Characterization of D-17 Canine Osteosarcoma Cell Line and Evaluation of Its Ability to Response to Infective Stressor Used as Alternative Anticancer Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Reference Genes Selection
2.3. Gene Expression Analysis
2.4. Immunocytochemical Assay
2.5. Innate Immune Response to Infective Stressor
2.5.1. Experiment 1: Bacterial Invasion
2.5.2. Experiment 2: Cell Viability after Treatment
2.5.3. Experiment 3: Modulation of Innate Immune Responses
2.6. Sequencing of the Key Gene CXCR4
2.7. Statistical Analyses
3. Results
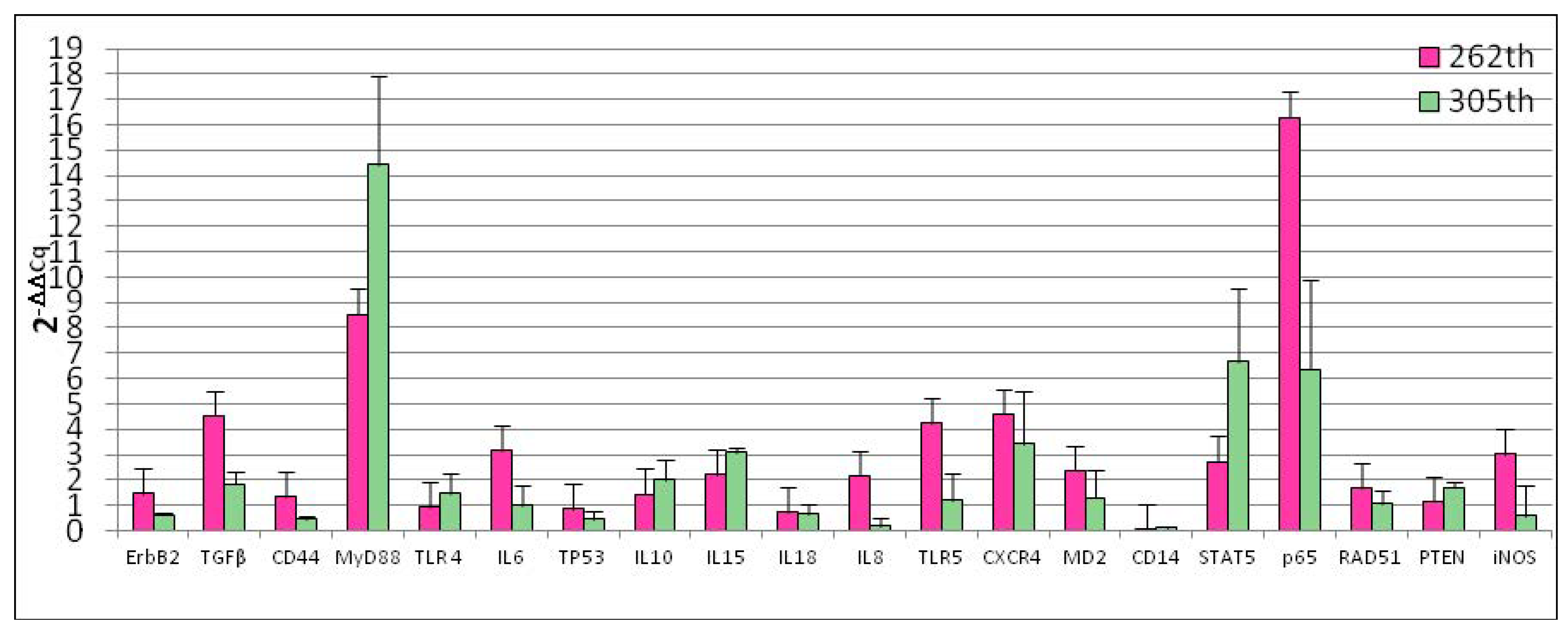
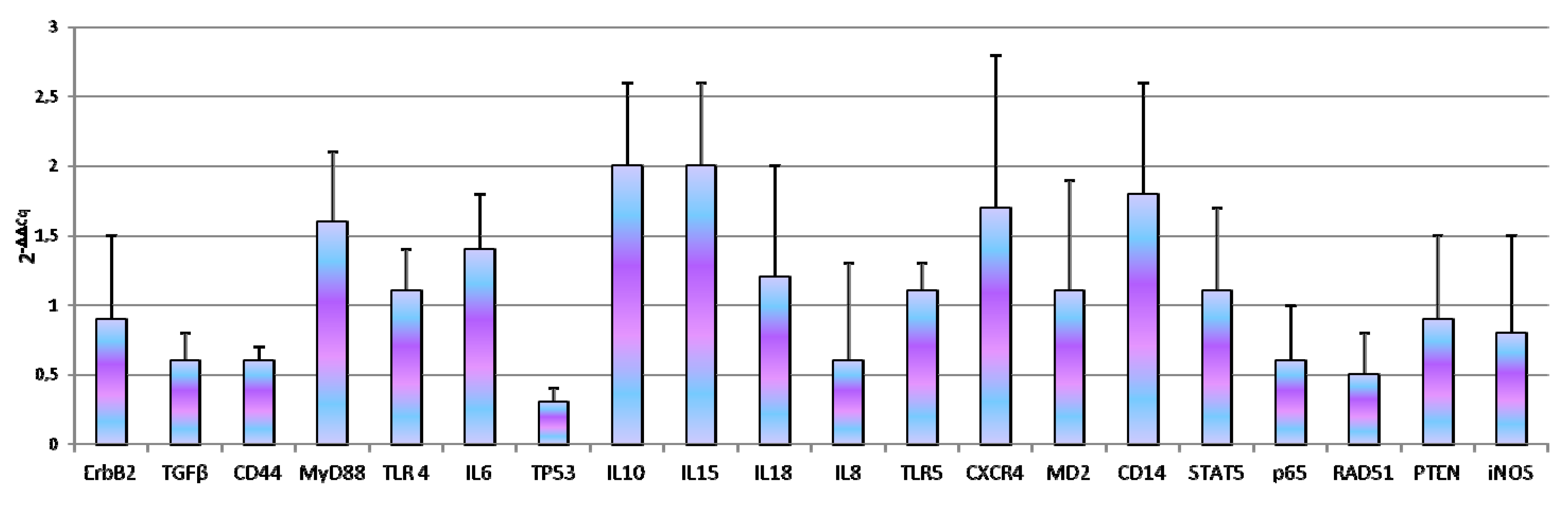
3.1. Reference Gene Selection and Basal Gene Expression
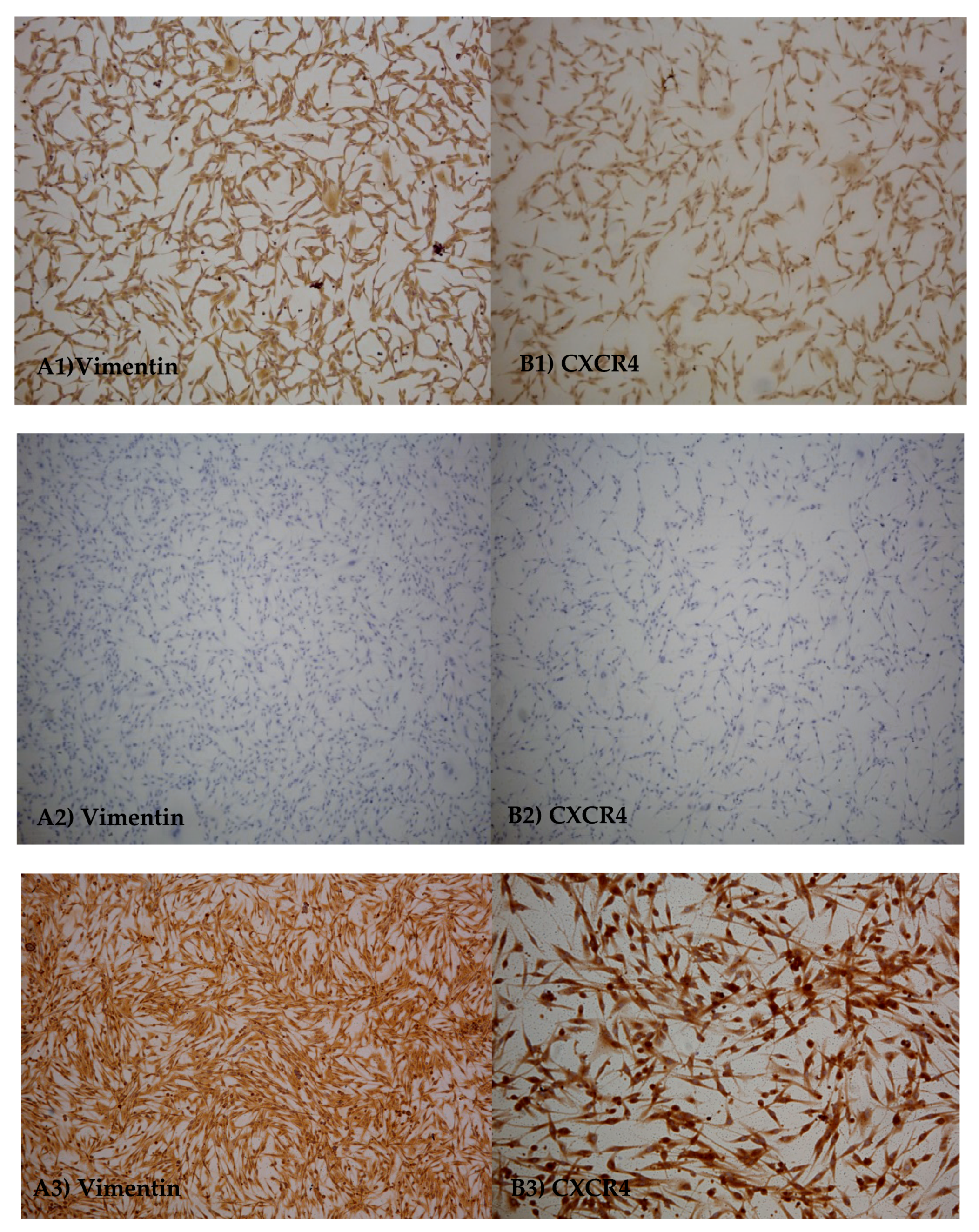
3.2. Immuno-Cytochemicals Assay
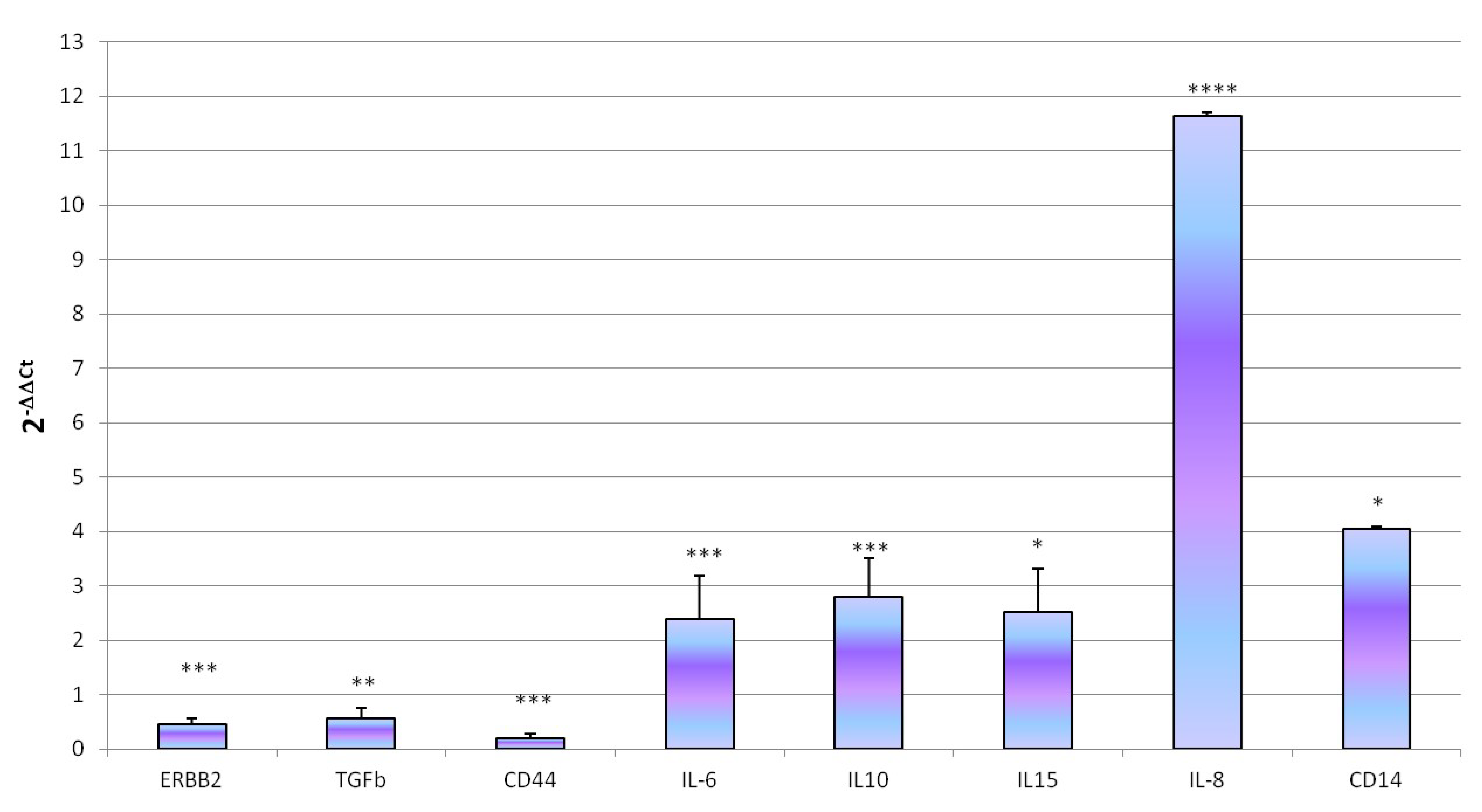
3.3. Innate Immune Response to Infective Stressor
3.4. Sequencing of the Key Gene CXCR4
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gorlick, R. Current concepts on the molecular biology of osteosarcoma. Cancer Treat. Res. 2009, 152, 467–478. [Google Scholar] [CrossRef]
- Wilson, H.; Heulsmeyer, M.; Chun, R.; Young, K.M.; Friedrichs, K.; Argyle, D.J. Isolation and characterization of cancer stem cells from canine osteosarcoma. Vet. J. 2008, 175, 69–75. [Google Scholar] [CrossRef]
- Makielski, K.M.; Mills, L.J.; Sarver, A.L.; Henson, M.S.; Spector, L.G.; Naik, S.; Modiano, J.F. Risk Factors for Development of Canine and Human Osteosarcoma: A Comparative Review. Vet. Sci. 2019, 6, 48. [Google Scholar] [CrossRef] [Green Version]
- Simpson, S.; Dunning, M.D.; de Brot, S.; Grau-Roma, L.; Mongan, N.P.; Rutland, C.S. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet. Scand. 2017, 59, 71. [Google Scholar] [CrossRef]
- Fenger, J.M.; London, C.A.; Kisseberth, W.C. Canine Osteosarcoma: A Naturally Occurring Disease to Inform Pediatric Oncology. ILAR J. 2014, 55, 69–85. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Rédini, F.; Gouin, F.; Cherrier, B.; Heymann, D. Osteosarcoma: Current status of immunotherapy and future trends. Oncol. Rep. 2006, 15, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henklewska, M.; Pawlak, A.; Kutkowska, J.; Pruchnik, H.; Rapak, A.; Obminska-Mrukowicz, B. In Vitro effects of the activity of novel platinum (II) complex in canine and human cell lines. Vet. Comp. Oncol. 