Interacting Proteins, Polymorphisms and the Susceptibility of Animals to SARS-CoV-2
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. SARS-CoV-2 and Viral Entry into Host Cells
1.2. Animals Which Have Become Infected by SARS-CoV-2
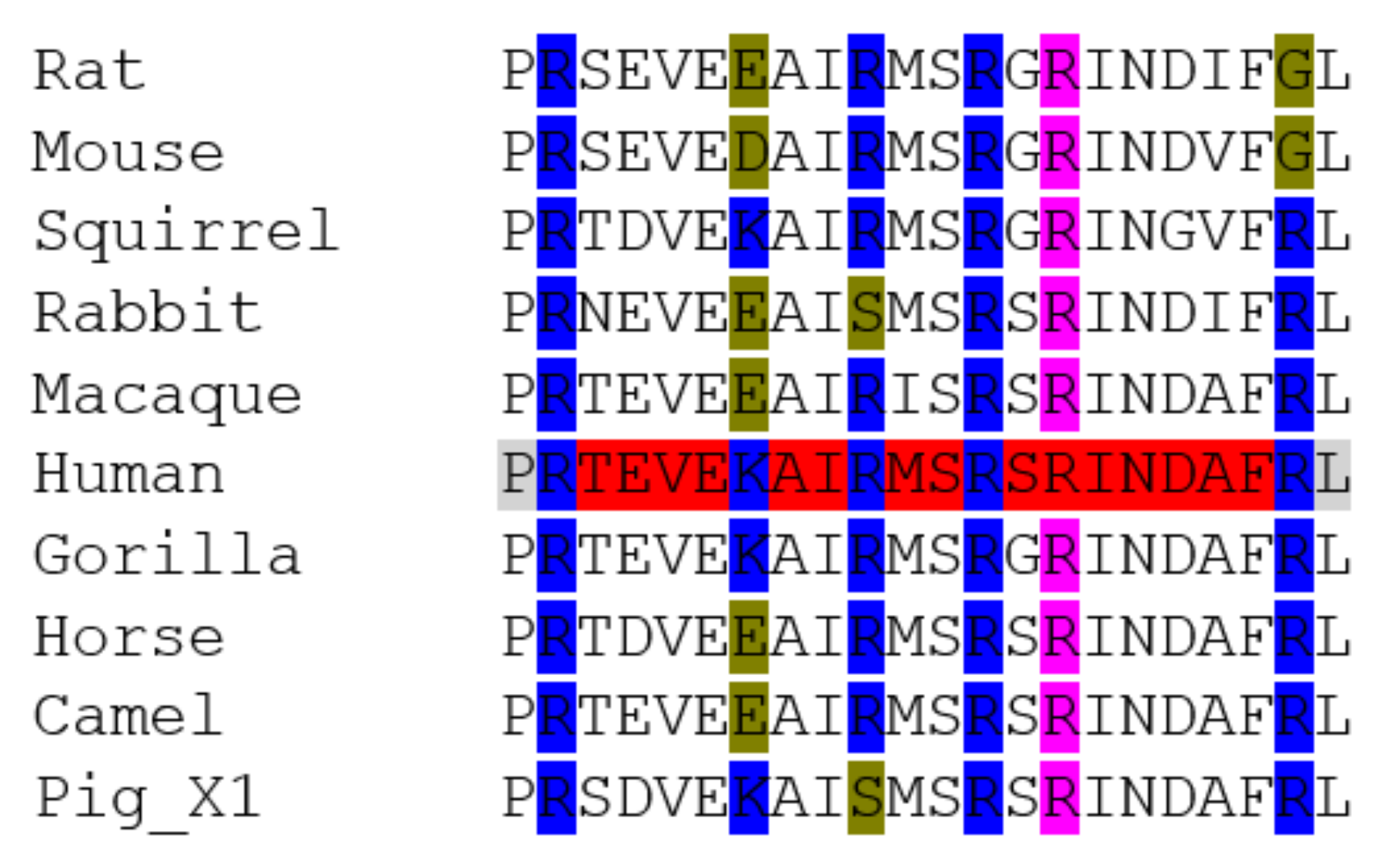
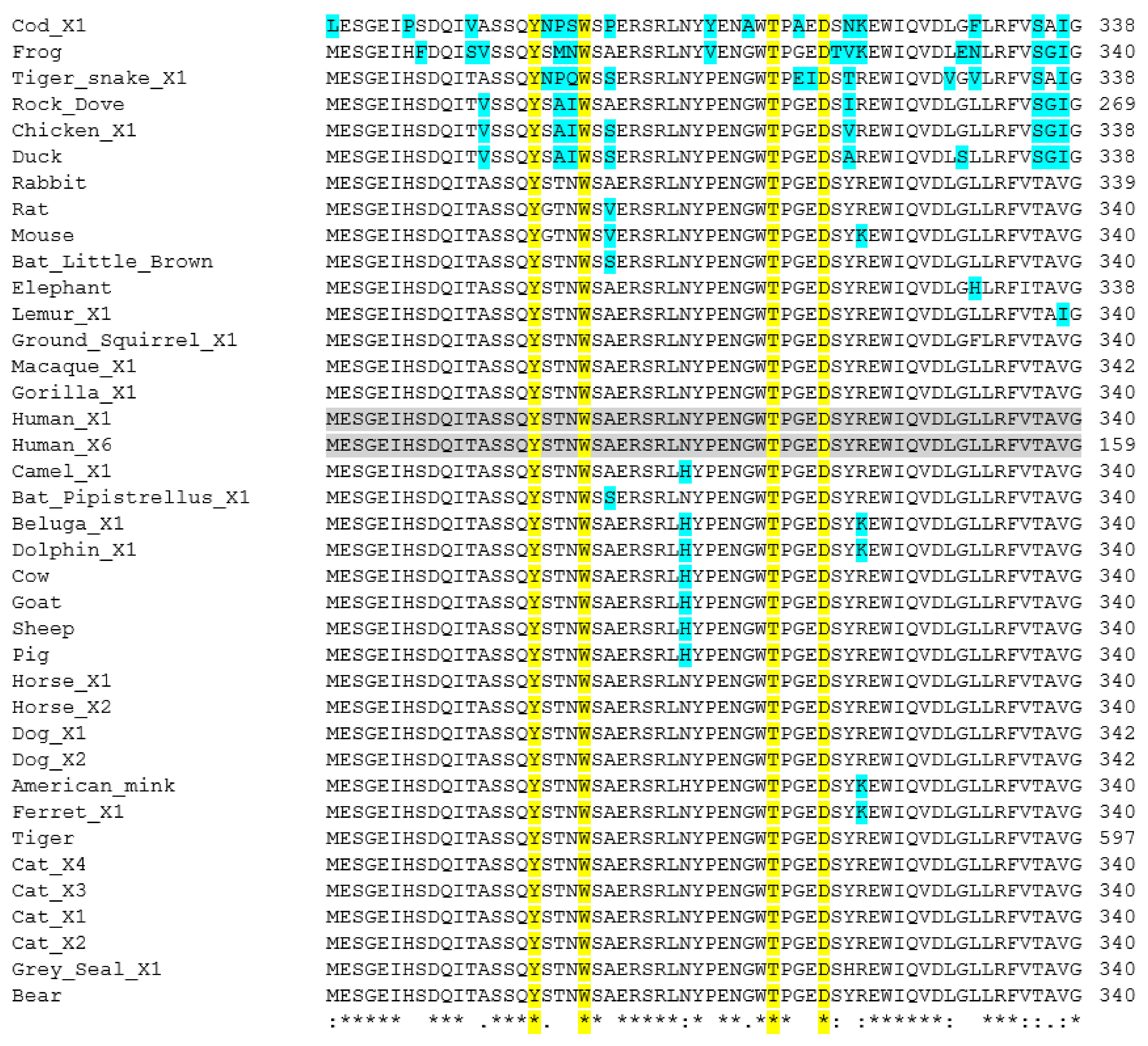
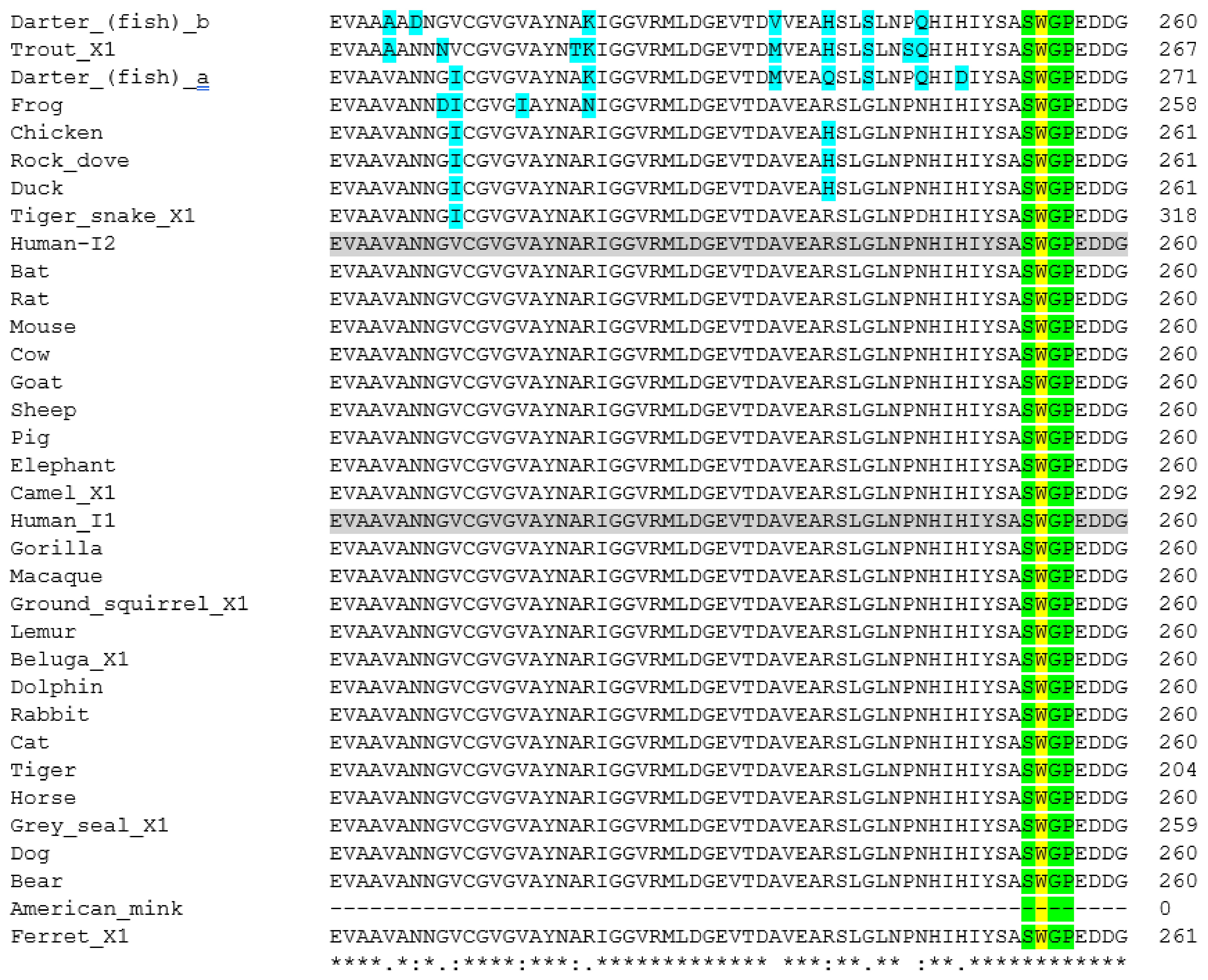
2. Methods Used for Sequence Analysis
3. Sequence Analysis
3.1. ACE2
3.2. TMPRSS2
3.3. Neuropilin-1
3.4. Furin
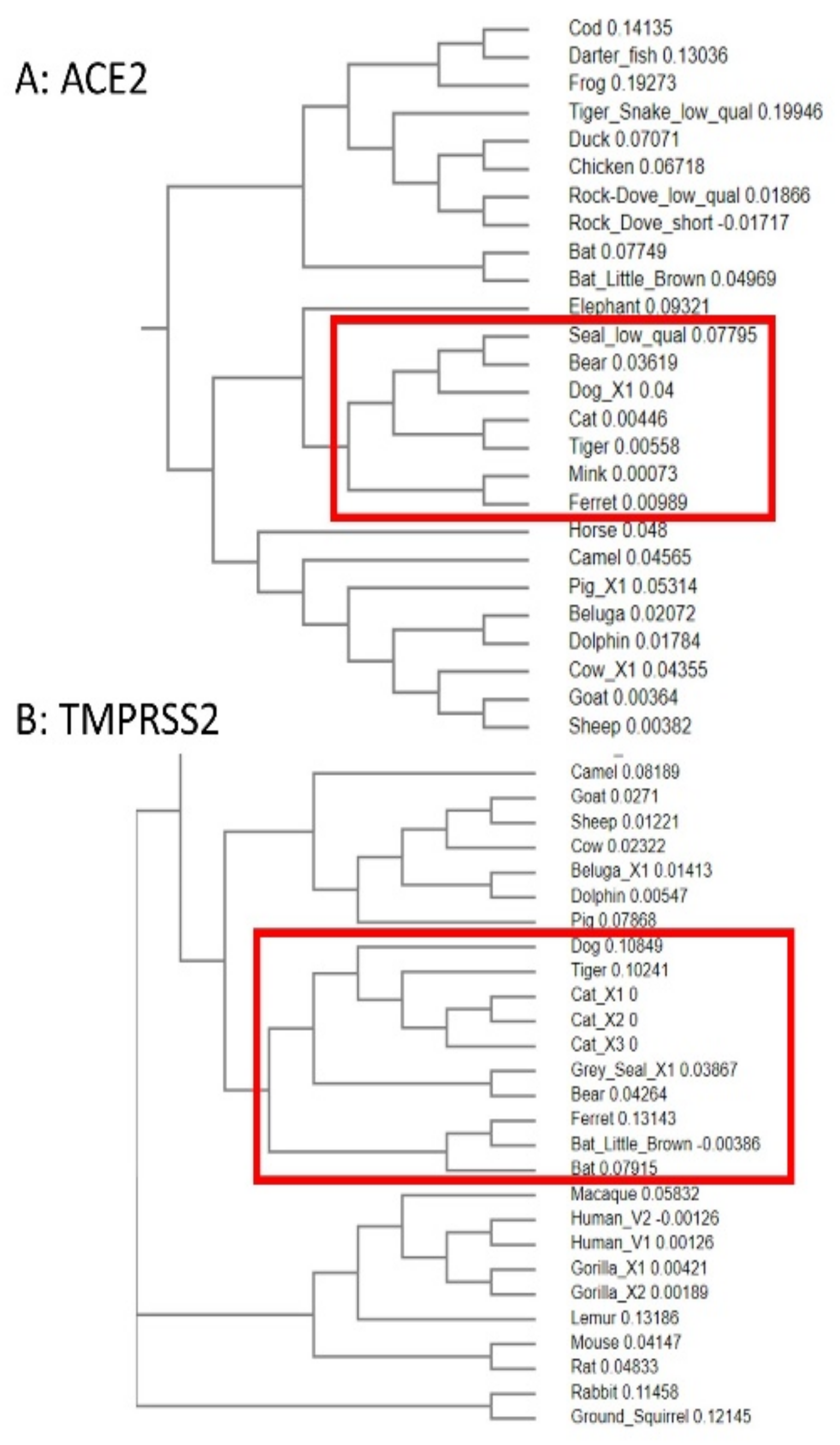
3.5. Phylogenetic Analysis of ACE2 and TMPRSS2 Sequences
4. Discussion
5. Final Thoughts and Future Perspectives
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.-C.; Chen, C.-S.; Chan, Y.-J. The outbreak of COVID-19: An overview. J. Chin. Med. Assoc. 2020, 83, 217–220. [Google Scholar] [CrossRef]
- Tizaoui, K.; Zidi, I.; Lee, K.H.; Ghayda, R.A.; Hong, S.H.; Li, H.; Smith, L.; Koyanagi, A.; Jacob, L.; Keonbichler, A.; et al. Update of the current knowledge on genetics, evolution, immunopathogenesis, and transmission for coronavirus disease 19 (COVID-19). Int. J. Biol. Sci. 2020, 16, 2906–2923. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, M.; Chen, X.; Montaner, L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020. [Google Scholar] [CrossRef]
- Blair, R.V.; Vaccari, M.; Doyle-Meyers, L.A.; Roy, C.J.; Russell-Lodrigue, K.; Fahlberg, M.; Monjure, C.J.; Beedingfield, B.; Plante, K.S.; Plante, J.A.; et al. ARDS and cytokine storm in SARS-CoV-2 infected Caribbean vervets. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef] [PubMed]
