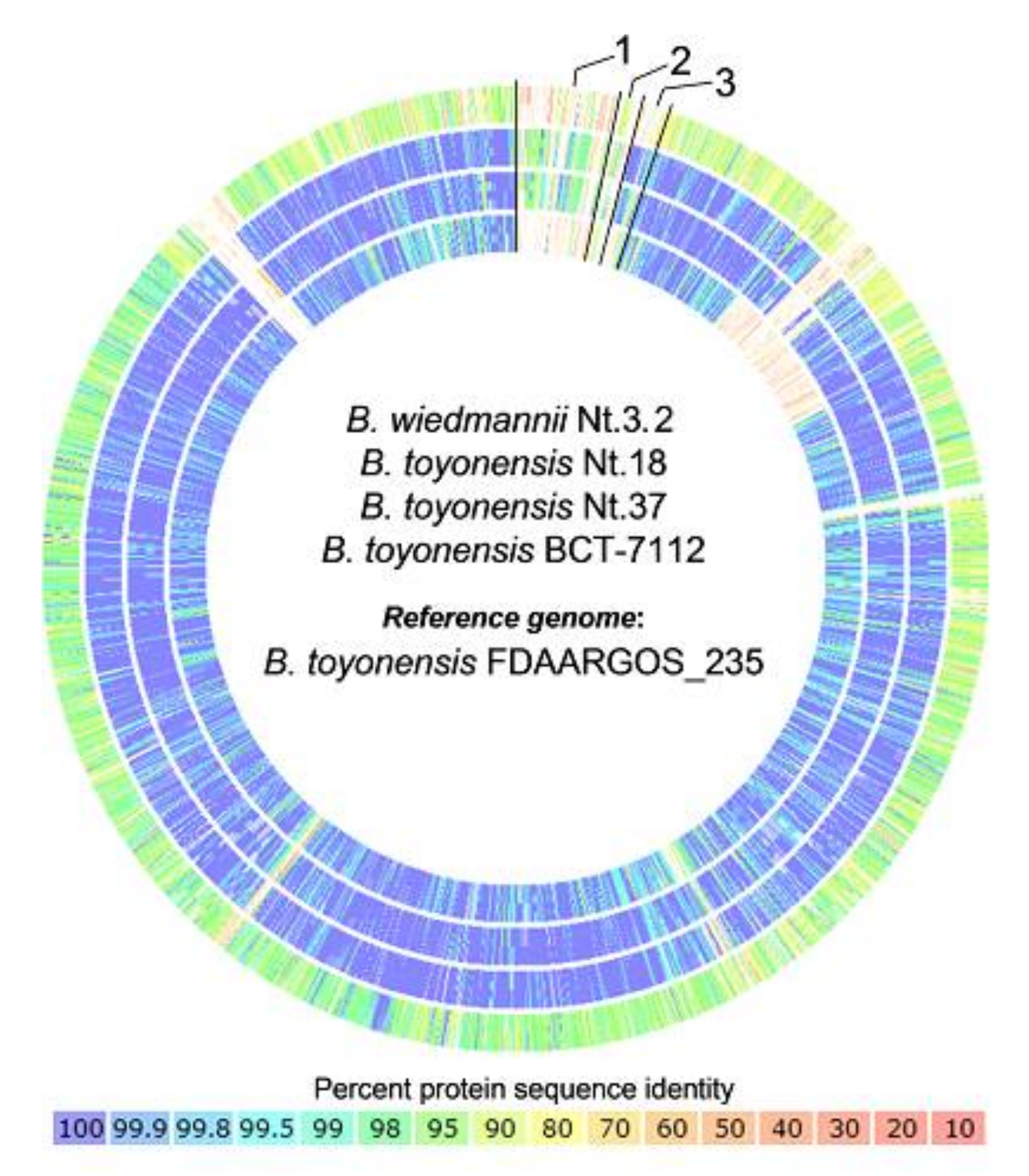
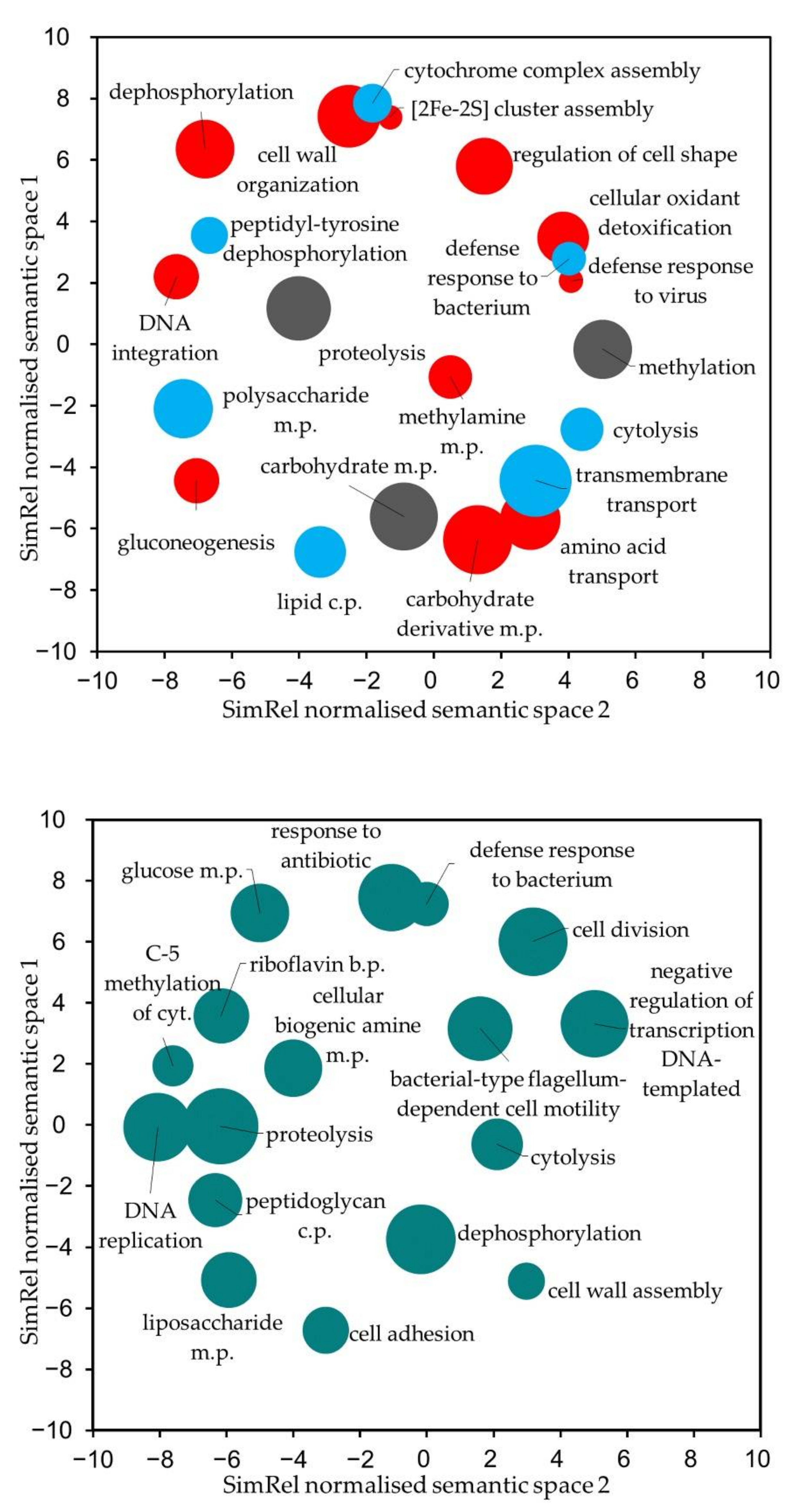
3.4. Comparative Genome Analysis of Closely Related B. toyonensis Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Clade No. a | Bacterial Strain | Accession b | Accession Length, bp | Sequence Identity, % | Reference |
|---|---|---|---|---|---|
| 1 A | Bacillus thuringiensis IAM 12077 | NR_043403.1 | 1486 | 97.3–99.9 | [106] |
| 1 B | Bacillus mobilis MCCC 1A05942 | NR_157731.1 | 1509 | 97.1–99.8 | [107] |
| 1 C | Bacillus mycoides DSM 11821 | NR_024697.1 | 1531 | 97.8–100.0 | [108] |
| 2 | Bacillus marisflavi TF-11 | NR_025240.1 | 1506 | 98.1–98.9 | [109] |
| 3 | Bacillus simplex NBRC 15720 = DSM 1321 | NR_112726.1 | 1476 | 95.4–99.7 | d.s. |
| 4 | Bacillus aryabhattai B8W22 | NR_115953.1 | 1533 | 95.0–97.5 | [110] |
| 5 | Bacillus pumilus NBRC 12092 | NR_112637.1 | 1474 | 99.89 | d.s. |
| 6 | Pseudomonas koreensis Ps 9-14 | NR_025228.1 | 1455 | 99.5 | [111] |
| Parameter | Isolate | ||
|---|---|---|---|
| Nt.18 | Nt.37 | Nt.3.2 | |
| Illumina demultiplexing and read mapping statistics | |||
| Number of read pairs | 2,468,194 | 2,298,751 | 2,950,725 |
| Yield (mbp) | 666 | 596 | 797 |
| Average quality | 36.03 | 36.23 | 36.14 |
| Total reads | 4,936,388 | 4,597,502 | 5,901,450 |
| Mapped reads | 4,899,758 | 4,571,302 | 5,869,378 |
| Average coverage | 119.6 | 107.16 | 135.82 |
| Insert size median | 317 | 272 | 300 |
| De novo short-read assembly statistics | |||
| Genome Length (bp) | 5,574,424 | 5,586,422 | 5,891,672 |
| GC Content (%) | 35.0626 | 35.090523 | 34.933907 |
| Contigs N50 (bp) | 564,507 | 276,397 | 324,644 |
| Number of scaffolds | 52 | 90 | 91 |
| Average scaffold size (bp) | 107,200 | 62,071 | 64,743 |
| Max scaffold size (bp) | 1,307,898 | 934,026 | 1,427,494 |
| Min scaffold size (bp) | 311 | 587 | 335 |
| Number of gaps | 3 | 5 | 5 |
| Genome quality statistics | |||
| Completeness (%) | 99.34 | 99.43 | 99.34 |
| Contamination (%) | 0.23 | 0.11 | 0.1 |
| Closest placement taxonomic assignment | |||
| Species | Bacillus toyonensis | Bacillus toyonensis | Bacillus wiedmannii |
| GTDB species representative | BCT-7112 | BCT-7112 | FSL W8-0169 |
| GTDB reference | GCF_000496285.1 | GCF_000496285.1 | GCF_001583695.1 |
| TYGS accession | NCIMB 14858 | NCIMB 14858 | FSL W8-0169 |
| ANI (%) | 99.39 | 99.33 | 96.49 |
| Alignment fraction (%) | 0.95 | 0.95 | 0.91 |
| Annotation statistics | |||
| Features | 5853 | 5905 | 6424 |
| Coding gene | 5761 | 5814 | 6217 |
| RNAs | 92 | 91 | 96 |
| Distinct functions | 4049 | 4072 | 4136 |
| Non-hypothetical proteins | 3707 | 3729 | 3933 |
| Hypothetical proteins | 2054 | 2085 | 2284 |
| Query | Reference | ANI Estimate | Matches | Total |
|---|---|---|---|---|
| Nt.18 | Nt.37 | 99.554 | 1740 | 1833 |
| Nt.37 | Nt.18 | 99.482 | 1746 | 1814 |
| Nt.18 | Nt.3.2 | 91.356 | 1527 | 1833 |
| Nt.37 | Nt.3.2 | 91.328 | 1529 | 1814 |
| Nt.3.2 | Nt.18 | 91.242 | 1553 | 1923 |
| Nt.3.2 | Nt.37 | 91.222 | 1528 | 1923 |
| Parameter | Isolate | |
|---|---|---|
| Nt.18 | Nt.37 | |
| Annotation statistics | ||
| Features | 5853 | 5905 |
| Coding gene | 5761 | 5814 |
| RNAs | 92 | 91 |
| Distinct functions | 4049 | 4072 |
| Non-hypothetical proteins | 3707 | 3729 |
| Hypothetical proteins | 2054 | 2085 |
| Comparative analysis statistics | ||
| Genes in core homolog families | 5576 | 5594 |
| Genes in singletons | 185 (60) a | 220 (20) a |
| Genes with ≥90% identity | 5425 | |
| Genes with <90% identity | 151 (52) a | |
| Symbol | Genome Feature No. | Gene Name | Id., % | Pfam | |||
|---|---|---|---|---|---|---|---|
| Nt.18 | Nt.37 | Domain | Reference | E Value | |||
| Cluster number 36; Cluster type LAP | |||||||