2019, 17, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Chirullo, B.; Ammendola, S.; Leonardi, L.; Falcini, R.; Petrucci, P.; Pistoia, C.; Vendetti, S.; Battistoni, A.; Pasquali, P. Attenuated mutant strain of Salmonella Typhimurium lacking the ZnuABC transporter contrasts tumor growth promoting anti-cancer immune response. Oncotarget 2015, 6, 17648–17660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvarajah, G.T.; Bonestroo, F.A.S.; Timmermans Sprang, E.P.M.; Kirpenstejin, J.; Mol, J.A. Reference gene validation for gene expression normalization in canine osteosarcoma: A geNorm algorithm approach. BMC Vet. Res. 2017, 13, 354–361. [Google Scholar] [CrossRef] [Green Version]
- Weinmann, M.A.; Fischer, J.A.; Jacobs, D.C.; Goodall, C.P.; Bracha, S.; Chappell, P.E. Autocrine production of reproductive axis neuropeptides affects proliferation of canine osteosarcoma In Vitro. BMC Cancer 2019, 19, 158. [Google Scholar] [CrossRef]
- Ayers, D.; Clements, D.N.; Salway, F.; Day, P.J.R. Expression stability of commonly used reference genes in canine articular connective tissues. BMC Vet. Res. 2007, 3, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, A.; Oliveira, J.T.; Gärtner, F.; Kohn, B.; Gruber, A.D.; Klopfleisch, R. Potential markers for detection of circulating canine mammary tumor cells in the peripheral blood. Vet. J. 2011, 190, 165–168. [Google Scholar] [CrossRef]
- Maissen-Villiger, C.A.; Schweighauser, A.; Dorland, H.A.; Morel, C.; Bruckmaier, R.M.; Zurbriggen, A.; Francey, T. Expression Profile of Cytokines and Enzymes mRNA in Blood Leukocytes of Dogs with Leptospirosis and Its Associated Pulmonary Hemorrhage Syndrome. PLoS ONE 2016, 11, e0148029. [Google Scholar] [CrossRef] [Green Version]
- Capellini, F.M.; Vencia, W.; Amadori, M.; Mignone, G.; Parisi, E.; Masiello, L.; Vivaldi, B.; Ferrari, A.; Razzuoli, E. Characterization of MDCK cells and evaluation of their ability to respond to infectious and non-infectious stressors. Cytotechnology 2020, 72, 97–109. [Google Scholar] [CrossRef]
