KAT2B Gene Polymorphisms Are Associated with Body Measure Traits in Four Chinese Cattle Breeds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Animals, DNA Sampling, and Phenotype Data Collection
2.3. RNA Extraction, cDNA Synthesis, and Real-Time Quantitative PCR (qPCR)
2.4. Detection of KAT2B SNPs by DNA Pool Sequencing
2.5. Genotyping for KAT2B SNPs by PCR–RFLP
2.6. Statistical Analysis
3. Results
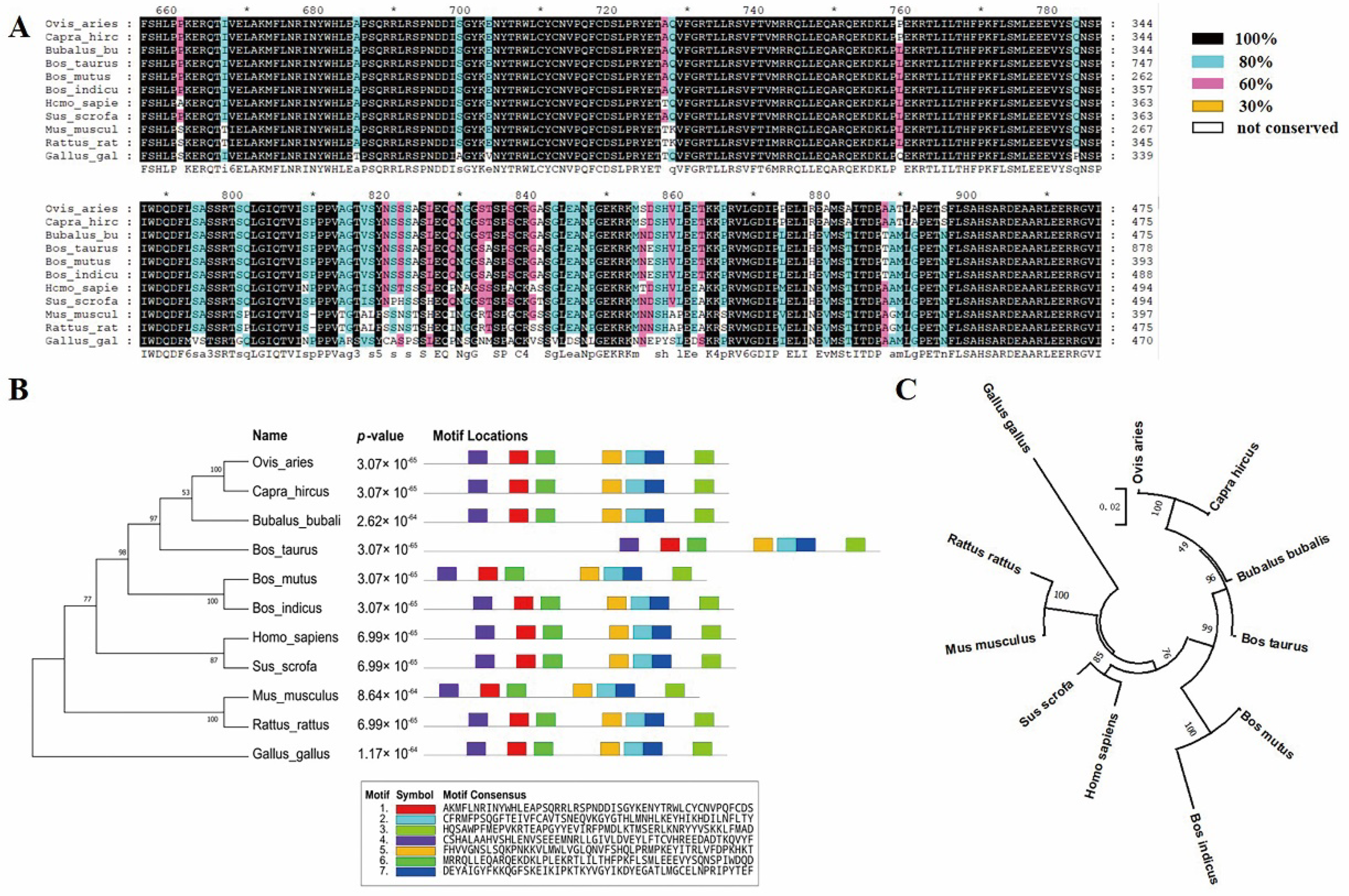
3.1. Biological Evolution and Conservation Analysis of KAT2B
3.2. Bovine KAT2B Gene Expression Profile
3.3. Identification of KAT2B SNPs
3.4. Correlation Analysis of Bovine KAT2B Gene SNPs and Body Measure Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, C.; Raza, S.H.A.; Khan, R.; Sabek, A.; Khan, S.; Ullah, I.; Memon, S.; El-Aziz, A.H.A.; Shah, M.A.; Shijun, L.; et al. Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle. Genomics 2020, 112, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Peng, W.; Cao, X.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Differential Expression of KCNJ12 Gene and Association Analysis of Its Missense Mutation with Growth Traits in Chinese Cattle. Animals 2019, 9, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Zhang, C.; Lai, X.; Xue, J.; Lan, X.; Lei, C.; Jia, Y.; Chen, H. Associations between polymorphisms in the NICD domain of bovine NOTCH1 gene and growth traits in Chinese Qinchuan cattle. J. Appl. Genet. 2017, 58, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Shijun, L.; Khan, R.; Schreurs, N.M.; Manzari, Z.; Abd El-Aziz, A.H.; Ullah, I.; Kaster, N.; Shah, M.A.; Zan, L. Polymorphism of the PLIN1 gene and its association with body measures and ultrasound carcass traits in Qinchuan beef cattle. Genome 2020, 63, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Schaeffer, L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Zhang, L.; Li, G.P. Progress in the molecular and genetic modification breeding of beef cattle in China. Yi Chuan 2017, 39, 984–1015. [Google Scholar] [PubMed]
- Abd El-Hack, M.E.; Abdelnour, S.A.; Swelum, A.A.; Arif, M. The application of gene marker-assisted selection and proteomics for the best meat quality criteria and body measurements in Qinchuan cattle breed. Mol. Biol. Rep. 2018, 45, 1445–1456. [Google Scholar] [CrossRef]