- Ghaebi, M.; Osali, A.; Valizadeh, H.; Roshangar, L.; Ahmadi, M. Vaccine development and therapeutic design for 2019-nCoV/SARS-CoV-2: Challenges and chances. J. Cell Physiol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Koirala, A.; Jin Joo, Y.; Khatami, A.; Chiu, C.; Britton, P.N. Vaccines for COVID-19: The current state of play. Paediatr. Respir. Rev. 2020, 35, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Zhang, J.; Ma, X.; Tan, J.; Chen, L.; Liu, S.; Xin, Y.; Zhuang, L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020, 131, 110678. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Jia, H.; Yue, X.; Lazartigues, E. ACE2 mouse models: A toolbox for cardiovascular and pulmonary research. Nat. Commun. 2020, 11, 5165. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens 2020, 9, 529. [Google Scholar] [CrossRef]
- Hoffman, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell entry Depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef]
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A potential therapeutic target for COVID-19. iScience 2020, 23, 101642. [Google Scholar] [CrossRef]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Li, X.; Gao, X.; Dong, Q. Natural polymorphisms are present in the Furin cleavage site of the SARS-CoV-2 spike glycoprotein. Front. Genet. 2020, 11, 783. [Google Scholar] [CrossRef] [PubMed]
- Da Cunda, J.P.C.; Galante, P.A.F.; de Souza, J.E.; de Souza, R.F.; Carvalho, P.M.; Ohara, D.T.; Moura, R.P.; Oba-Shinia, S.M.; Marie, S.K.N.; Silva, W.A., Jr.; et al. Bioinformatics construction of the human cell surfaceome. Proc. Natl. Acad. Sci. USA 2009, 106, 16752–16757. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 216. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/ny-zoo-covid-19 (accessed on 16 November 2020).