- Cavalcanti, A.S.; Ribeiro-Alves, M.; Pereira, L.O.; Mestre, G.L.; Ferreira, A.B.; Morgado, F.N.; Boité, M.C.; Cupolillo, E.; Moraes, M.O.; Porrozzi, R. Parasite load induces progressive spleen architecture breakage and impairs cytokine mRNA expression in Leishmania infantum-naturally infected dogs. PLoS ONE 2015, 10, e0123009. [Google Scholar] [CrossRef] [PubMed]
- Peeters, D.; Peters, I.R.; Farnir, F.; Clercx, C.; Day, M.J. Real-time RT-PCR quantification of mRNA encoding cytokines and chemokines in histologically normal canine nasal, bronchial and pulmonary tissue. Vet. Immunol. Immunopathol. 2005, 104, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.W.; Shin, I.S.; Bhang, D.H.; Lee, D.-H.; Bae, B.-K.; Kang, M.-S.; Kim, D.-Y.; Hwang, C.-Y.; Lee, C.-W.; Youn, H.-Y. Hormonal change and cytokine mRNA expression in peripheral blood mononuclear cells during the development of canine autoimmune thyroiditis. Clin. Exp. Immunol. 2006, 146, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.; Leitão, S.; Henriques, S.; Kowalewski, M.P.; Hoffmann, B.; Ferreira-Dias, G.; Lopes da Costa, L.; Mateus, L. Gene transcription of TLR2, TLR4, LPS ligands and prostaglandin synthesis enzymes are up-regulated in canine uteri with cystic endometrial hyperplasia-pyometra complex. J. Reprod. Immunol. 2010, 84, 66–74. [Google Scholar] [CrossRef]
- Ishikawa, S.; Takemitsu, H.; Li, G.; Mori, N.; Yamamoto, I.; Arai, T. Short communication: Molecular characterization of dog and cat p65 subunits of NF-kappaB. Res. Vet. Sci. 2015, 99, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Kurata, K.; Iwata, A.; Masuda, K.; Sakaguchi, M.; Ohno, K.; Tsujimoto, H. Identification of CpG oligodeoxynucleotide sequences that induce IFN-gamma production in canine peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 2004, 102, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Turchetti, A.P.; da Costa, L.F.; Romão Ede, L.; Fujiwara, R.T.; da Paixão, T.A.; Santos, R.L. Transcription of innate immunity genes and cytokine secretion by canine macrophages resistant or susceptible to intracellular survival of Leishmania infantum. Vet. Immunol. Immunopathol. 