| Core biosynthetic genes | |||||||
| HP | 5593 | Hypothetical protein | YcaO | PF02624.16 | 4 × 10−58 | ||
| HP | 5594 | Hypothetical protein | Nitroreductase | PF00881.24 | 5.3 × 10−11 | ||
| Cluster number 20; Cluster Type: saccharide, EPS related cluster | |||||||
| Core biosynthetic genes | |||||||
| UDPGE | 3491 | 2631 | UDP-glucose 4-epimerase (EC 5.1.3.2) | 100 | Epimerase | PF01370.21 | 1.5 × 10−57 |
| MAEP | 3503 | Multi antimicrobial extrusion protein (Na(+)/drug antiporter), MATE family of MDR efflux pumps | Polysacc synt Polysacc synt C | PF01943.17 PF14667.6 | 9.6 × 10−16 2 × 10−9 | ||
| HP | 3506 | Hypothetical protein | Glycos transf 1 Glyco transf 4 | PF00534.20 PF13439.6 | 9.1 × 10−28 8.6 × 10−9 | ||
| UFS | 3508 | UDP-N-acetyl-L-fucosamine synthase (EC 5.1.3.28) | Epimerase 2 | PF02350.19 | 1.9 × 10−88 | ||
| CPBP | 3509 | Capsular polysaccharide synthesis enzyme Cap5F | Epimerase GPI | PF01370.21 PF06560.11 | 4.4 × 10−20 4.7 × 10−5 | ||
| UGADH | 3510 | UDP-N-acetylglucosamine 4,6-dehydratase (EC 4.2.1.135) | Polysacc synt 2 Polysacc syn 2C | PF02719.15 PF08485.10 | 3 × 10−103 9.2 × 10−23 | ||
| CPBP | 3511 | Capsular polysaccharide biosynthesis protein Cps4F | Glycos transf 1 Glyco trans 4 4 | PF00534.20 PF13579.6 | 3.2 × 10−18 5.4 × 10−13 | ||
| UPGPT | 3512 | 2613 | Undecaprenyl-phosphate galactosephosphotransferase (EC2.7.8.6) | 40 | Bac transf | PF02397.16 | 2.1 × 10−62 |
| PPBP | 3513 | Probable polysaccharide biosynthesis protein EpsC | Polysacc synt 2 CoA binding 3 | PF02719.15 PF13727.6 | 7 × 10−124 6 × 10−20 | ||
| UGP | 3514 | 2612 | UTP--glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) | 96 | NTP transferase | PF00483.23 | 2.2 × 10−35 |
| HP | 2615 | hypothetical protein | 19 | Glycos transf 1 | PF00534.20 | 2.1 × 10−13 | |
| HP | 2616 | Hypothetical protein | Polysacc synt | PF01943.17 | 4.5 × 10−15 | ||
| GTF | 2620 | Glycosyltransferase | 22 | Glyco trans 1 4 | PF13692.6 | 3 × 10−13 | |
| EPGTF | 2621 | Exopolysaccharide biosynthesis glycosyltransferase EpsF (EC 2.4.1.) | Glyco transf 4 Glycos transf 1 | PF13439.6 PF00534.20 | 1.4 × 10−19 1.6 × 10−24 | ||
| GTF | 2622 | Glycosyltransferase | Glyco transf 4 Glycos transf 1 | PF13439.6 PF00534.20 | 1.3 × 10−8 3.2 × 10−26 | ||
| SPGT | 2623 | Sugar-phosphate guanylyltransferase/Sugar-phosephate isomerase | NTP transferase MannoseP isomer | PF00483.23 PF01050.18 | 2 × 10−32 2.3 × 10−19 | ||
| Additional biosynthetic and transport-related genes | |||||||
| CPCT | 3494 | 2628 | Choline-phosphate cytidylyltransferase (EC 2.7.7.15)/Choline kinase (EC 2.7.1.32) | 99 | HTH 24 NTP transf 3 Choline kinase | PF13412.6 PF12804.7 PF01633.20 | 1.3 × 10−11 3.5 × 10−9 9.5 × 10−34 |
| UMDH | 2614 | UDP-N-acetyl-D-mannosamine dehydrogenase (EC 1.1.1.336) | UDPG MGDP dh N UDPG MGDP dh UDPG MGDP dh C | PF03721.14 PF00984.19 PF03720.15 | 5 × 10−58 5.8 × 10−28 8.1 × 10−20 | ||
| HP | 2618 | Hypothetical protein | Acyl transf 3 | PF01757.22 | 1.3 × 10−31 | ||
| HP | 3493 | 2629 | Hypothetical protein | 100 | EamA | PF00892.20 | 9.3 × 10−12 |
| Other genes | |||||||
| HP | 3507 | Hypothetical protein | |||||
| HP | 3505 | Hypothetical protein | O-ag pol Wzy | PF14296.6 | 4.7 × 10−23 | ||
| HP | 3504 | Hypothetical protein | MAF flag10 | PF01973.18 | 6.7 × 10−14 | ||
| KDSB | 3502 | 3-deoxy-manno-octulosonate cytidylyltransferase (EC 2.7.7.38) | CTP transf 3 | PF02348.19 | 2.9 × 10−55 | ||
| KDOPS | 3501 | 2-Keto-3-deoxy-D-manno-octulosonate-8-phosphate synthase (EC 2.5.1.55) | DAHP synth 1 | PF00793.20 | 1 × 10−71 | ||
| API | 3500 | D-arabinose-5-phosphate isomerase (EC 5.3.1.13) | SIS CBS | PF01380.22 PF00571.28 | 1.1 × 10−28 1.3 × 10−8 | ||
