On Gram-Positive- and Gram-Negative-Bacteria-Associated Canine and Feline Skin Infections: A 4-Year Retrospective Study of the University Veterinary Microbiology Diagnostic Laboratory of Naples, Italy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Informed Consent
2.2. Ethics Statement
2.3. Sample Collection
2.4. Data Management
2.5. Statistical Analysis
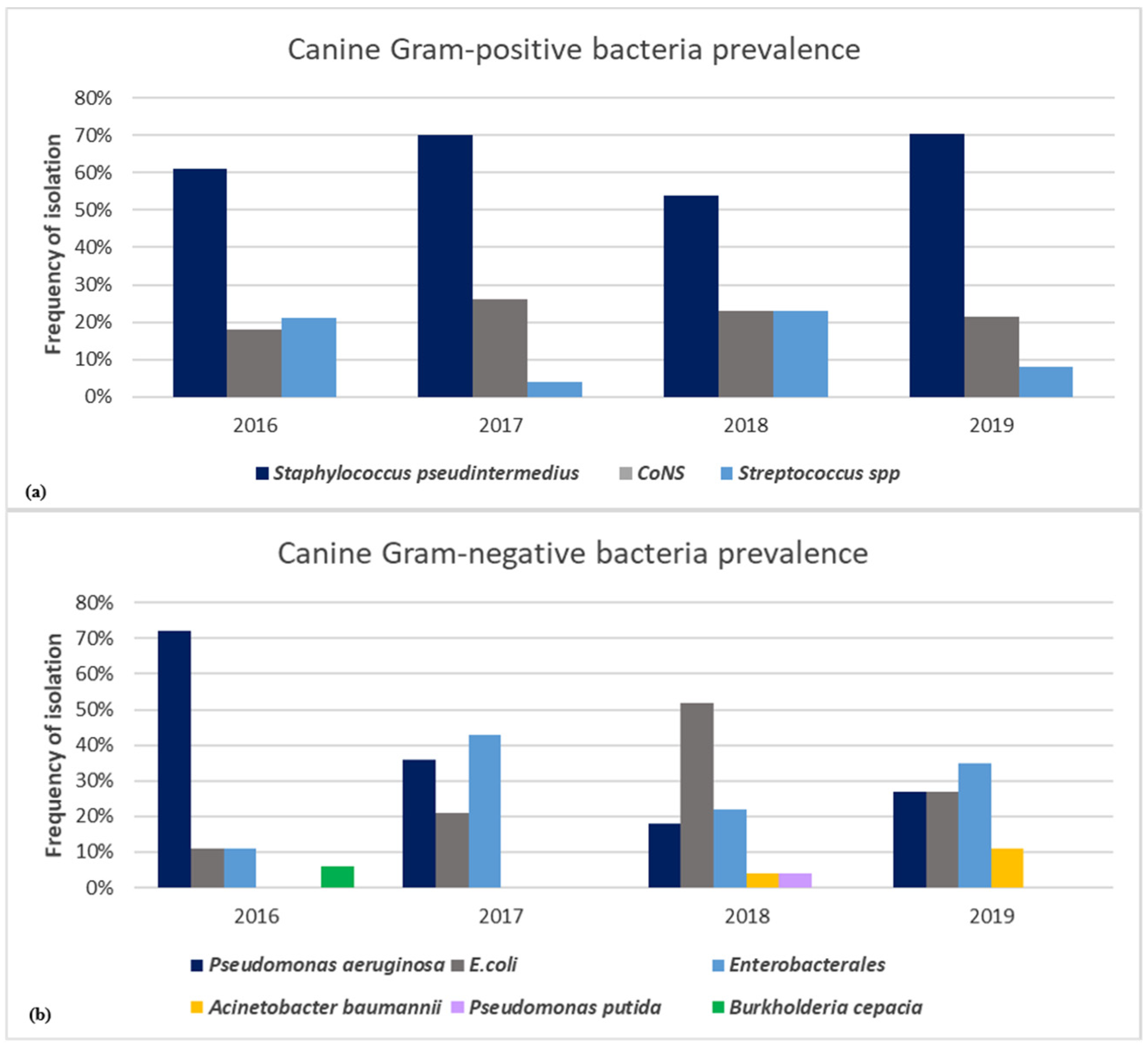
3. Results
3.1. Gram-Positive- and Gram-Negative-Bacteria-Associated Pet Animal Skin Infections
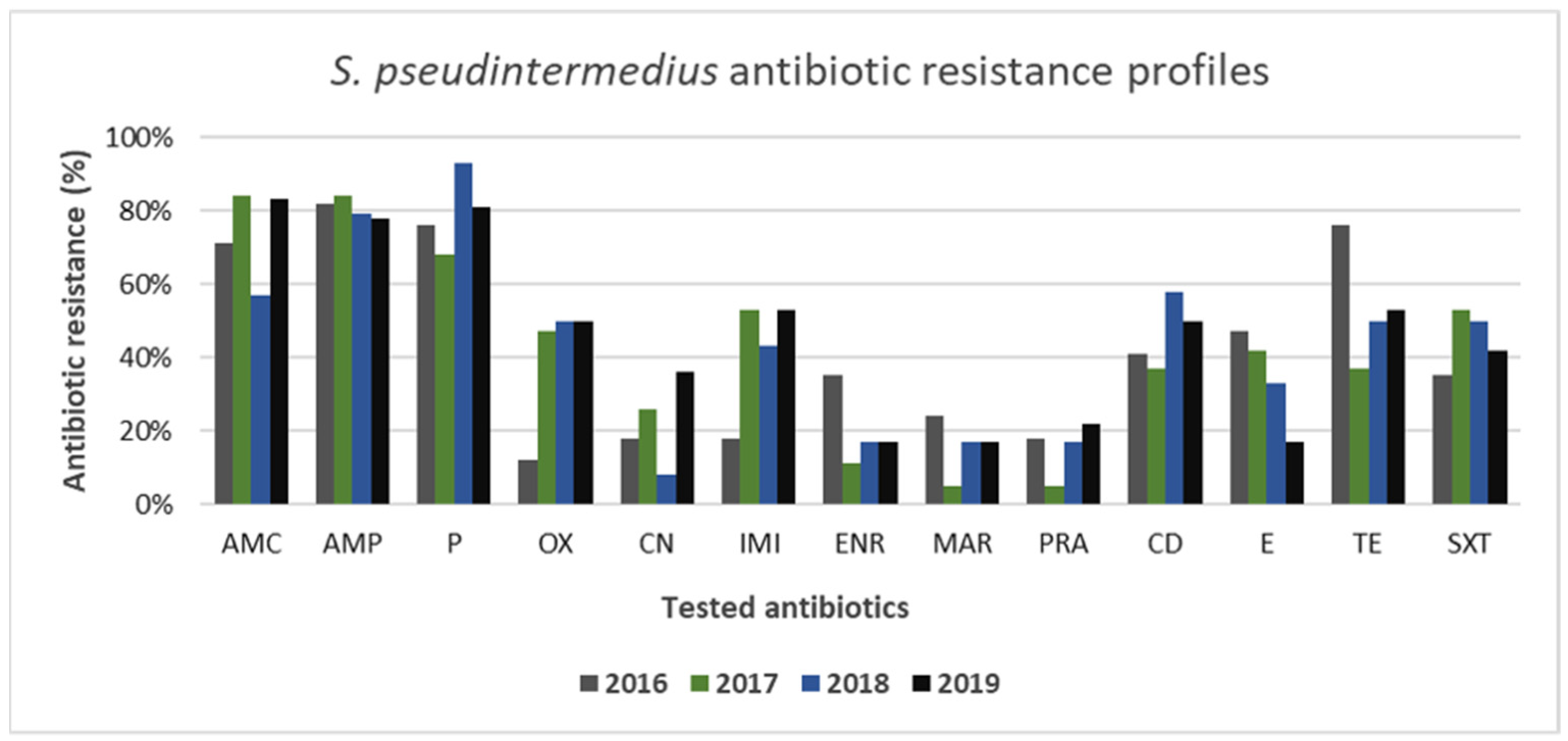
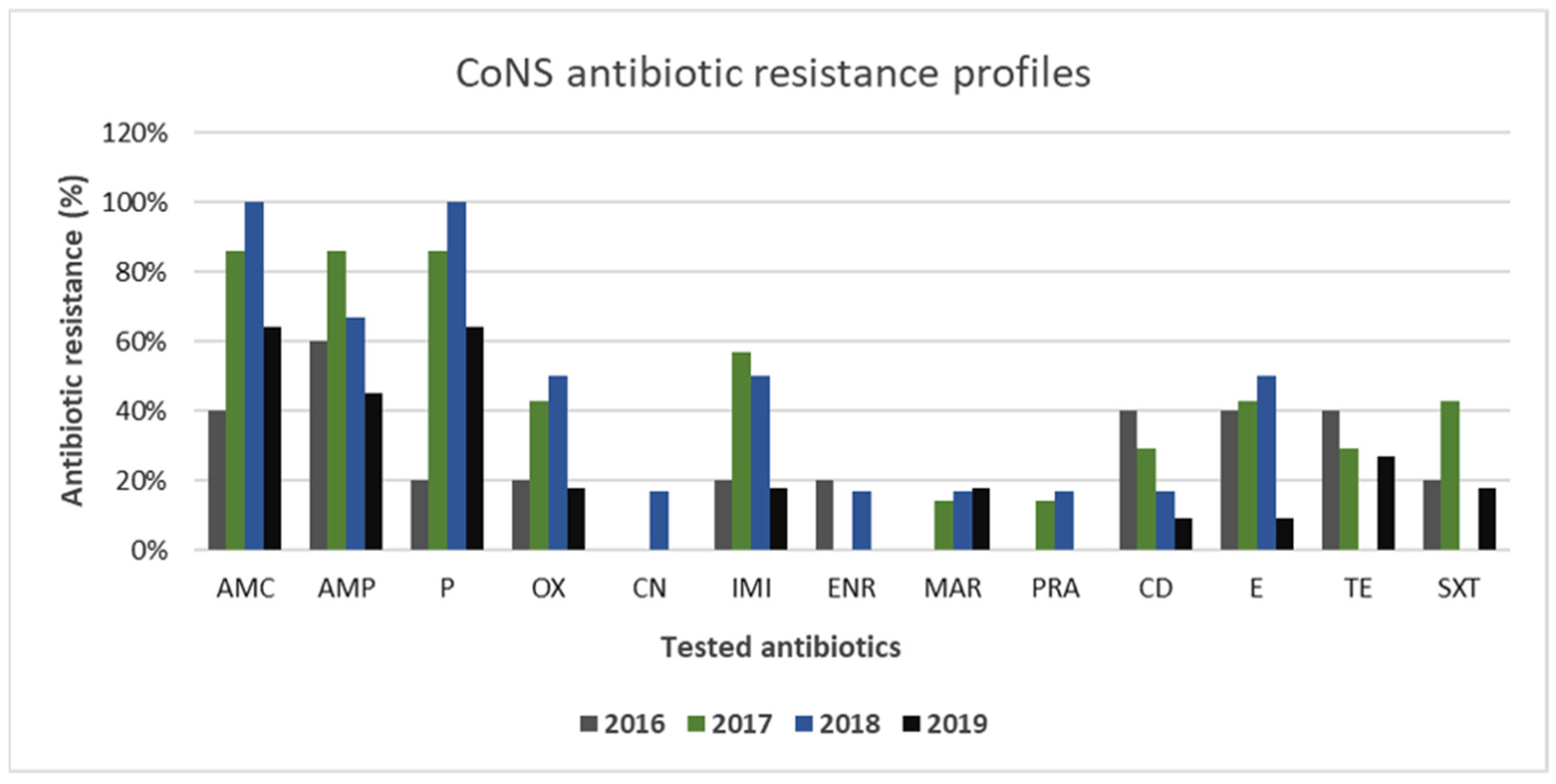
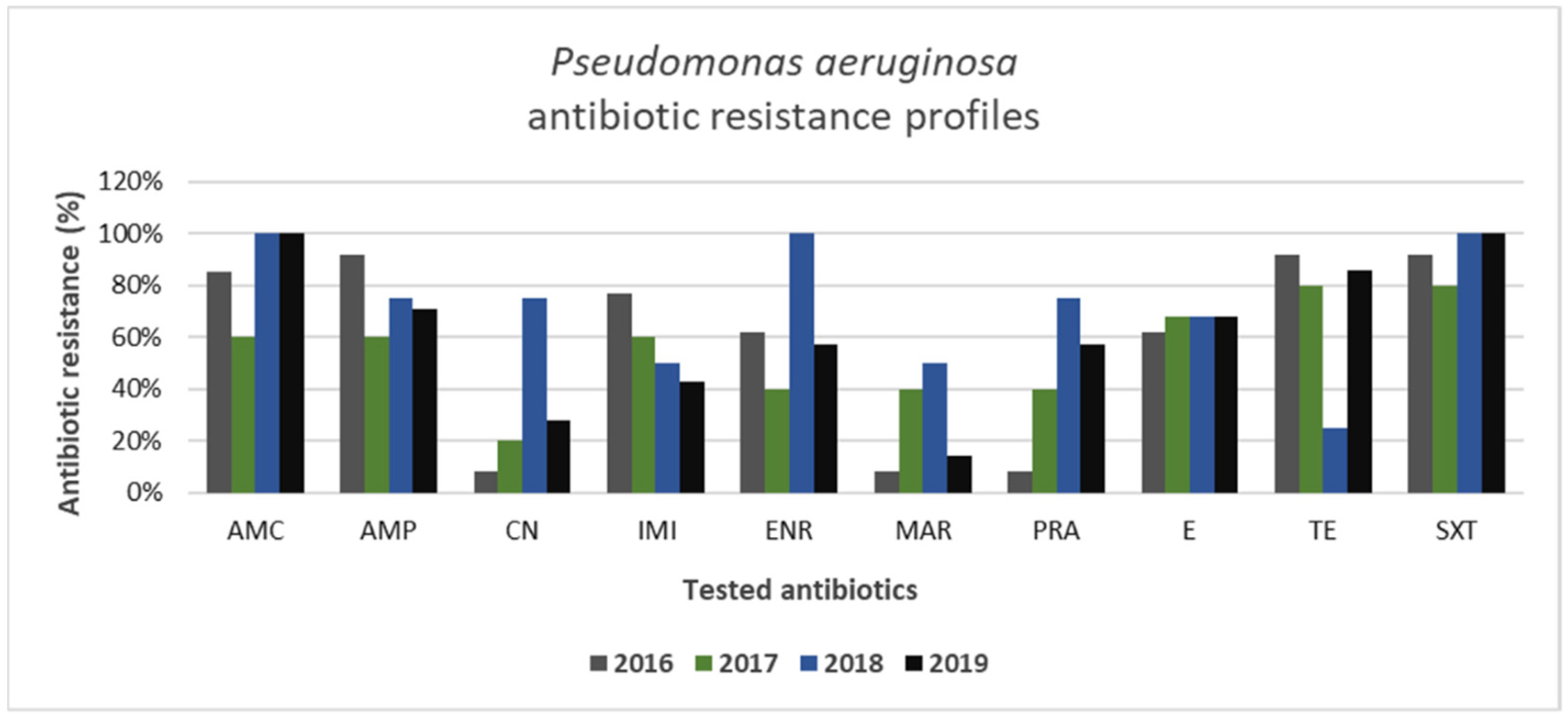
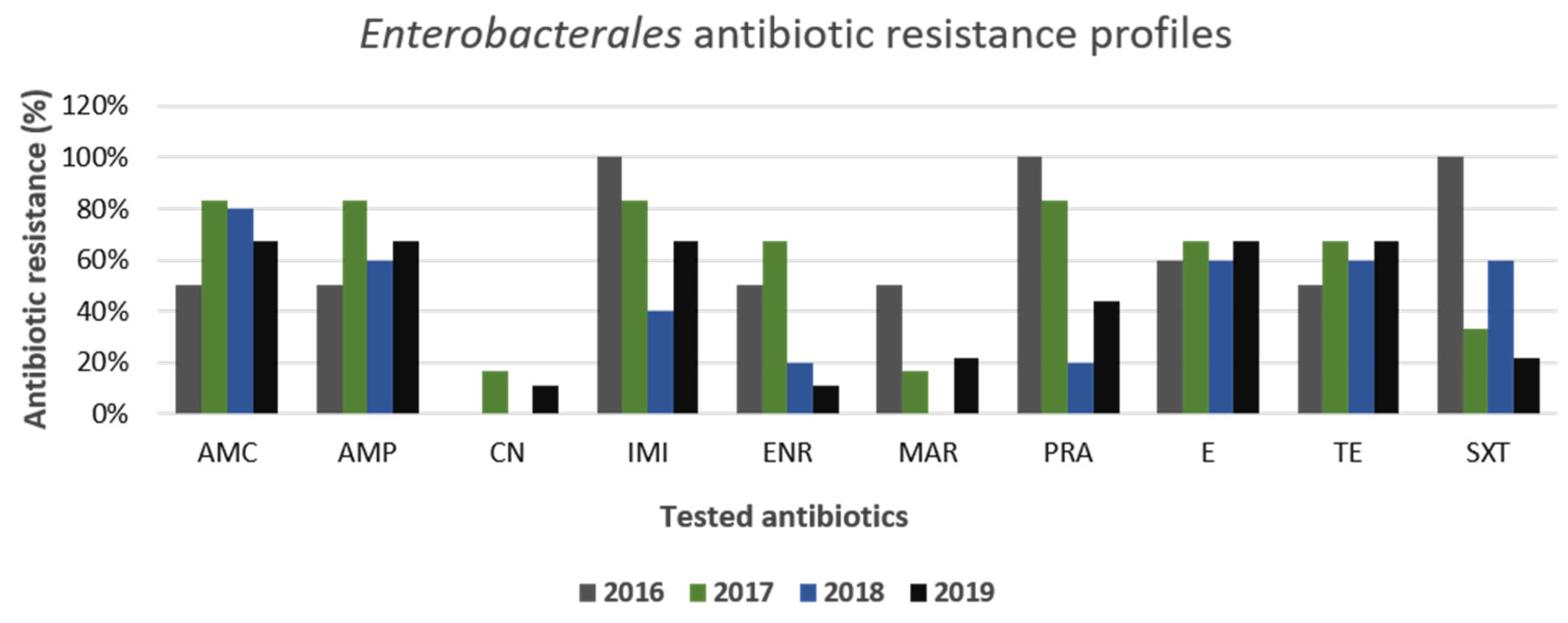
3.2. Gram-Positive and Gram-Negative Bacteria Antibiotic Resistance Profiles
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marsella, R.; De Benedetto, A. Atopic Dermatitis in Animals and People: An Update and Comparative Review. Vet. Sci. 2017, 4, 37. [Google Scholar] [CrossRef]
- Marsella, R. Advances in our understanding of canine atopic dermatitis. Vet. Dermatol. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Nakaminami, H.; Okamura, Y.; Tanaka, S.; Wajima, T.; Murayama, N.; Noguchi, N. Prevalence of antimicrobial-resistant staphylococci in nares and affected sites of pet dogs with superficial pyoderma. J. Vet. Med. Sci. 2021, 83, 214–219. [Google Scholar] [CrossRef]
- De Martino, L.; Nocera, F.P.; Mallardo, K.; Nizza, S.; Masturzo, E.; Fiorito, F.; Iovane, G.; Catalanotti, P. An update on microbiological causes of canine otitis externa in Campania Region, Italy. Asian Pac. J. Trop. Biomed. 2016, 6, 384–389. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Fernández, R.; Durán, I.; Molina-López, R.A.; Darwich, L. Antimicrobial resistance in bacteria isolated from cats and dogs from the Iberian Peninsula. Front. Microbiol. 2021, 11, 621597. [Google Scholar] [CrossRef]
- Chan, W.Y.; Hickey, E.E.; Page, S.W.; Trott, D.J.; Hill, P.B. Biofilm production by pathogens associated with canine otitis externa, and the antibiofilm activity of ionophores and antimicrobial adjuvants. J. Vet. Pharmacol. Ther. 2019, 42, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.F.; Boysen, L.; Jessen, L.R.; Guardabassi, L.; Damborg, P. Diversity of Staphylococcus pseudintermedius in carriage sites and skin lesions of dogs with superficial bacterial folliculitis: Potential implications for diagnostic testing and therapy. Vet. Dermatol. 2018, 29, 291-e100. [Google Scholar] [CrossRef]