2015, 163, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Kaim, U.; Moritz, A.; Failing, K.; Baumgärtner, W. The regression of a canine Langerhans cell tumour is associated with increased expression of IL-2, TNF-alpha, IFN-gamma and iNOS mRNA. Immunology 2006, 4, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Klopfleisch, R.; Gruber, A.D. Derlin-1 and stanniocalcin-1 are differentially regulated in metastasizing canine mammary adenocarcinomas. J. Comp. Pat. 2009, 141, 113–120. [Google Scholar] [CrossRef]
- Kanae, Y.; Endoh, D.; Yokota, H.; Taniyama, H.; Hayashi, M. Expression of the PTEN tumor suppressor gene in malignant mammary gland tumors of dogs. Am. J. Vet. Res. 2006, 67, 127–133. [Google Scholar] [CrossRef]
- Razzuoli, E.; Amadori, M.; Lazzara, F.; Bilato, D.; Ferraris, M.; Vito, G.; Ferrari, A. Salmonella serovar-specific interaction with jejunal epithelial cells. Vet. Microbiol. 2017, 207, 219–225. [Google Scholar] [CrossRef]
- Razzuoli, E.; Villa, R.; Sossi, E.; Amadori, M. Reverse transcription real-time PCR for detection of porcine interferon α and β genes. Scand. J. Immunol. 2011, 74, 412–418. [Google Scholar] [CrossRef]
- Paoloni, M.; Khanna, C. Translation of new cancer treatments from pet dogs to humans. Nat. Rev. Cancer 2008, 8, 147–156. [Google Scholar] [CrossRef]
- Russell, D.S.; Jaworski, L.; Kisseberth, W.C. Immunohistochemical detection of p53, PTEN, Rb, and p16 in canine osteosarcoma using tissue microarray. J. Vet. Diagn. Investig. 2018, 30, 504–509. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Zhou, Y.; Ma, B.; Chen, X.; Wen, Y.; Liu, Y.; Fan, Q.; Qiu, X. The clinical value of CXCR4, HER2 and CD44 in human osteosarcoma: A pilot study. Oncol. Lett. 2012, 3, 797–801. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genßler, S.; Schönfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2015, 108. [Google Scholar] [CrossRef] [PubMed]
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, function, and association with the malignant process. Adv. Cancer Res. 1997, 71, 241–319. [Google Scholar] [CrossRef]