- Wang, L.; Mitchell, P.K.; Calle, P.P.; Bartlett, S.L.; McAloose, D.; Killian, M.L.; Yuan, F.; Fang, Y.; Goodman, L.B.; Fredrickson, R.; et al. Complete genome sequence of SARS-CoV-2 in a tiger from a U.S. zoological collection. Microbiol. Resour. Announc. 2020, 9, e00468-20. [Google Scholar] [CrossRef]
- Temmam, S.; Barbarino, A.; Maso, D.; Behillil, S.; Enouf, V.; Huon, C.; Jaraud, A.; Chevallier, L.; Backovic, M.; Pérot, P.; et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health 2020, 10, 100164. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.-I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 2020, 1. [Google Scholar] [CrossRef] [PubMed]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Nat. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Available online: https://www.bbc.co.uk/news/world-europe-55229433 (accessed on 9 December 2020).
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. Bull. Eur. Mal. Transm. 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Jumping back and forth: Anthropozoonotic and zoonotic transmission of SARS-CoV-2 on mink farms. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dyer, O. Covid-19: Denmark to kill 17 million minks over mutation that could undermine vaccine effort. BMJ 2020, 371, m4338. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound. Emerg. Dis. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_one_health/sars-cov-2-animals-us (accessed on 22 February 2021).