| KDOPP | 3499 | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (EC 3.1.3.45) | Hydrolase 3 | PF08282.12 | 2.6 × 10−9 | ||
| HP | 3498 | Hypothetical protein | HIT | PF01230.23 | 3.1 × 10−7 | ||
| LCP | 3497 | 2625 | Cell envelope-associated transcriptional attenuator LytR-CpsA-Psr, subfamily F2 | 75 | LytR cpsA psr | PF03816.14 | 2 × 10−50 |
| EPSX | 3496 | 2626 | EPSX protein | 99 | |||
| HP | 3495 | 2627 | Hypothetical protein | 100 | LicD | PF04991.13 | 3.5 × 10−6 |
| CPCT | 3492 | 2630 | Choline-phosphate cytidylyltransferase (EC 2.7.7.15) | 98 | NTP transferase | PF00483.23 | 1.8 × 10−12 |
| HP | 2617 | Hypothetical protein | Hepar II III N Hepar II III | PF16889.5 PF07940.13 | 3.2 × 10−16 5.4 × 10−21 | ||
| HP | 2619 | Hypothetical protein | |||||
| CPCT | 2624 | Mannose-6-phosphate isomerase (EC 5.3.1.8) | PMI typeI | PF01238.21 | 5.8 × 10−47 | ||


References
- Schulz, B.; Boyle, C. What are endophytes? In Microbial root Endophytes Soil Biology; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 9, pp. 1–13. [Google Scholar]
- Abreu-Tarazi, M.F.; Navarrete, A.A.; Andreote, F.D.; Almeida, C.V.; Tsai, S.M.; Almeida, M. Endophytic bacteria in long-term in vitro cultivated “axenic” pineapple microplants revealed by PCR–DGGE. World J. Microbiol. Biotechnol. 2010, 26, 555–560. [Google Scholar] [CrossRef]
- Miliute, I.; Buzaite, O.; Baniulis, D.; Stanys, V. Bacterial endophytes in agricultural crops and their role in stress tolerance: A review. Zemdirbyste 2015, 102, 465–478. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Krug, L.; Yang, H.; Li, H.; Yang, M.; Berg, G.; Cernava, T. Nicotiana tabacum seed endophytic communities share a common core structure and genotype-specific signatures in diverging cultivars. Comput. Struct. Biotechnol. J. 2020, 18, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, L.; Zhang, J.; Zhang, X.; Xue, Y.; Liu, J.; Zou, X. Characterization of the core microbiome in tobacco leaves during aging. Microbiologyopen 2020, 9, e984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Huang, L.; Xia, Z.; Zhao, X.; Xu, S.; Lin, L.; Ren, Z.; Jin, S.; Wang, M. Species diversity characteristics of endophytic bacteria in tobacco at different regions of Yunnan Province. SW China J. Agric. Sci. 2015, 28, 857–861. [Google Scholar]
- Gao, L.; Kong, F.; Feng, C.; Wang, J.; Gao, J.; Shen, G.; Zhang, C. Isolation, characterization, and growth promotion of phosphate-solubilizing bacteria associated with Nicotiana tabacum (tobacco). Pol. J. Environ. Stud. 2016, 25, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Read, P.E.; Preece, J.E. Cloning: Plants—Micropropagation/tissue culture. In Encyclopedia of Agriculture and Food Systems; Van Alfen, N.K., Ed.; Academic Press: San Diego, CA, USA, 2014; Volume 2, pp. 317–336. [Google Scholar]
- Bhatia, S.; Sharma, K. Micropropagation. In Modern Applications of Plant Biotechnology in Pharmaceutical Sciences; Bhatia, S., Sharma, K., Dahiya, R., Bera, T., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 361–368. [Google Scholar] [CrossRef]
- Kukkurainen, S.; Leino, A.; Vähämiko, S.; Kärkkäinen, H.R.; Ahanen, K.; Sorvari, S.; Rugienius, R.; Toldi, O. Occurrence and location of endophytic bacteria in garden and wild strawberry. HortScience 2005, 40, 348. [Google Scholar] [CrossRef] [Green Version]
- Leone, G.F.; Andrade, P.A.M.; de Almeida, C.V.; de Almeida, C.V.; Andreote, F.D.; de Almeida, M. Use of antibiotics to control endophytic bacterial growth migration onto culture medium in Eucalyptus cloeziana F.Muell.: A micropropagation approach. In Vitro Cell Dev. Biol. Plant 2019, 55, 421–432. [Google Scholar] [CrossRef]