- Menandro, M.L.; Dotto, G.; Mondin, A.; Martini, M.; Ceglie, L.; Pasotto, D. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius from symptomatic companion animals in Northern Italy: Clonal diversity and novel sequence types. Comp. Immunol. Microbiol. Infect. Dis. 2019, 66, 101331. [Google Scholar] [CrossRef] [PubMed]
- Nocera, F.P.; Parisi, A.; Corrente, M.; De Martino, L. Evidence of new sequence types of methicillin-resistant Staphylococcus pseudintermedius in Italy. Pol. J. Vet. Sci. 2020, 23, 465–468. [Google Scholar]
- Rampacci, E.; Bottinelli, M.; Stefanetti, V.; Hyatt, D.R.; Sgariglia, E.; Coletti, M.; Passamonti, F. Antimicrobial susceptibility survey on bacterial agents of canine and feline urinary tract infections: Weight of the empirical treatment. J. Glob. Antimicrob. Resist. 2018, 13, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, P.; Tsagkaraki, E.; Athanasaki, A.; Tsioutis, C.; Gikas, A. Gram-negative bacteria as emerging pathogens affecting mortality in skin and soft tissue infections. Hippokratia 2018, 22, 23–28. [Google Scholar]
- Hillier, A.; Alcorn, J.R.; Cole, L.K.; Kowalski, J.J. Pyoderma caused by Pseudomonas aeruginosa infection in dogs: 20 cases. Vet. Dermatol. 2006, 17, 432–439. [Google Scholar] [CrossRef]
- Banovic, F.; Koch, S.; Robson, D.; Jacob, M.; Olivry, T. Deep pyoderma caused by Burkholderia cepacia complex associated with ciclosporin administration in dogs: A case series. Vet. Dermatol. 2015, 26, 287-e64. [Google Scholar] [CrossRef]
- Escher, M.; Vanni, M.; Intorre, L.; Caprioli, A.; Tognetti, R.; Scavia, G. Use of antimicrobials in companion animal practice: A retrospective study in a veterinary teaching hospital in Italy. J. Antimicrob. Chemother. 2011, 66, 920–927. [Google Scholar] [CrossRef] [Green Version]
- Robbins, S.N.; Goggs, R.; Lhermie, G.; Lalonde-Paul, D.F.; Menard, J. Antimicrobial prescribing practices in small animal emergency and critical care. Front. Vet. Sci. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Chirollo, C.; Nocera, F.P.; Piantedosi, D.; Fatone, G.; Della Valle, G.; De Martino, L.; Cortese, L. Data on before and after the traceability system of Veterinary antimicrobial prescriptions in small animals at the University Veterinary Teaching Hospital of Naples. Animals 2021, 11, 913. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Beltrán, D.A.; Villar, D.; López-Osorio, S.; Ferguson, D.; Monsalve, L.K.; Chaparro-Gutiérrez, J.J. Prevalence of antimicrobial resistance in bacterial isolates from dogs and cats in a Veterinary Diagnostic Laboratory in Colombia from 2016–2019. Vet. Sci. 2020, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Nocera, F.P.; Mancini, S.; Najar, B.; Bertelloni, F.; Pistelli, L.; De Filippis, A.; Fiorito, F.; De Martino, L.; Fratini, F. Antimicrobial activity of some essential oils against methicillin-susceptible and methicillin-resistant Staphylococcus pseudintermedius-associated pyoderma in dogs. Animals 2020, 10, 1782. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency (EMA). Reflection Paper on the Risk of Antimicrobial Resistance Transfer from Companion Animals; Committee for Medicinal Products for Veterinary Use (CVMP): Amsterdam, The Netherlands, 2015; Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-risk-antimicrobial-resistance-transfer-companion-animals_en.pdf (accessed on 15 January 2015).