- Fan, T.M.; Barger, A.M.; Fredrickson, R.L.; Fitzsimmons, D.; Garrett, L.D. Investigating CXCR4 Expression in Canine Appendicular Osteosarcoma. J. Vet. Intern. Med. 2008, 22, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.J.; Gibson, J.T. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Heishima, K.; Meuten, T.; Yoshida, K.; Mori, T.; Thamm, D.H. Prognostic significance of circulating microRNA-214 and -126 in dogs with appendicular osteosarcoma receiving amputation and chemotherapy. Vet. Res. 2019, 15, 39. [Google Scholar] [CrossRef]
- Hoddinott, K.; Oblak, M.L.; Wood, G.A.; Boston, S.; Mutsaers, A.J. Evaluation of effects of radiation therapy combined with either pamidronate or zoledronate on canine osteosarcoma cells. Can. J. Vet. Res. 2019, 83, 3–10. [Google Scholar] [PubMed]
- Morrow, J.J.; Khanna, C. Osteosarcoma Genetics and Epigenetics: Emerging Biology and Candidate Therapies. Crit. Rev. Oncog. 2015, 20, 173–197. [Google Scholar] [CrossRef] [Green Version]
- Swiatkowska, A.; Zydowicz, P.; Sroka, J.; Ciesiołka, J. The role of the 5’ terminal region of p53 mRNA in the p53 gene expression. Rev. Acta Biochim. Pol. 2016, 63, 645–651. [Google Scholar] [CrossRef]
- Hopkins, B.D.; Hodakoski, C.; Barrows, D.; Mense, S.M.; Parsons, R.E. PTEN function: The long and the short of it. Rev. Trends Biochem. Sci. 2014, 39, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Levine, R.A.; Forest, T.; Smith, C. Tumor suppressor PTEN is mutated in canine osteosarcoma cell lines and tumors. Vet. Pathol. 2002, 39, 372–378. [Google Scholar] [CrossRef]
- Koch, A.E.; Polverini, P.J.; Kunkel, S.L.; Harlow, L.A.; DiPietro, L.A.; Elner, V.M.; Elner, S.G.; Strieter, R.M. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science 1992, 258, 1798–1801. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, X.; Miao, W.; Wang, B.; Qiu, Y. CXCL8 promotes the invasion of human osteosarcoma cells by regulation of PI3K/Akt signaling pathway. APMIS 2017, 125, 773–780. [Google Scholar] [CrossRef]
- Du, L.; Han, X.; Tu, B.; Wang, M.; Qiao, H.; Zhang, S.; Fan, Q.; Tang, T. CXCR1/Akt signaling activation induced by mesenchymal stem cell-derived IL-8 promotes osteosarcoma cell anoikis resistance and pulmonary metastasis. Cell Death Dis. 2018, 9, 714. [Google Scholar] [CrossRef] [PubMed]