- Kulkarni, A.A.; Kelkar, S.; Watve, M.; Krishnamurthy, K. Characterization and control of endophytic bacterial contaminants in in vitro cultures of Piper spp., Taxus baccata subsp. wallichiana, and Withania somnifera. Can. J. Microbiol. 2007, 53, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Pirttilä, A.M.; Podolich, O.; Koskimäki, J.J.; Hohtola, E.; Hohtola, A. Role of origin and endophyte infection in browning of bud-derived tissue cultures of Scots pine (Pinus sylvestris L.). Plant Cell Tiss. Org. Cult. 2008, 95, 47–55. [Google Scholar] [CrossRef]
- Thomas, P.; Swarna, G.K.; Patil, P.; Rawal, R.D. Ubiquitous presence of normally non-culturable endophytic bacteria in field shoot-tips of banana and their gradual activation to quiescent cultivable form in tissue cultures. Plant Cell Tiss. Org. Cult. 2008, 93, 39–54. [Google Scholar] [CrossRef]
- Ray, S.S.; Ali, M.N.; Mukherjee, S.; Chatterjee, G.; Banerjee, M. Elimination and molecular identification of endophytic bacterial contaminants during in vitro propagation of Bambusa balcooa. World J. Microbiol. Biotechnol. 2017, 33, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, K.; Stauber, T.; Ewald, D. Paenibacillus—A predominant endophytic bacterium colonising tissue cultures of woody plants. Plant Cell Tiss. Org. Cult. 2008, 93, 347–351. [Google Scholar] [CrossRef]
- Botta, A.L.; Santacecilia, A.; Ercole, C.; Cacchio, P.; Gallo, M.D. In vitro and in vivo inoculation of four endophytic bacteria on Lycopersicon esculentum. New Biotechnol. 2013, 30, 666–674. [Google Scholar] [CrossRef]
- Salomon, M.V.; Bottini, R.; de Souza Filho, G.A.; Cohen, A.C.; Moreno, D.; Gil, M.; Piccoli, P. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant 2014, 151, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Quambusch, M.; Brümmer, J.; Haller, K.; Winkelmann, T.; Bartsch, M. Dynamics of endophytic bacteria in plant in vitro culture: Quantification of three bacterial strains in Prunus avium in different plant organs and in vitro culture phases. Plant Cell Tiss. Org. Cult. 2016, 126, 305–317. [Google Scholar] [CrossRef]
- Maggini, V.; Mengoni, A.; Gallo, E.R.; Biffi, S.; Fani, R.; Firenzuoli, F.; Bogani, P. Tissue specificity and differential effects on in vitro plant growth of single bacterial endophytes isolated from the roots, leaves and rhizospheric soil of Echinacea purpurea. BMC Plant Biol. 2019, 19, 284. [Google Scholar] [CrossRef] [Green Version]
- Dias, A.C.F.; Costa, F.E.C.; Andreote, F.D.; Lacava, P.T.; Teixeira, M.A.; Assumpção, L.C.; Araújo, W.L.; Azevedo, J.L.; Melo, I.S. Isolation of micropropagated strawberry endophytic bacteria and assessment of their potential for plant growth promotion. World J. Microbiol. Biotechnol. 2009, 25, 189–195. [Google Scholar] [CrossRef] [Green Version]
- Tamosiune, I.; Staniene, G.; Haimi, P.; Stanys, V.; Rugienius, R.; Baniulis, D. Endophytic Bacillus and Pseudomonas spp. modulate apple shoot growth, cellular redox balance, and protein expression under in vitro conditions. Front. Plant Sci. 2018, 9, 889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassells, A.C.; Curry, R.F. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: Implications for micropropagators and genetic engineers. Plant Cell Tiss. Org. Cult. 2001, 64, 145–157. [Google Scholar] [CrossRef]
- Gaspar, T.; Franck, T.; Bisbis, B.; Kevers, C.; Jouve, L.; Hausman, J.F.; Dommes, J. Concepts in plant stress physiology. Application to plant tissue cultures. Plant Growth. Regul. 2002, 37, 263–285. [Google Scholar] [CrossRef]
- Desjardins, Y.; Dubuc, J.F.; Badr, A. In vitro culture of plants: A stressful activity! Acta Hortic 2009, 812, 29–50. [Google Scholar] [CrossRef]