- Damborg, P.; Broens, E.M.; Chomel, B.B.; Guenther, S.; Pasmans, F.; Wagenaar, J.A.; Weese, J.S.; Wieler, L.H.; Windahl, U.; Vanrompay, D.; et al. Bacterial zoonoses transmitted by household pets: State-of-the-art and future perspectives for targeted research and policy actions. J. Comp. Path. 2016, 155, S27–S40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI. Performance Standard for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 3rd ed.; Clinical Laboratory and Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1. 2017. Available online: http://www.eucast.org (accessed on 10 March 2017).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Thomson, K.H.; Rantala, M.H.; Viita-Aho, T.K.; Vainio, O.M.; Kaartinen, L.A. Condition-based use of antimicrobials in cats in Finland: Results from two surveys. J. Feline Med. Surg. 2009, 11, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.A.; Williams, N.; Clegg, P.; Callaby, R.; Nuttall, T.; Coyne, K.; Pinchbeck, G.; Dawson, S. Cross-sectional survey of antimicrobial prescribing patterns in UK small animal veterinary practice. Prev. Vet. Med. 2012, 104, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, A.; Linek, M.; Moodley, A.; Guardabassi, L.; Sung, J.M.; Winkler, M.; Weiss, R.; Lloyd, D.H. First report of multiresistant, mecA-positive Staphylococcus intermedius in Europe: 12 cases from a veterinary dermatology referral clinic in Germany. Vet. Dermatol. 2007, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.W.; Vogelnest, L.J. Feline superficial pyoderma: A retrospective study of 52 cases (2001–2011). Vet. Dermatol. 2012, 23, 448-e86. [Google Scholar] [CrossRef]
- Patel, A.; Llyod, H.D.; Howell, S.A.; Noble, W.C. 2002. Investigation into the potential pathogenicity of Staphylococcus felis in a cat. Vet. Rec. 2002, 150, 668–669. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.B.; Lo, A.; Eden, C.A.; Huntley, S.; Morey, V.; Ramsey, S.; Richardson, C.; Smith, D.J.; Sutton, C.; Taylor, M.D.; et al. Survey of the prevalence, diagnosis and treatment of dermatological conditions in small animals in general practice. Vet. Rec. 2006, 158, 533–539. [Google Scholar] [CrossRef]
- Icen, H.; Yesilmen, S. Staphylococcal pyoderma in a cat: A case report. J. Anim. Vet. Adv. 2008, 7, 1332–1334. [Google Scholar]
- Misic, A.M.; Davis, M.F.; Tyldsley, A.S.; Hodkinson, B.P.; Tolomeo, P.; Hu, B.; Nachamkin, I.; Lautenbach, E.; Morris, D.O.; Griceet, E.A. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qekwana, D.N.; Oguttu, J.W.; Sithole, F.; Odoi, A. Burden and predictors of S. aureus and S. pseudintermedius infections among dogs presented at an academic veterinary hospital in South Africa (2007–2012). PeerJ 2017, 5, e3198. [Google Scholar] [CrossRef] [Green Version]
- Awosile, B.B.; Mcclure, J.T.; Saab, M.E.; Heider, L.C. Antimicrobial resistance in bacteria isolated from cats and dogs from the Atlantic Provinces, Canada from 1994–2013. Can. Vet. J. 2018, 59, 885–893. [Google Scholar]
- Jeffers, J.G. Topical therapy for drug-resistant pyoderma in small animals. Vet. Clin. N. Am. Small. Anim. Pract. 2013, 43, 41–50. [Google Scholar] [CrossRef]
- Perego, R.; Proverbio, D.; Bagnagatti De Giorgi, G.; Della Pepa, A.; Spada, E. Prevalence of otitis externa in stray cats in northern Italy. J. Feline Med. Surg. 2014, 16, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Hiblu, M.A.; Ellraiss, O.M.; Karim, E.S.; Elmishri, R.A.; Duro, E.M.; Altaeb, A.A.; Bennour, E.M. Otodectic and bacterial etiology of feline otitis externa in Tripoli, Libya. Open Vet. J. 2021, 10, 377–383. [Google Scholar] [CrossRef]
- Petersen, A.D.; Walker, R.D.; Bowman, M.M.; Schott, H.C., 2nd; Rosser, E.J., Jr. Frequency of isolation and antimicrobial susceptibility patterns of Staphylococcus intermedius and Pseudomonas aeruginosa isolates from canine skin and ear samples over a 6-year period (1992–1997). J. Am. Anim. Hosp. Assoc. 2002, 38, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Lyskova, P.; Vydrzalova, M.; Mazurova, J. Identification and antimicrobial susceptibility of bacteria and yeasts isolated from healthy dogs and dogs with otitis externa. J. Vet. Med. 2007, 54, 559–563. [Google Scholar] [CrossRef]