- Pickering, B.S.; Smith, G.; Pinette, M.M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect Dis. 2021, 27, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Morozov, I.; Trujillo, J.D.; Gaudreault, N.N.; Bold, D.; Carossino, M.; Artiaga, B.L.; Indran, S.V.; Kwon, T.; Balaraman, V.; et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 2278–2288. [Google Scholar] [CrossRef] [PubMed]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Kumar, A.; Pandey, S.N.; Pareek, V.; Narayan, R.K.; Faig, M.A.; Kumari, C. Predicting susceptibility for SARS-CoV-2 infection in domestic and wildlife animals using ACE2 protein sequence homology. Zoo Biol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.-P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Nat. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, X.; Wei, X.; Li, J.; Yang, J.; Tan, H.; Zhu, J.; Zhang, Q.; Wu, J.; Liu, L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020, 92, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Cui, W.; Tian, B.-P. The potential intermediate hosts for SARS-CoV-2. Front. Microbiol. 2020, 11, 580137. [Google Scholar] [CrossRef]
- Zhai, X.; Sun, J.; Yan, Z.; Zhang, J.; Zhao, J.; Zhao, Z.; Gao, Q.; He, W.T.; Veit, M.; Su, S. Comparison of severe acute respiratory syndrome Coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020, 94, e00831-e20. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [Green Version]
- Gonnet, G.H.; Cohen, M.A.; Benner, S.A. Exhaustive matching of the entire protein sequence database. Science 1992, 256, 1443–1445. [Google Scholar] [CrossRef]
- GnomAD Browser. Available online: https://gnomad.broadinstitute.org/ (accessed on 2 November 2020).