- Achek, A.; Yesudhas, D.; Choi, S. Toll-like receptors: Promising therapeutic targets for inflammatory diseases. Arch. Pharm. Res. 2016, 39, 1032–1049. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Csakai, A.; Jin, J.; Zhang, F.; Yin, H. Therapeutic Developments Targeting Toll-like Receptor-4-Mediated Neuroinflammation. ChemMedChem 2016, 11, 154–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henneke, P.; Takeuchi, O.; Malley, R.; Lien, E.; Ingalls, R.R.; Freeman, M.W.; Mayadas, T.; Nizet, V.; Akira, S.; Kasper, D.L.; et al. Cellular activation, phagocytosis, and bactericidal activity against group B streptococcus involve parallel myeloid differentiation factor 88-dependent and independent signaling pathways. J. Immunol. 2002, 169, 3970–3977. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr. Med. Chem. 2019, 26, 3026–3041. [Google Scholar] [CrossRef]
- Payne, D.; Drinkwater, S.; Baretto, R.; Duddridge, M.; Browning, M.J. Expression of chemokine receptors CXCR4, CXCR5 and CCR7 on B and T lymphocytes from patients with primary antibody deficiency. Clin. Exp. Immunol. 2009, 156, 254–262. [Google Scholar] [CrossRef] [Green Version]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. CXCR4 signaling in health and disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef]
- Hampton, H.R.; Bailey, J.; Tomura, M.; Brink, R.; Chtanova, T. Microbe-dependent lymphatic migration of neutrophils modulates lymphocyte proliferation in lymph nodes. Nat. Commun. 2015, 6, 139. [Google Scholar] [CrossRef]
- Chirullo, B.; Pesciaroli, M.; Drumo, R.; Ruggeri, J.; Razzuoli, E.; Pistoia, C.; Petrucci, P.; Martinelli, N.; Cucco, L.; Moscati, L.; et al. Salmonella Typhimurium exploits inflammation to its own advantage in piglets. Front. Microbiol. 2015, 6, 985. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primers | Efficiency | R2 | Slope | References/ Designed |
|---|---|---|---|---|---|
| RPS5 | F: TCACTGGTGAGAACCCCCT R: CCTGATTCACACGGCGTAG | 106.9% | 0.998 | −3.167 | [9] |
| B2M | F: TCCTCATCCTCCTCGCT R: TTCTCTGCTGGGTGTCG | 111.8% | 0.990 | −3.000 | [9] |
| GUSB | F: AGACGTTCCAAGTACCCC R: AGGTGTGGTGTAGAGGAGCAC | 99.7% | 0.998 | −3.328 | [9] |
| GAPDH | F: CTGGGGCTCACTTGAAAGG R: GGAGGCATTGCTGACAATC | 96.7% | 0.999 | −3.404 | Designed |
| HPRT1 | F: CTGAAGAGCTACTGTAATGACCAGTC R: CTTTTCACCAGCAAGCTTGCAACC | 101.6% | 0.987 | −3.284 | Designed |