- Budgen, D.L. Identification and antibiotic susceptibility of bacterial isolates from dogs with otitis externa in Australia. Austr. Vet. J. 2013, 91, 43–46. [Google Scholar]
- Bloom, P. Canine superficial bacterial folliculitis: Current understanding of its etiology, diagnosis and treatment. Vet. J. 2014, 199, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Perry, L.R.; MacLennan, B.; Korven, R.; Rawlings, T.A. Epidemiological study of dogs with otitis externa in Cape Breton, Nova Scotia. Can. Vet. J. 2017, 58, 168–174. [Google Scholar] [PubMed]
- Loeffler, A.; Lloyd, D.H. What has changed in canine pyoderma? A narrative review. Vet. J. 2018, 235, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourély, C.; Cazeau, G.; Jarrige, N.; Leblond, A.; Madec, J.Y.; Haenni, M.; Gay, E. Antimicrobial resistance patterns of bacteria isolated from dogs with otitis. Epidemiol. Infect. 2019, 147, e121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nocera, F.P.; Meroni, G.; Fiorito, F.; De Martino, L.; Martino, P.A. Occurrence and antimicrobial susceptibility patterns of canine Staphylococcus pseudintermedius strains isolated from two different Italian university veterinary hospitals. Vet. Ital. 2020, 56, 263–269. [Google Scholar]
- Ferreira, J.P.; Anderson, K.L.; Correa, M.T.; Lyman, R.; Ruffin, F.; Reller, L.B.; Fowler, V.G., Jr. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS ONE 2011, 6, e26978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Sanz, E.; Torres, C.; Lozano, C.; Zarazaga, M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 83–94. [Google Scholar] [CrossRef]
- Dos Santos, T.P.; Damborg, P.; Moodley, A.; Guardabassi, L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: Inference of population structure from Multilocus sequence typing data. Front. Microbiol. 2016, 7, 1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Kolk, J.H.; Endimiani, A.; Graubner, C.; Gerber, V.; Perreten, V. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2019, 16, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Nocera, F.P.; Addante, L.; Capozzi, L.; Bianco, A.; Fiorito, F.; De Martino, L.; Parisi, A. Detection of a novel clone of Acinetobacter baumannii isolated from a dog with otitis externa. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101471. [Google Scholar] [CrossRef] [PubMed]
- Nocera, F.P.; Attili, A.R.; De Martino, L. Acinetobacter baumannii: Its clinical significance in Human and Veterinary Medicine. Pathogens 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Rafei, R.; Hamze, M.; Pailhoriès, H.; Eveillard, M.; Marsollier, L.; Joly-Guillou, M.-L.; Dabboussi, F.; Kempf, M. Extrahuman epidemiology of Acinetobacter baumannii in Lebanon. Appl. Environ. Microbiol. 2015, 81, 2359–2367. [Google Scholar] [CrossRef] [Green Version]
- Gentilini, F.; Turba, M.E.; Pasquali, F.; Mion, D.; Romagnoli, N.; Zambon, E.; Terni, D.; Peirano, G.; Pitout, J.D.D.; Parisi, A.; et al. Hospitalized pets as a source of carbapenem-resistance. Front. Microbiol. 2018, 9, 2872. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; de Moraes, N.A.; Châtre, P.; Médaille, C.; Moodley, A.; Madec, J.Y. Characterisation of clinical canine meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius in France. J. Glob. Antimicrob. Resist. 2014, 2, 119–123. [Google Scholar] [CrossRef]
- Osland, A.M.; Vestby, L.K.; Fanuelsen, H.; Schau Slettemeås, J.; Sunde, M. Clonal diversity and biofilm-forming ability of methicillin-resistant Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 2012, 67, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Moodley, A.; Damborg, P.; Nielsen, S.S. Antimicrobial resistance in methicillin susceptible and methicillin resistant Staphylococcus pseudintermedius of canine origin: Literature review from 1980 to 2013. Vet. Microbiol. 2014, 171, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Stefanetti, V.; Bietta, A.; Pascucci, L.; Marenzoni, M.L.; Coletti, M.; Franciosini, M.P.; Passamonti, F.; Casagrande Proietti, P. Investigation of the antibiotic resistance and biofilm formation of Staphylococcus pseudintermedius strains isolated from canine pyoderma. Vet. Ital. 2017, 53, 289–296. [Google Scholar] [PubMed]