- Helmlinger, D.; Tora, L. Sharing the SAGA. Trends Biochem. Sci. 2017, 42, 850–861. [Google Scholar] [CrossRef]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef]
- Bondy-Chorney, E.; Denoncourt, A.; Sai, Y.; Downey, M. Nonhistone targets of KAT2A and KAT2B implicated in cancer biology. Biochem. Cell Biol. 2019, 97, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Fournier, M.; Orpinell, M.; Grauffel, C.; Scheer, E.; Garnier, J.M.; Ye, T.; Chavant, V.; Joint, M.; Esashi, F.; Dejaegere, A.; et al. KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun. 2016, 7, 13227. [Google Scholar] [CrossRef] [PubMed]
- Moisa, S.J.; Shike, D.W.; Meteer, W.T.; Keisler, D.; Faulkner, D.B.; Loor, J.J. Yin yang 1 and adipogenic gene network expression in longissimus muscle of beef cattle in response to nutritional management. Gene Regul. Syst. Biol. 2013, 7, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, J.; Li, C.; Wang, Y.; Li, L.; Cai, H.; Lan, X.; Lei, C.; Zhao, X.; Chen, H. Characterization of transcriptional complexity during adipose tissue development in bovines of different ages and sexes. PLoS ONE 2014, 9, e101261. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell., D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Zhang, D.; Xu, J.; Yang, P.; Wen, Y.; He, H.; Li, J.; Liang, J.; Zheng, Y.; Zhang, Z.; Wang, X.; et al. Genetic variant of SPARC gene and its association with growth traits in Chinese cattle. Arch. Anim. Breed. 2020, 63, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Liu, M.; Li, B.; Zhou, Y.; Huang, Y.; Lan, X.; Qu, W.; Qi, X.; Bai, Y.; Chen, H. Polymorphisms of FLII implicate gene expressions and growth traits in Chinese cattle. Mol. Cell. Probes 2016, 30, 266–272. [Google Scholar] [CrossRef]
- Gui, L.S.; Raza, S.H.A.; Garcia, M.; Sun, Y.G.; Ullah, I.; Han, Y.C. Genetic variants in the SIRT6 transcriptional regulatory region affect gene activity and carcass quality traits in indigenous Chinese beef cattle (Bos taurus). BMC Genom. 2018, 19, 785. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Jia, C.; Jiang, A.; Zhang, H.; Wang, Y.; Liu, F.; Yang, L.; Sun, Y.; Lv, R.; Song, X. Analysis of Association between MGMT and p53 Gene Single Nucleotide Polymorphisms and Laryngeal Cancer. Anticancer Res. 2017, 37, 4399–4403. [Google Scholar]