- Senapati, S.; Banerjee, P.; Bhagavatula, S.; Kushwaha, P.P.; Kumar, S. Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction in COVID-19. OSF 2020. preprints. [Google Scholar] [CrossRef]
- Guruprasad, L. Human coronavirus spike protein-host receptor recognition. Prog. Biophys. Mol. Biol. 2020, in press. [Google Scholar] [CrossRef]
- Sun, J.; He, W.-T.; Wang, L.; Lai, A.; Ji, X.; Zhai, X.; Li, G.; Suchard, M.A.; Tian, J.; Zhou, J.; et al. COVID-19: Epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020, 26, 483–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calcagnile, M.; Forgez, P.; Iannelli, A.; Bucci, C.; Alifano, M.; Alifano, P. ACE2 polymorphisms and individual susceptibility to SARS-CoV-2 infection: Insights from an in silico study. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Melin, A.D.M.; Janiak, M.C.J.; Ill, F.M.I. Comparative ACE2 variation and primate Covid-19 risk. Commun. Biol. 2020, 3, 641. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stawiski, E.W.; Diwanji, D.; Suryamohan, K.; Gupta, R.; Fellouse, F.A.; Sathirapongsasuti, F.; Liu, J.; Jiang, Y.-P.; Ratan, A.; Mis, M.; et al. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Abiko, K.; Mandai, M.; Yaegashi, N.; Konishi, I. Highly conserved binding region of ACE2 as a receptor for SARS-CoV-2 between humans and mammals. Vet. Q. 2020, 40, 243–249. [Google Scholar] [CrossRef]
- MacGowan, S.A.; Barton, G.J. Missense variants in ACE2 are predicted to encourage and inhibit interaction with SARS-CoV-2 Spike and contribute to genetic risk in COVID-19. BioRxiv 2020. [Google Scholar] [CrossRef]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.; Jabeen, N.; Raza, F.; Shabbir, S.; Baig, A.A.; Amanullah, A.; Aziz, B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, F.; Elserafy, M.; Alkordi, M.; Amin, M. ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity. BioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Dahms, S.O.; Arciniega, M.; Steinmetzer, T.; Huber, R.; Than, M.E. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Nat. Acad. Sci. USA 2016, 113, 11196–11201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Maio, F.; Lo Cascio, E.; Babini, G.; Sali, M.; Della Longa, S.; Tilocca, B.; Roncada, P.; Arcovito, A.; Sanguinetti, M.; Scambia, G.; et al. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020, 22, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Gkogkou, E.; Barnasas, G.; Vougas, K.; Trougakos, I.P. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020, 36, 101615. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, I.; Haider, S.; Malik, M.Z.; Ponnusamy, K.; Rai, E. ACE2 homo-dimerization human genomic variants and interaction of host proteins explain high population specific differences in outcomes of COVID19. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J. Virol. 2020, 94, e00127-e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef]
- Mathavarajah, S.; Dellaire, G. Lions, tigers and kittens too: ACE2 and susceptibility to COVID-19. Evol. Med. Public Health 2020, 109–113. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X.; Wen, F.; Huang, X.; Ning, G.; Wang, W. Comparative genetic analysis of the novel coronavirus 2019-nCoV/SARS-CoV-2 receptor ACE2 in different populations. Cell Discov. 2020, 61, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Senapati, S.; Kumar, S.; Singh, A.K.; Banerjee, P.; Bhagavatula, S. Assessment of risk conferred by coding and regulatory variations from TMPRSS2 and CD26 in susceptibility of SARS-CoV-2 infection in human. J. Genet. 2020, 99, 53. [Google Scholar] [CrossRef]
- Hu, T.; Liu, Y.; Zhao, M.; Zhuang, Q.; Xu, L.; He, Q. A comparison of COVID-19, SARS and MERS. PeerJ 2020, 8, e9725. [Google Scholar] [CrossRef] [PubMed]
- Gryseels, S.; De Bruyn, L.; Gryselings, R.; Calvignac-Spencer, S.; Leendertz, F.H.; Leirs, H. Risk of human-to-wildlife transmission of SARS-CoV-2. Mammal Rev. 2020. [Google Scholar] [CrossRef]
- Salajegheh Tazerji, S.; Magalhães Duarte, P.; Rahimi, P.; Shahabinejad, F.; Dhakal, S.; Singh Malik, Y.; Shehata, A.A.; Lama, J.; Klein, J.; Safdar, M.; et al. Transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to animals: An updated review. J. Transl. Med. 2020, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, C.; Das, C.; Ghosh, A.; Singh, A.K.; Mukherjee, S.; Majumder, P.P.; Basu, A.; Biswas, N.K. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Liu, K.; Tan, S.; Niu, S.; Wang, J.; Wu, L.; Sun, H.; Zhang, Y.; Pan, X.; Qu, X.; Du, P.; et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. USA 2021, 118, e2020216118. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Ji, X.; He, W.; Dellicour, S.; Wang, S.; Li, G.; Zhang, L.; Gilbert, M.; Zhu, H.; Xing, G.; et al. Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Mol. Biol. Evol. 2020, 37, 2641–2654. [Google Scholar] [CrossRef]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef] [Green Version]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef]
- Kostov, I. Public health risk analysis due to the spread of the Covid-19 virus from the free living minks and mink farms in Bulgaria. Int. J. Vet. Sci. Anim. Husb. 2020, 5, 89–92. [Google Scholar]
- Enserink, M. Coronavirus rips through Dutch mink farms, triggering culls. Science 2020, 368, 1169. [Google Scholar] [CrossRef]
- Conti, P.; Younes, A. Coronavirus COV-19/SARS-CoV-2 affects women less than men: Clinical response to viral infection. J. Biol. Regul. Homeost. Agents 2020, 34, 339–343. [Google Scholar] [CrossRef]
- Pareek, M.; Bangash, M.N.; Pareek, N.; Pan, D.; Sze, S.; Minhas, J.S.; Hanif, W.; Khunti, K. Ethnicity and COVID-19: An urgent public health research priority. Lancet 2020, 395, 1421–1422. [Google Scholar] [CrossRef]
- Available online: https://www.sciencemag.org/news/2021/01/captive-gorillas-test-positive-coronavirus (accessed on 22 January 2021).