- Grönthal, T.; Eklund, M.; Thomson, K.; Piiparinen, H.; Sironen, T.; Rantala, M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J. Antimicrob. Chemother. 2017, 72, 1021–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, N.C.; Moodley, A.; Ghibaudo, G.; Guardabassi, L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: Indirect evidence of zoonotic transmission. Zoonoses Public Health 2011, 58, 533–539. [Google Scholar] [CrossRef] [PubMed]
- van Duijkeren, E.; Kamphuis, M.I.C.; Mije, V.; Laarhoven, L.M.; Duim, B.; Wagenaar, J.A.; Houwers, D.J. Staphylococcus pseudintermedius between infected dogs and cats and contact pets, humans and the environment in households and veterinary clinics. Vet. Microbiol. 2011, 150, 338–343. [Google Scholar] [CrossRef]
- Saber, H.; Jasni, A.S.; Jamaluddin, T.Z.M.T.; Ibrahim, R. A Review of Staphylococcal Cassette Chromosome mec (SCCmec) Types in Coagulase-Negative Staphylococci (CoNS) Species. Malays J. Med. Sci. 2017, 24, 7–18. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [Green Version]
- Mekić, S.; Matanović, K.; Šeol, B. Antimicrobial susceptibility of Pseudomonas aeruginosa isolates from dogs with otitis externa. Vet. Rec. 2011, 169, 125. [Google Scholar] [CrossRef]
- Hariharan, H.; Coles, M.; Poole, D.; Lund, L.; Page, R. Update on antimicrobial susceptibilities of bacterial isolates from canine and feline otitis externa. Can. Vet. J. 2006, 47, 253–255. [Google Scholar] [PubMed]
- Rubin, J.; Walker, R.D.; Blickenstaff, K.; Bodeis-Jones, S.; Zhao, S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet. Microbiol. 2008, 131, 164–172. [Google Scholar] [CrossRef]
- Ludwig, C.; de Jong, A.; Moyaert, H.; El Garch, F.; Janes, R.; Klein, U.; Morrissey, I.; Thiry, J.; Youala, M. Antimicrobial susceptibility monitoring of dermatological bacterial pathogens isolated from diseased dogs and cats across Europe (ComPath results). J. Appl. Microbiol. 2016, 121, 1254–1267. [Google Scholar] [CrossRef]
- Dowling, P.M. Antimicrobial therapy of skin and ear infections. Can. Vet. J. 1996, 37, 695–699. [Google Scholar]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Johnson, A.P.; Woodford, N. Global spread of antibiotic resistance: The example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 2013, 62, 499–513. [Google Scholar] [CrossRef]
- WHO (World Health Organization). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently. 2017. Available online: http://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 18 June 2018).
- Kroemer, S.; El Garch, F.; Galland, D.; Petit, J.L.; Woehrle, F.; Boulouis, H.J. Antibiotic susceptibility of bacteria isolated from infections in cats and dogs throughout Europe (2002–2009). Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 97–108. [Google Scholar] [CrossRef]
- European Law 2017 No. 167 of 20 November 2017 on Traceability of Veterinary Medicines and Medicated Feeds for the Achievement of the Objectives Stated in Directives 2001/82/EC and 90/167/EEC; European Commission: Brussels, Belgium, 2017.





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nocera, F.P.; Ambrosio, M.; Fiorito, F.; Cortese, L.; De Martino, L. On Gram-Positive- and Gram-Negative-Bacteria-Associated Canine and Feline Skin Infections: A 4-Year Retrospective Study of the University Veterinary Microbiology Diagnostic Laboratory of Naples, Italy. Animals 2021, 11, 1603. https://doi.org/10.3390/ani11061603
Nocera FP, Ambrosio M, Fiorito F, Cortese L, De Martino L. On Gram-Positive- and Gram-Negative-Bacteria-Associated Canine and Feline Skin Infections: A 4-Year Retrospective Study of the University Veterinary Microbiology Diagnostic Laboratory of Naples, Italy. Animals. 2021; 11(6):1603. https://doi.org/10.3390/ani11061603
Chicago/Turabian StyleNocera, Francesca Paola, Monica Ambrosio, Filomena Fiorito, Laura Cortese, and Luisa De Martino. 2021. "On Gram-Positive- and Gram-Negative-Bacteria-Associated Canine and Feline Skin Infections: A 4-Year Retrospective Study of the University Veterinary Microbiology Diagnostic Laboratory of Naples, Italy" Animals 11, no. 6: 1603. https://doi.org/10.3390/ani11061603