- Zhou, Y.; Li, C.; Cai, H.; Xu, Y.; Lan, X.; Lei, C.; Chen, H. Novel polymorphisms of the APOA2 gene and its promoter region affect body traits in cattle. Gene 2013, 531, 288–293. [Google Scholar] [CrossRef]
- Nei, M.; Roychoudhury, A.K. Sampling variances of heterozygosity and genetic distance. Genetics 1974, 76, 379–390. [Google Scholar] [CrossRef]
- Wang, J.; Shete, S. Testing Departure from Hardy-Weinberg Proportions. Methods Mol. Biol. 2012, 850, 77–102. [Google Scholar]
- Nagy, Z.; Tora, L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 2007, 26, 5341–5357. [Google Scholar] [CrossRef] [Green Version]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [Green Version]
- Clements, A.; Rojas, J.R.; Trievel, R.C.; Wang, L.; Berger, S.L.; Marmorstein, R. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 1999, 18, 3521–3532. [Google Scholar] [CrossRef]
- Carré, C.; Szymczak, D.; Pidoux, J.; Antoniewski, C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell. Biol. 2005, 25, 8228–8238. [Google Scholar] [CrossRef] [Green Version]
- Fang, M.; Zhang, M.; Wang, Y.; Wei, F.; Wu, J.; Mou, X.; Zhang, Y.; Liang, X.; Tang, J. Long Noncoding RNA AFAP1-AS1 Is a Critical Regulator of Nasopharyngeal Carcinoma Tumorigenicity. Front. Oncol. 2020, 10, 601055. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yamauchi, J.; Kuwata, T.; Tamura, T.; Yamashita, T.; Bae, N.; Westphal, H.; Ozato, K.; Nakatani, Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 11303–11306. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, A.; Minamide, R.; Iwata, J. The role of acetyltransferases for the temporal-specific accessibility of beta-catenin to the myogenic gene locus. Sci. Rep. 2018, 8, 15057. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.K.; Aparicio-Sánchez, J.J.; Buxton, S.; Ketley, A.; Mohamed, T.; Rutland, C.S.; Loughna, S.; Brook, J.D. Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb development. J. Mol. Cell. Cardiol. 2018, 114, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, N.; Denechaud, P.D.; Gromada, X.; Hannou, S.A.; Zhang, H.; Rashid, T.; Salas, E.; Durand, E.; Sand, O.; Bonnefond, A.; et al. KAT2B Is Required for Pancreatic Beta Cell Adaptation to Metabolic Stress by Controlling the Unfolded Protein Response. Cell Rep. 2016, 15, 1051–1061. [Google Scholar] [CrossRef] [Green Version]