- Balen, B.; Tkalec, M.; Pavoković, D.; Pevalek-Kozlina, B.; Krsnik-Rasol, M. Growth conditions in in vitro culture can induce oxidative stress in Mammillaria gracilis tissues. J. Plant Growth. Regul. 2009, 28, 36–45. [Google Scholar] [CrossRef]
- Rojas-Martinez, L.; Visser, R.G.F.; de Klerk, G.J. The hyperhydricity syndrome: Waterlogging of plant tissues as a major cause. Propag. Ornam. 2010, 10, 169–175. [Google Scholar]
- Zhang, P.; Li, X.; Yuan, X.L.; Du, Y.M.; Wang, B.G.; Zhang, Z.F. Antifungal prenylated diphenyl ethers from Arthrinium arundinis, an endophytic fungus isolated from the leaves of tobacco (Nicotiana tabacum L.). Molecules 2018, 23, 3179. [Google Scholar] [CrossRef] [Green Version]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madden, T.L.; Tatusov, R.L.; Zhang, J. Applications of network BLAST server. Methods Enzymol. 1996, 266, 131–141. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 1825. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. nTaxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef] [Green Version]
- Nikolenko, S.I.; Korobeynikov, A.I.; Alekseyev, M.A. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genom. 2013, 14, S7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [Green Version]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome. Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The united states department of energy systems Biology knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome. Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Toronen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlicker, A.; Domingues, F.S.; Rahnenfuhrer, J.; Lengauer, T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinform. 2006, 7, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- Van Bloois, L.v.d.G.; Wagenaar, J.A.; Zomer, A.L. RFPlasmid: Predicting plasmid sequences from short read assembly data using machine learning. bioRxiv 2020, 2020.07.31.230631. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; González-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marvasi, M.; Visscher, P.T.; Casillas Martinez, L. Exopolymeric substances (EPS) from Bacillus subtilis: Polymers and genes encoding their synthesis. FEMS Microbiol. Lett. 2010, 313, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Gordon, R.E. One hundred and seven years of the genus Bacillus. In The Aerobic Endospore-Forming Bacteria: Classification and Identification; Berkeley, R.C., Goodfellow, M., Eds.; Academic Press: London, UK; New York, NY, USA, 1981; pp. 1–15. [Google Scholar]
- Cho, W.-I.; Chung, M.-S. Bacillus spores: A review of their properties and inactivation processing technologies. Food Sci. Biotechnol. 2020, 29, 1447–1461. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Prakash, A.; Johri, B. Bacillus as PGPR in crop ecosystem. In Bacteria in Agrobiology: Crop Ecosystems; Maheshwari, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 37–59. [Google Scholar] [CrossRef]
- Lopes, R.; Tsui, S.; Gonçalves, P.J.R.O.; De Queiroz, M.V. A look into a multifunctional toolbox: Endophytic Bacillus species provide broad and underexploited benefits for plants. World J. Microbiol. Biotechnol. 2018, 34, 94. [Google Scholar] [CrossRef]