- Gibbons, A. Ape researchers mobilize to save primates from coronavirus. Science 2020, 368, 566. [Google Scholar] [CrossRef]
- Mupatsi, N. Observed and potential environmental impacts of COVID -19 in Africa. Preprints 2020, 2020080442. [Google Scholar] [CrossRef]
- Sharif, S.; Arshad, S.S.; Hair-Bejo, M.; Omar, A.R.; Zeenathul, N.A.; Alazawy, A. Diagnostic methods for feline coronavirus: A review. Vet. Med. Int. 2010, 809480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preistnall, S.L. Canine respiratory coronavirus: A naturally occurring model of COVID-19? Vet. Pathol. 2020, 57, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.A.; Wu, G.; St Leger, J.; Nordhausen, R.W.; Wang, D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [Green Version]
- Nabi, G. Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 2020, 188, 109732. [Google Scholar] [CrossRef]
- Quilliam, R.S.; Weidmann, M.; Moresco, V.; Purshouse, H.; O’Hara, Z.; Oliver, D.M. COVID-19: The environmental implications of shedding SARS-CoV-2 in human faeces. Environ. Int. 2020, 140, 105790. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Hewson, I. Coronaviruses in the sea. Front. Microbiol. 2020, 11, 1795. [Google Scholar] [CrossRef] [PubMed]
- Maal-Bared, R.; Sobsey, M.; Bibby, K.; Sherchan, S.P.; Fitzmorris, K.B.; Munakata, N.; Gerba, C.; Schaefer, S.; Swift, J.; Gary, L.; et al. Letter to the Editor regarding Mathavarajah; et al. (2020) Pandemic danger to the deep: The risk of marine mammals con-tracting SARS-CoV-2 from wastewater. Sci. Total Environ. 2020, 2020, 144855. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Stoddart, A.K.; Gagnon, G.A.; Dellaire, G. Pandemic danger to the deep: The risk of marine mammals contracting SARS-CoV-2 from wastewater. Sci. Total Environ. 2021, 760, 143346. [Google Scholar] [CrossRef]
- Olival, K.J.; Cryan, P.M.; Amman, B.R.; Baric, R.S.; Blehert, D.S.; Brook, C.E.; Calisher, C.H.; Castle, K.T.; Coleman, J.T.H.; Daszak, P.; et al. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: A case study of bats. PLoS Pathog. 2020, 16, e1008758. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, D.; Rocha, R. Guidelines for communicating about bats to prevent persecution in the time of COVID-19. Biol. Conserv. 2020, 248, 108650. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Guo, Y.; Xu, J.; Li, H.; Fuller, A.; Tait, R.G., Jr.; Wu, X.L.; Bauck, S. Comparing SNP panels and statistical methods for estimating genomic breed composition of individual animals in ten cattle breeds. BMC Genet. 2018, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, H.; Liu, Z. Expression of ACE2 in airways: Implication for COVID-19 risk and disease management in patients with chronic inflammatory respiratory diseases. Clin. Exp. Allergy 2020, in press. [Google Scholar] [CrossRef]
- Hikmet, F.; Méar, L.; Edvinsson, A.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef] [PubMed]
- Parry, N.M.A. COVID-19 and pets: When pandemic meets panic. Forensic Sci. Int. Rep. 2020, 2, 100090. [Google Scholar] [CrossRef]
- Available online: https://news.sky.com/story/covid-19-snow-leopard-in-louisville-tests-positive-for-coronavirus-12158797N (accessed on 17 December 2020).
- Available online: https://www.bbc.co.uk/news/science-environment-55309269 (accessed on 17 December 2020).
- Leroy, E.M.; Gouilh, M.A.; Brugère-Picoux, J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a one-health strategy to control the COVID-19 pandemic. One Health 2020, 100133. [Google Scholar] [CrossRef] [PubMed]


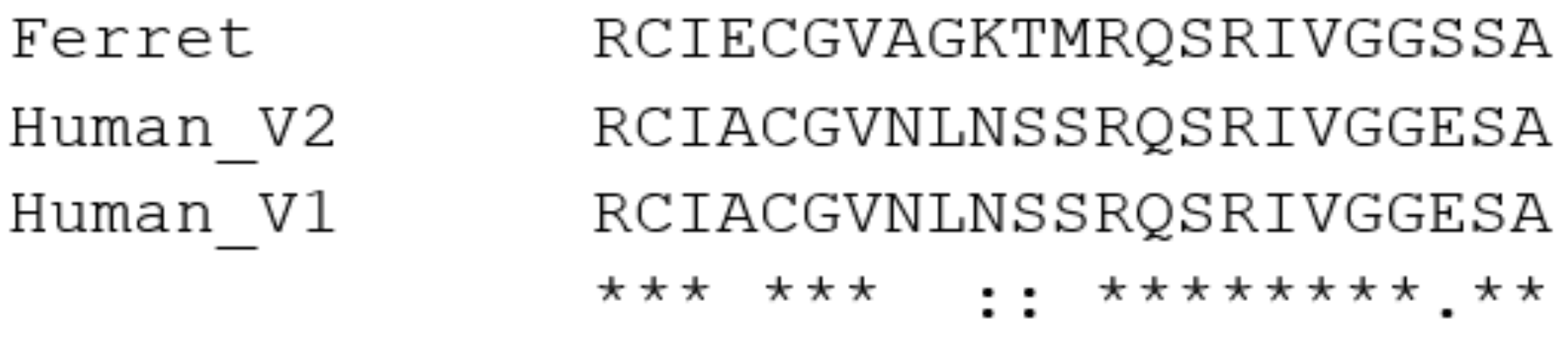
| Common Name | Animal | ACE2 | TMPRSS2 | Neuropilin-1 | Furin |
|---|---|---|---|---|---|
| Human | Homo sapiens | BAB40370.1 | V1: NM_001135099.1 | X1: NM_003873.7 | I1: NP_001276753.1 |
| V2: NM_005656.4 | X6: XM_017016866.2 | I2: NP_001369551.1 | |||
| Gorilla | Gorilla gorilla | XP_018874749.1 | X1: XM_019017901.2 | X1: XM_004049248.3 | XP_018866183.1 |
| X2: XM_004062839.3 | |||||
| Macaque | Macaca nemestrina | XP_024647450.1 | XM_011725990.2 | X1: XM_011738246.1 | XP_011750505.1 |
| Lemur (Grey mouse) | Microcebus murinus | XP_020140826.1 | XM_012749836.2 | X1: XP_020139780.1 | XP_012618739.1 |
| Camel | Camelus ferus | XP_006194263.1 | XM_032461838.1 | X1: XP_032329703.1 | X1: XP_032325184.1 |