| RPL13A | F: GGGGCAGGTCCTGGTGCTCG R: CCAGGTACTTCAACTTGTTTCTGTAG | 97.7% | 0.999 | −3.378 | Designed |
| RPS19 | F: CCTTCCTCAAAAAGTCTGGG R: GCTGTGGAAGCAGCTCGC | 97.0% | 0.999 | −3.396 | [10] Designed |
| SDHA | F: GGTGGCACTTCTACGACACC R: CCATAATTCTCCAGCTCTACC | 103.7% | 1.000 | −3.237 | [11] Designed |
| Gene | Primer | Product Length (bp) | Accession Number | Source | |
|---|---|---|---|---|---|
| ErbB2 | Forward | CTGAGGGCCGATATACCTTC | 113 | NM_001003217.2 | [13] |
| Reverse | TCACCTCTTGGTTGTTCAGG | ||||
| TGFβ | Forward | CAAGTAGACATTAACGGGTTCAGTTC | 70 | L34956 | [14] |
| Reverse | GGTCGGTTCATGCCATGAAT | ||||
| CD44 | Forward | CAAGGCTTTCAACAGCACCC | 191 | NM_001197022.1 | [15] |
| Reverse | TACGTGTCGTACTGGGAGGT | ||||
| IL6 | Forward | TCCAGAACAACTATGAGGGTGA | 99 | NM_001003300.1 | [16] |
| Reverse | TCCTGATTCTTTACCTTGCTCTT | ||||
| IL10 | Forward | CGACCCAGACATCAAGAACC | 100 | NM_001003077.1 | [17] |
| Reverse | CACAGGGAAGAAATCGGTGA | ||||
| IL15 | Forward | ACTTGCATCCAGTGCTACTT | 270 | AF479882.1 | [18] |
| Reverse | CGAGCGAGATAACACCTAAC | ||||
| IL8 | Forward | TGATTGACAGTGGCCCACATTGTG | 355 | D14285.1 | [15] |
| Reverse | GTCCAGGCACACCTCATTTC | ||||
| CD14 | Forward | GCCGGGCCTCAAGGTACT | 60 | XM_843653.4 | [19] |
| Reverse | TCGTGCGCAGGAAAAAGC | ||||
| p65 | Forward | TGTAAAGAAGCGGGACCTGG | 249 | AB930129.1 | [20] |
| Reverse | AGAGTTTCGGTTCACTCGGC | ||||
| IL18 | Forward | CTCTCCTGTAAGAACAAAACTATTTCCTT | 99 | NM_001003169.1 | [21] |
| Reverse | GAACACTTCTCTGAAAGAATATGATGTCA | ||||
| TLR4 | Forward | GCTGGATGGTAAACCGTGGA | 157 | NM_001002950.2 | [15] |
| Reverse | AGCACAGTGGCAGGTACATC | ||||
| TLR5 | Forward | CCAGGACCAGACGTTCAGAT | 108 | EU551146.1 | [22] |
| Reverse | GCCCAGGAAGATGGTGTCTA | ||||
| TP53 | Forward | CGTTTGGGGTTCCTGCATTC | 231 | NM_001003210.1 | [15] |
| Reverse | CACTACTGTCAGAGCAGCGT | ||||
| MD2 | Forward | GGGAATACGATTTTCTAAGGGACAA | 91 | XM_848045.3 | [19] |
| Reverse | CGGTAAAATTCAAACAAAAGAGCTT | ||||
| MyD88 | Forward | GAGGAGATGGGCTTCGAGTA | 159 | XM_534223.5 | [15] |
| Reverse | GTTCCACCAACACGTCGTC | ||||
| STAT5 | Forward | TTGACTCTCCTGACCGCAAC | 181 | XM_548091.5 | [15] |