- Azizoglu, U. Bacillus thuringiensis as a biofertilizer and biostimulator: A mini-review of the little-known plant growth-promoting properties of Bt. Curr. Microbiol. 2019, 76, 1379–1385. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd_Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Schisler, D.A.; Slininger, R.J.; Behle, R.W.; Jackson, M.A. Formulation of Bacillus spp. for biological control of plant diseases. Phytopathology 2004, 94, 1267–1271. [Google Scholar] [CrossRef] [Green Version]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Asaf, S.; Lee, I.J. What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front. Plant Sci. 2018, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, L.K.; Jackson, M.A. Clarification of the taxonomy of Bacillus mycoides. Int. J. Syst. Evol. Microbiol. 1995, 45, 46–49. [Google Scholar] [CrossRef] [Green Version]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a taxonomic nomenclature for the Bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. MBio 2020, 11, e00034-20. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, G.; Urdiain, M.; Cifuentes, A.; López-López, A.; Blanch, A.R.; Tamames, J.; Kämpfer, P.; Kolstø, A.-B.; Ramón, D.; Martínez, J.F.; et al. Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst. Appl. Microbiol. 2013, 36, 383–391. [Google Scholar] [CrossRef] [Green Version]
- Guan, P.; Ai, P.; Dai, X.; Zhang, J.; Xu, L.; Zhu, J.; Li, Q.; Deng, Q.; Li, S.; Wang, S.; et al. Complete genome sequence of Bacillus thuringiensis serovar Sichuansis strain MC28. J. Bacteriol. 2012, 194, 6975. [Google Scholar] [CrossRef] [Green Version]
- Gagne-Bourgue, F.; Aliferis, K.A.; Seguin, P.; Rani, M.; Samson, R.; Jabaji, S. Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. J. Appl. Microbiol. 2013, 114, 836–853. [Google Scholar] [CrossRef]
- Herrera, S.D.; Grossi, C.; Zawoznik, M.; Groppa, M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 2016, 186, 37–43. [Google Scholar] [CrossRef]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Asaf, S.; Lee, I.J. Plant growth-promoting endophytic bacteria versus pathogenic infections: An example of Bacillus amyloliquefaciens RWL-1 and Fusarium oxysporum f. sp lycopersici in tomato. PeerJ 2017, 5, e3107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras-Perez, M.; Hernandez-Salmeron, J.; Rojas-Solis, D.; Rocha-Granados, C.; Orozco-Mosqueda, M.D.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Santoyo, G. Draft genome analysis of the endophyte, Bacillus toyonensis COPE52, a blueberry (Vaccinium spp. var. Biloxi) growth-promoting bacterium. 3 Biotech 2019, 9, 370. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Solis, D.; Vences-Guzman, M.A.; Sohlenkamp, C.; Santoyo, G. Bacillus toyonensis COPE52 modifies lipid and fatty acid composition, exhibits antifungal activity, and stimulates growth of tomato plants under saline conditions. Curr. Microbiol. 2020, 77, 2735–2744. [Google Scholar] [CrossRef]
- Rocha, F.Y.O.; de Oliveira, C.M.; da Silva, P.R.A.; de Melo, L.H.V.; do Carmo, M.G.F.; Baldani, J.I. Taxonomical and functional characterization of Bacillus strains isolated from tomato plants and their biocontrol activity against races 1, 2 and 3 of Fusarium oxysporum f. sp Lycopersici. Appl. Soil. Ecol. 2017, 120, 8–19. [Google Scholar] [CrossRef]
- Lopes, R.; Cerdeira, L.; Tavares, G.S.; Ruiz, J.C.; Blom, J.; Horacio, E.C.A.; Mantovani, H.C.; de Queiroz, M.V. Genome analysis reveals insights of the endophytic Bacillus toyonensis BAC3151 as a potentially novel agent for biocontrol of plant pathogens. World J. Microbiol. Biotechnol. 2017, 33, 185. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Beno, S.M.; Kent, D.J.; Carroll, L.M.; Martin, N.H.; Boor, K.J.; Kovac, J. Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int. J. Syst. Evol. Microbiol. 2016, 66, 4744–4753. [Google Scholar] [CrossRef]
- Osman, N.I.; Shixue, Y. Isolation and characterization of pea plant (Pisum sativum L.) growth-promoting Rhizobacteria. Afr. J. Microbiol. Res. 2018, 12, 820–828. [Google Scholar] [CrossRef]