| Horse | Equus caballus | XP_001490241.1 | X1: XM_005606160.3 | X1: XM_023632110.1 | XP_023505390.1 |
| X2: XM_014736385.2 | X2: XM_001915381.4 | ||||
| African elephant | Loxodonta africana | XP_023410960.1 | XM_023559111.1 | XM_023539786.1 | XM_003413876.3 |
| Goat | Capra hircus | AHI85757.1 | X1: XM_013966670.2 | X1: XP_017912559.1 | XP_017921808.1 |
| Cow | Bos taurus * | X1: XP_024843618.1 | X1: XM_015473655.2 | X1: XP_024856535.1 | XP_027377940.1 |
| Sheep | Ovis aries | XP_011961657.1 | XM_027960704.1 | X1: XP_027832377.1 | CAJ29337.1 |
| Pig | Sus scrofa | X1: XP_020935033.1 | NM_001386131.1 | XP_020920629.1 | X1: XP_020954578.1 |
| Black rat | Rattus rattus | XP_032746145.1 | XM_032900351.1 | XP_032743292.1 | XP_032749231.1 |
| Mouse | Mus musculus | EDL40761.1 | NM_015775.2 | AAH60129.1 | CAA37988.1 |
| Rabbit | Oryctolagus cuniculus | QHX39726.1 | NM_001386128.1 | XP_008272962.1 | XP_002721548.2 |
| Cat (domestic) | Felis catus | AAX59005.1 | X1: XM_023238709.1 | X1: XP_006933414.1 | XP_023110662.1 |
| X2: XM_023238710.1 | X2: XP_003988264.1 | ||||
| X3: XM_023238711.1 | X3: XP_003988265.1 | ||||
| X4: XP_006933415.1 | |||||
| Bear (grizzly) | Ursus arctos horribilis | XP_026333866.1 | X1: XM_026499970.1 | X1: XM_026483469.1 | XM_026510505.1 |
| Tiger (Siberian) | Panthera tigris altaica | XP_007090142.1 | XM_015541202.1 | XP_007092922.2 | XP_015391216.1 |
| Dog (domestic) | Canis lupus familiaris | X1: XP_013966804.1 | XM_022413273.1 | X1: XP_005617003.1 | XP_022272656.1 |
| X2: XP_535142.3 | |||||
| American mink | Neovison vison | CCP86723.1 | N/A | CCP78142.1 | CCP82836.1 |
| Ferret | Mustela putorius furo | BAE53380.1 | XM_013061267.1 | X1: XP_004774343.2 | X1: XP_004763758.1 |
| Ground squirrel | Ictidomys tridecemlineatus | XP_005316051.3 | XM_021725256.1 | X1: XM_005330824.1 | X1: XP_021578591.1 |
| Kuhl’s pipistrelle bat | Pipistrellus kuhlii | XP_036295422.1 | XM_036446834.1 | X1: XP_036309963.1 | XP_036271438.1 |
| Little brown bat | Myotis lucifugus | X1: XP_023609438.1 | XM_006104440.3 | XP_023607944.1 | N/A |
| Beluga whale | Delphinapterus leucas | XP_022418360.1 | X1: XM_022552959.1 | X1: XP_030617644.1 | X1: XP_022419719.1 |
| Grey seal | Halichoerus grypus | XP_035963182.1 | X1: XM_036108025.1 | X1: XP_035974500.1 | X1: XP_035951550.1 |
| Bottlenosed dolphin | Tursiops truncatus | XP_019781177.2 | XM_033856384.1 | X1: XP_033708297.1 | XP_019801399.1 |
| Cod (fish) | Gadus morhua | XP_030232530.1 | N/A | X1: XP_030204747.1 | N/A |
| Arkansas darter (fish) | Etheostoma cragini | XP_034732342.1 | XM_034889577.1 | N/A | a: XP_034734947.1 |
| b: XP_034734346.1 | |||||
| Rainbow trout (fish) | Oncorhynchus mykiss | N/A | X1: XM_021618133.2 | N/A | X1: XP_021428901.2 |
| Chicken (domestic) | Gallus gallus domesticus | QEQ50331.1 | X1: XM_416737.6 | X1: XP_015136776.1 | NP_990046.1 |
| Duck | Anas platyrhynchos | XP_012949915.2 | X1: XM_027448371.1 | X1: XP_027306331.1 | X1: XP_027321808.1 |
| Rock dove | Columba livia | XP_021154486.1 | XM_021282586.1 | PKK32102.1 | XP_021155000.1 |
| PKK30539.1 | |||||
| Tiger snake | Notechis scutatus | XP_026530754.1 | X1: XM_026673992.1 | X1: XP_026528008.1 | X1: XP_026540426.1 |
| African clawed frog | Xenopus laevis | XP_018104311.1 | V1: NM_001087958.1 | NP_001081380.1 | AAW83022.1 |
| Sloth | Folivora | N/A | N/A | N/A | N/A |
| African lion | Panthera leo | N/A | N/A | N/A | N/A |
| Indian elephant | Elephas maximus indicus | N/A | N/A | N/A | N/A |
| Protein | Reference(s)/Comments | List of Amino Acids Thought to Be Important for Viral/Host Interactions | Polymorphisms of Interest | Comments and Refs for PMs |
|---|---|---|---|---|
| ACE2 | [50] | S19, Q24, T27, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L45, L79, M82, Y83, N330, K353, G354, D355, R357, R393 | S19P, T27A, K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, E329G, G352V, D355N, Q388L, P389H, D509Y | Decreased S protein affinity: [49] and refs within |
| [51] | Q24, K31, H34, E35, D38, Y41, Q42, N53, L79, M82, Y83, N90, N322, Q325, E329, N330, K353, R652, R710 | I21V, E23K, K26R, T27A, N64K, T92I, Q102P, D206G, G211R, R219C, G326E, K341R, H378R, V447F, I468V, A501T, R559S | Increased S protein affinity: [49] and refs within | |