- Fragomeni, B.O.; Lourenco, D.A.L.; Legarra, A.; VanRaden, P.M.; Misztal, I. Alternative SNP weighting for single-step genomic best linear unbiased predictor evaluation of stature in US Holsteins in the presence of selected sequence variants. J. Dairy Sci. 2019, 102, 10012–10019. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, M.; Wang, S.; Xu, Y.; Lan, X.; Li, Z.; Lei, C.; Yang, D.; Jia, Y.; Chen, H. Association analysis of bovine Foxa2 gene single sequence variant and haplotype combinations with growth traits in Chinese cattle. Gene 2014, 536, 385–392. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, E.; Wang, K.; Zhang, Y.; Yan, H.; Qu, L.; Chen, H.; Lan, X.; Pan, C. Two Insertion/Deletion Variants within SPAG17 Gene Are Associated with Goat Body Measurement Traits. Animals 2019, 9, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Li, B.; Shi, T.; Huang, Y.; Liu, G.E.; Lan, X.; Lei, C.; Chen, H. Copy number variation of bovine SHH gene is associated with body conformation traits in Chinese beef cattle. J. Appl. Genet. 2019, 60, 199–207. [Google Scholar] [CrossRef]

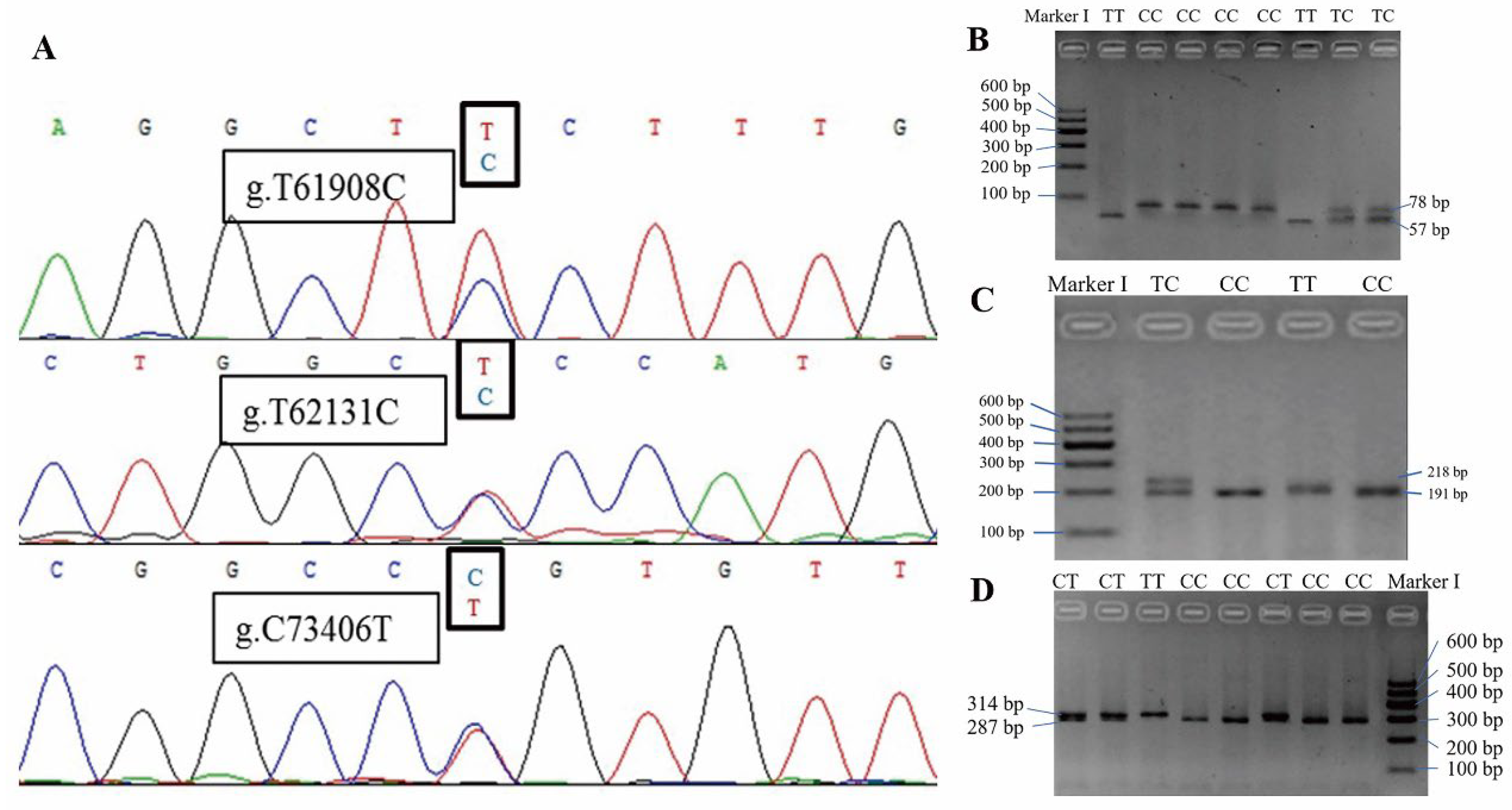
| Targeted SNP or Gene | Accession Number | Primer Pair | Tm (°C) | Name | Position | Sequence (5′-3′) |
|---|---|---|---|---|---|---|
| g.T61908C | AC_000158.1 | P1 | 68 | F1 | 61,883–61,907 | GGGTTCCCACTGCACAGGCCAAGCT |
| R1 | 61,936–61,960 | GTCCATCAGACGCCCCCACACAGAG | ||||
| g.T62131C | AC_000158.1 | P2 | 64 | F2 | 62,105–62,129 | ACCTTCAAGGCCTTTTACATGCGGG |
| R2 | 62,298–62,322 | TCAAAGAGGAATGGACACAGGCAGA | ||||
| g.C73406T | AC_000158.1 | P3 | 61 | F3 | 73,379–73,403 | CTCTTCCCAGTCTCACTTTTGTGGG |
| R3 | 73,668–73,692 | AGGCACACTGTTTGATGAGTTTCTA | ||||
| KAT2B | XM_019966941.1 | P | 60 | F | 7–26 | CGGTCTCTTGACCTTCGTGA |
| R | 158–177 | TTTGCCGGGTATGGAAGGAG | ||||
| β-actin | NM_173979.3 | Pr | 58 | Fr | 831–851 | GTCATCACCATCGGCAATGAG |
| Rr | 896–914 | AATGCCGCAGGATTCCATG |
| SNP | Breed | Genotype Frequency | Allele Frequency | χ2 (HWE) | PIC | He | Ne | |||
|---|---|---|---|---|---|---|---|---|---|---|
| g.T61908C | CC | TC | TT | C | T | |||||
| Qinchuan (658) | 0.13 | 0.61 | 0.26 | 0.43 | 0.57 | p > 0.05 | 0.37 | 0.49 | 1.96 | |
| Fu (52) | 0.23 | 0.56 | 0.21 | 0.51 | 0.49 | p > 0.05 | 0.37 | 0.50 | 2.00 | |
| Yak (48) | 0.27 | 0.58 | 0.15 | 0.56 | 0.44 | p > 0.05 | 0.37 | 0.49 | 1.97 | |