- Bach, E.; Seger, G.D.d.S.; Fernandes, G.d.C.; Lisboa, B.B.; Passaglia, L.M.P. Evaluation of biological control and rhizosphere competence of plant growth promoting bacteria. Appl. Soil. Ecol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Rajapaksha, R.M.C.P.; Herath, D.; Senanayake, A.P.; Senevirathne, M.G.T.L. Mobilization of rock phosphate phosphorus through bacterial inoculants to enhance growth and yield of wetland rice. Commun. Soil. Sci. Plant Anal. 2011, 42, 301–314. [Google Scholar] [CrossRef]
- Ambrosini, A.; Stefanski, T.; Lisboa, B.B.; Beneduzi, A.; Vargas, L.K.; Passaglia, L.M.P. Diazotrophic bacilli isolated from the sunflower rhizosphere and the potential of Bacillus mycoides B38V as biofertiliser. Ann. Appl. Biol. 2016, 168, 93–110. [Google Scholar] [CrossRef]
- Guetskyl, R.; Shtienberg, D.; Dinoor, A.; Elad, Y. Establishment, survival and activity of the biocontrol agents Pichia guilermondii and Bacillus mycoides applied as a mixture on strawberry plants. Biocontrol. Sci. Technol. 2002, 12, 705–714. [Google Scholar] [CrossRef]
- Malekzadeh, E.; Alikhani, H.A.; Savaghebi-Firoozabadi, G.R.; Zarei, M. Bioremediation of cadmium-contaminated soil through cultivation of maize inoculated with plant growth–promoting rhizobacteria. Bioremediat. J. 2012, 16, 204–211. [Google Scholar] [CrossRef]
- Thomas, P. Isolation of Bacillus pumilus from in vitro grapes as a long-term alcohol-surviving and rhizogenesis inducing covert endophyte. J. Appl. Microbiol. 2004, 97, 114–123. [Google Scholar] [CrossRef]
- Joo, G.-J.; Kim, Y.-M.; Lee, I.-J.; Song, K.-S.; Rhee, I.-K. Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macroides and Bacillus pumilus. Biotechnol. Lett. 2004, 26, 487–491. [Google Scholar] [CrossRef]
- De-Bashan, L.E.; Hernandez, J.-P.; Bashan, Y.; Maier, R. Bacillus pumilus ES4: Candidate plant growth-promoting bacterium to enhance establishment of plants in mine tailings. Environ. Exp. Bot. 2010, 69, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Akinrinlola, R.J.; Yuen, G.Y.; Drijber, R.A.; Adesemoye, A.O. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018, 2018, 5686874. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.K.; Bisht, S.C.; Ruwari, P.; Selvakumar, G.; Joshi, G.K.; Bisht, J.K.; Bhatt, J.C.; Gupta, H.S. Alleviation of cold stress in inoculated wheat (Triticum aestivum L.) seedlings with psychrotolerant Pseudomonads from NW Himalayas. Arch. Microbiol. 2011, 193, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, C.; Bakshi, U.; Mallick, I.; Mukherji, S.; Bera, B.; Ghosh, A. Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211. Front. Microbiol. 2017, 8, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Oliveira, A.L.M.; Urquiaga, S.; Döbereiner, J.; Baldani, J.I. The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil. 2002, 242, 205–215. [Google Scholar] [CrossRef]
- Tiwari, R.; Kalra, A.; Darokar, M.P.; Chandra, M.; Aggarwal, N.; Singh, A.K.; Khanuja, S.P. Endophytic bacteria from Ocimum sanctum and their yield enhancing capabilities. Curr. Microbiol. 2010, 60, 167–171. [Google Scholar] [CrossRef]
- Fan, D.; Subramanian, S.; Smith, D.L. Plant endophytes promote growth and alleviate salt stress in Arabidopsis thaliana. Sci. Rep. 2020, 10, 12740. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 2014, 27, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Cocq, K.L.; Gurr, S.J.; Hirsch, P.R.; Mauchline, T.H. Exploitation of endophytes for sustainable agricultural intensification. Mol. Plant Pathol. 2017, 18, 469–473. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, J.A.; Donia, M.S.; Schmidt, E.W. Insights into heterocyclization from two highly similar enzymes. J. Am. Chem. Soc. 2010, 132, 4089–4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.M.; Milne, J.C.; Madison, L.L.; Kolter, R.; Walsh, C.T. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science 1996, 274, 1188–1193. [Google Scholar] [CrossRef]