| [52] | S19, Q24, T27, F28, K31, H34, E35, E37, D38, Y41, Q42, L45, L79, M82, Y83, E329, N330, K353, G354, D355, R357 | E300Ter, A627V, N638S, L656Ter, S692P, N720D, L731I/F, E668L | Affinity not reported [49] and refs within | |
| [41]: glycosylation sites in red | S19, Q24, T27, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L45, N53, L79, M82, Y83, N90, N322, N330, K353, G354, D355, R357, R393 | M82I, E329G, D355N, R652K, R710C, R710H: No other significant polymorphisms found | from gnomAD | |
| [39]: concentrated on residues 36–53 AEDLFYQSSLA SWNYNTN | Q24, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L79, M82, Y83, K353, G354, D355, R357 | S19P (increases affinity), K26R (decreases affinity) | [53] | |
| [54] with input from [55] | Q24, D30, H34, E37, D38, Y41, Q42, M82, Y83, K353, D355, R357 | S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P, H378R | Increase [56] | |
| [49] | E23, Q24, K26, T27, D30, K31, H34, E35, E37, D38, Y41, Q42, K68, L79, M82, Y83, D206, G211, R219, K317, G326, E329, K341, G352, K353, D355, R357, P389, V447, I468, R559, D442, N437, T478, F486, | K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L, D509Y | Decrease [56] | |
| [57] | 353-KGDFR-357 | E37K, G352V, D355N (38 others mentioned) | Decrease [58] | |
| [59] | R and K residues within 697–716 needed for cleavage | T27R, G326E | Increase [58] | |
| S19P, E329G | Decrease [60] | |||
| K26R (decrease). Increasing affinity in order: I468V, R219C, K341R, D206G, G211R | [61] | |||
| TMPRSS2 | [20] | D435 | V160M, G181R, R240C, G259S, P335L, G432A, D435Y | [20] |
| [49] | G190, N192, P191, F195, Y189, S234, K399, D396, N395, T324, A280, C278, R277, F251, I279, F394, Y232, N284, V283, Q290, L285, P325, N488, S287, S288, I489, N286, T393 | G8V, R255S, S441G | Decreased S protein affinity: [49] and refs within | |
| V197M/V160M, A65T/A28T | Increased S protein affinity: [49] and refs within | |||
| G190R, P191Q, Y189C, S234G, D396N, T324N, A280D, R277H, R277P, R277C, I279T, I279V, F394S, N284K, N284S, P325T, I489T, N286Y, | from gnomAD | |||
| Neuropilin-1 | [14] | Y297, W301, T316, D320, S346, E348, T249, Y353 | T249S (rare), V179A, R563Q, D592N (non-canonical), V733I | from gnomAD |
| No polymorphisms matching other highlighted amino acids | ||||
| Furin | [62] | H194, W254, N295, S368 (b-strand at S253-P256), T309, S316 | T309I (rare) | from gnomAD |
| No polymorphisms matching other highlighted amino acids |
| Protein of Interest | Comments and Conclusions |
|---|---|
| ACE2 |
|
| TMPRSS2 |
|
| Neuropilin-1 |
|
| Furin |
|
| Animal Group | Susceptibility Predicted | Real-Life Outcomes Reported/Comments |
|---|---|---|
| Non-human primates (especially great apes) | Very high—sequences from several species very similar to humans | |
| Marine mammals e.g., beluga whale (Delphinapterus leucas), dolphin (Tursiops truncates), killer whale (Orcinus orca), seal species (Pinnepedia), Atlantic Walrus (Odobenus rosmarus) | High |
|
| Cats Including domestic (Felis catus) and big cats, e.g., Malayan tigers (Panthera tigris jacksoni), Amur tigers (Panthera tigris altaica) and African lions (Panthera leo krugeri) | Medium | |
| Dogs Including domestic (Canis lupus familiaris) | Medium | |
| Mustelids Including mink (Neovison vison) | Low | |
| Marine mammals e.g., California sea lion (Zalophus californianus) and West Indian manatee (Trichechus manatus), Sowerby’s beaked whale (Mesoplodon bidens) | Low |
|
| Bats including fruit bat (Rousettus aegytiacus), Chinese rufous horseshoe bat (Rhinolophus sinicus), great roundleaf bat (Hipposideros armiger), big brown bat (Eptesicus fuscus) | Low/very low |
|
| Birds/reptiles/amphibians/fish | Very low |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hancock, J.T.; Rouse, R.C.; Stone, E.; Greenhough, A. Interacting Proteins, Polymorphisms and the Susceptibility of Animals to SARS-CoV-2. Animals 2021, 11, 797. https://doi.org/10.3390/ani11030797
Hancock JT, Rouse RC, Stone E, Greenhough A. Interacting Proteins, Polymorphisms and the Susceptibility of Animals to SARS-CoV-2. Animals. 2021; 11(3):797. https://doi.org/10.3390/ani11030797
Chicago/Turabian StyleHancock, John T., Ros C. Rouse, Emma Stone, and Alexander Greenhough. 2021. "Interacting Proteins, Polymorphisms and the Susceptibility of Animals to SARS-CoV-2" Animals 11, no. 3: 797. https://doi.org/10.3390/ani11030797