| Reverse | TCCGTCTACTGCTTTAGCGA | ||||
| iNOS | Forward | AGACACACTTCACCACAAGG | 284 | AF07782.1 | [23] |
| Reverse | TGCTTGGTGGCGAAGATGAGC | ||||
| CXCR4 | Forward | GCGTCTGGATACCTGCTCTC | 163 | NM_001048026.1 | [15] |
| Reverse | GATACCCGGCAGGATAAGGC | ||||
| RAD51 | Forward | GGAGAAGGAAAGGCCATGTA | 147 | NM_001003043.1 | [24] |
| Reverse | GGGTCTGGTGGTCTGTGTT | ||||
| PTEN | Forward | GTGAAGCTGTACTTCACAA | 135 | NM_001003192.1 | [25] |
| Reverse | CTGGGTCAGAGTCAGTGGTG | ||||
| Primer | Position | Product Lenght (bp) | Accession Number | Source | |
|---|---|---|---|---|---|
| CXCR4 F | TCT GTG GCA GAC CTC CTC TT | F 266–285 R 611–630 | 364 | NM_001048026.1 | [33] |
| CXCR4 R | TGA AAC TGG AAC ACC ACC AA | ||||
| CXCR4 F7 | TGA CTC CAT GAA GGA ACC CTG | F 88–108 R 971–990 | 902 | NM_001048026.1 | This paper |
| CXCR4 R2 | CTG CTC ACA GAG GTG AGT GC | ||||
| CXCR4 Fow 3a | GTC ATC CTG TCC TGC TAC TG | F 665–684 R 296–314 | Sequencing 902 bp | NM_001048026.1 | This Paper |
| CXCR4 Rev 3b | CAA CTG CCC AGA AGG GAA G | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Modesto, P.; Castrillo Fernandez, J.L.; Martini, I.; Zoccola, R.; Pugliano, M.C.; De Ciucis, C.G.; Goria, M.; Ferrari, A.; Razzuoli, E. Characterization of D-17 Canine Osteosarcoma Cell Line and Evaluation of Its Ability to Response to Infective Stressor Used as Alternative Anticancer Therapy. Animals 2020, 10, 1981. https://doi.org/10.3390/ani10111981
Modesto P, Castrillo Fernandez JL, Martini I, Zoccola R, Pugliano MC, De Ciucis CG, Goria M, Ferrari A, Razzuoli E. Characterization of D-17 Canine Osteosarcoma Cell Line and Evaluation of Its Ability to Response to Infective Stressor Used as Alternative Anticancer Therapy. Animals. 2020; 10(11):1981. https://doi.org/10.3390/ani10111981
Chicago/Turabian StyleModesto, Paola, Jordi Leonardo Castrillo Fernandez, Isabella Martini, Roberto Zoccola, Maria Concetta Pugliano, Chiara Grazia De Ciucis, Maria Goria, Angelo Ferrari, and Elisabetta Razzuoli. 2020. "Characterization of D-17 Canine Osteosarcoma Cell Line and Evaluation of Its Ability to Response to Infective Stressor Used as Alternative Anticancer Therapy" Animals 10, no. 11: 1981. https://doi.org/10.3390/ani10111981