- Collin, F.; Maxwell, A. The microbial toxin microcin B17: Prospects for the development of new antibacterial agents. J. Mol. Biol. 2019, 431, 3400–3426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Molohon, K.J.; Melby, J.O.; Lee, J.; Evans, B.S.; Dunbar, K.L.; Bumpus, S.B.; Kelleher, N.L.; Mitchell, D.A. Structure determination and interception of biosynthetic intermediates for the plantazolicin class of highly discriminating antibiotics. ACS Chem. Biol. 2011, 6, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Banala, S.; Ensle, P.; Süssmuth, R.D. Total synthesis of the ribosomally synthesized linear szole-containing peptide plantazolicin A from Bacillus amyloliquefaciens. Angew. Chem. Int. Ed. 2013, 52, 9518–9523. [Google Scholar] [CrossRef]
- Wanke, A.; Malisic, M.; Wawra, S.; Zuccaro, A. Unraveling the sugar code: The role of microbial extracellular glycans in plant–microbe interactions. J. Exp. Bot. 2020, 72, 15–35. [Google Scholar] [CrossRef]
- Lemon, K.P.; Earl, A.M.; Vlamakis, H.C.; Aguilar, C.; Kolter, R. Biofilm development with an emphasis on Bacillus subtilis. Curr. Top. Microbiol. Immunol. 2008, 322, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Cech, D.L.; Markin, K.; Woodard, R.W. Identification of a d-arabinose-5-phosphate isomerase in the Gram-positive Clostridium tetani. J. Bacteriol. 2017, 199, e00246-17. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Yamasato, K. Phylogeny of spore-forming lactic acid bacteria based on 16S rRNA gene sequences. FEMS Microbiol. Lett. 1994, 115, 13–17. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Lai, Q.; Zeng, R.; Ye, D.; Xu, J.; Shao, Z. Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 2017, 67, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Omura, T.; Hara, Y.; Sadaie, Y. Application of the partial 16S rDNA sequence as an index for rapid identification of species in the genus Bacillus. J. Gen. Appl. Microbiol. 2000, 46, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.-H.; Kim, I.-G.; Kang, K.H.; Oh, T.-K.; Park, Y.-H. Bacillus marisflavi sp. nov. and Bacillus aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Shivaji, S.; Chaturvedi, P.; Begum, Z.; Pindi, P.K.; Manorama, R.; Padmanaban, D.A.; Shouche, Y.S.; Pawar, S.; Vaishampayan, P.; Dutt, C.B.S.; et al. Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 2009, 59, 2977–2986. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.W.; Kim, J.S.; Park, I.C.; Yoon, S.H.; Park, D.H.; Lim, C.K.; Go, S.J. Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 21–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andriūnaitė, E.; Tamošiūnė, I.; Aleksandravičiūtė, M.; Gelvonauskienė, D.; Vinskienė, J.; Rugienius, R.; Baniulis, D. Stimulation of Nicotiana tabacum L. In Vitro Shoot Growth by Endophytic Bacillus cereus Group Bacteria. Microorganisms 2021, 9, 1893. https://doi.org/10.3390/microorganisms9091893
Andriūnaitė E, Tamošiūnė I, Aleksandravičiūtė M, Gelvonauskienė D, Vinskienė J, Rugienius R, Baniulis D. Stimulation of Nicotiana tabacum L. In Vitro Shoot Growth by Endophytic Bacillus cereus Group Bacteria. Microorganisms. 2021; 9(9):1893. https://doi.org/10.3390/microorganisms9091893
Chicago/Turabian StyleAndriūnaitė, Elena, Inga Tamošiūnė, Monika Aleksandravičiūtė, Dalia Gelvonauskienė, Jurgita Vinskienė, Rytis Rugienius, and Danas Baniulis. 2021. "Stimulation of Nicotiana tabacum L. In Vitro Shoot Growth by Endophytic Bacillus cereus Group Bacteria" Microorganisms 9, no. 9: 1893. https://doi.org/10.3390/microorganisms9091893