| Chaidam (69) | 0.45 | 0.35 | 0.20 | 0.62 | 0.38 | p > 0.05 | 0.36 | 0.47 | 1.89 | |
| g.T62131C | CC | TC | TT | C | T | |||||
| Qinchuan (658) | 0.39 | 0.43 | 0.18 | 0.61 | 0.39 | p > 0.05 | 0.36 | 0.48 | 1.91 | |
| Fu (52) | 0.28 | 0.37 | 0.35 | 0.47 | 0.53 | p > 0.05 | 0.37 | 0.50 | 1.99 | |
| Yak (48) | 0.80 | 0.10 | 0.10 | 0.84 | 0.16 | p < 0.01 | 0.23 | 0.27 | 1.37 | |
| Chaidam (69) | 0.29 | 0.38 | 0.33 | 0.48 | 0.52 | p > 0.05 | 0.37 | 0.50 | 2.00 | |
| g.C73406T | CC | CT | TT | C | T | |||||
| Qinchuan (658) | 0.47 | 0.43 | 0.10 | 0.69 | 0.31 | p > 0.05 | 0.34 | 0.43 | 1.75 | |
| Fu (52) | 0.40 | 0.33 | 0.27 | 0.57 | 0.43 | p > 0.05 | 0.37 | 0.49 | 1.96 | |
| Yak (48) | 0.02 | 0.44 | 0.54 | 0.24 | 0.76 | p > 0.05 | 0.30 | 0.36 | 1.57 | |
| Chaidam (69) | 0.23 | 0.55 | 0.21 | 0.51 | 0.49 | p > 0.05 | 0.37 | 0.50 | 2.00 | |
| SNP | Breed | Growth Traits | Genotype | ||
|---|---|---|---|---|---|
| CC | TC | TT | |||
| g.T61908C | Fu | BL(cm) | 84.00 ± 7.06 a,b | 82.86 ± 8.22 b | 89.73 ± 10.04 a |
| g.T62131C | Fu | CCB(cm) | 11.6 ± 1.35 A | 10.26 ± 1.49 B | 11.39 ± 1.54 A |
| Qinchuan | BH(cm) | 127.3 ± 6.74 b | 130.35 ± 6.07 a | 129.26 ± 7.15 a,b | |
| CW(cm) | 36.52 ± 4.75 b | 37.91 ± 4.43 a,b | 39.48 ± 4.13 a | ||
| HIW(cm) | 40.67 ± 5.38 b | 43.06 ± 4.78 a | 43.5 ± 5.02 a | ||
| WH(cm) | 124.31 ± 6.47 b | 129.63 ± 10.81 a | 127 ± 7.05 a,b | ||
| g.C73406T | Yak | BM(cm) | 151.00 ± 0.11 B | 166.57 ± 9.53 B | 177 ± 15.34 A |
| Chaidam | BM(cm) | 246.36 ± 30.29 A,B | 272.27 ± 21.14 A | 228.9 ± 57.21 B | |
| CG(cm) | 152.64 ± 7.59 A | 141.92 ± 8.12 B | 140.2 ± 13.75 B | ||
| CCB(cm) | 16.09 ± 1.22 A | 14.69 ± 0.95 B | 14.3 ± 1.06 B | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Li, B.; Chen, Y.; Chen, H.; Liu, M. KAT2B Gene Polymorphisms Are Associated with Body Measure Traits in Four Chinese Cattle Breeds. Animals 2022, 12, 1954. https://doi.org/10.3390/ani12151954
Lin X, Li B, Chen Y, Chen H, Liu M. KAT2B Gene Polymorphisms Are Associated with Body Measure Traits in Four Chinese Cattle Breeds. Animals. 2022; 12(15):1954. https://doi.org/10.3390/ani12151954
Chicago/Turabian StyleLin, Xiaoding, Bo Li, Yuhan Chen, Hong Chen, and Mei Liu. 2022. "KAT2B Gene Polymorphisms Are Associated with Body Measure Traits in Four Chinese Cattle Breeds" Animals 12, no. 15: 1954. https://doi.org/10.3390/ani12151954






