An Isolated and Deeply Divergent Hynobius Species from Fujian, China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Sampling
2.2. Ethical Approval
2.3. DNA Preparation
2.4. Molecular Analyses
2.5. Morphometry
2.6. Egg and Larval Development
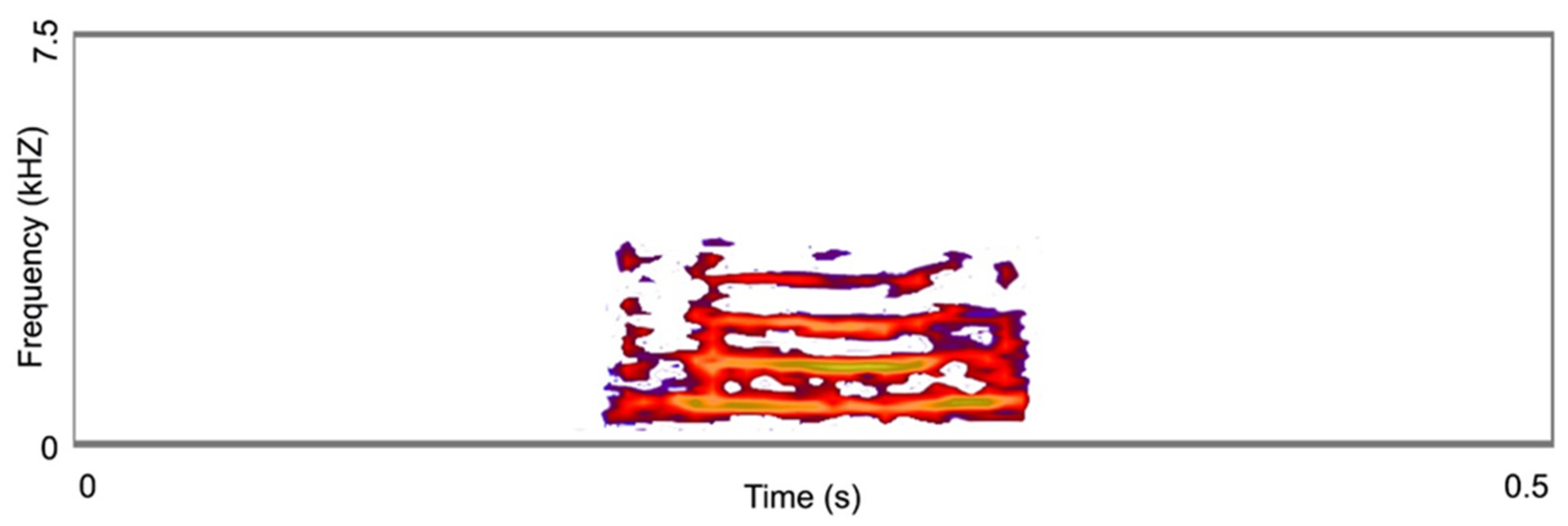
2.7. Acoustic Signal
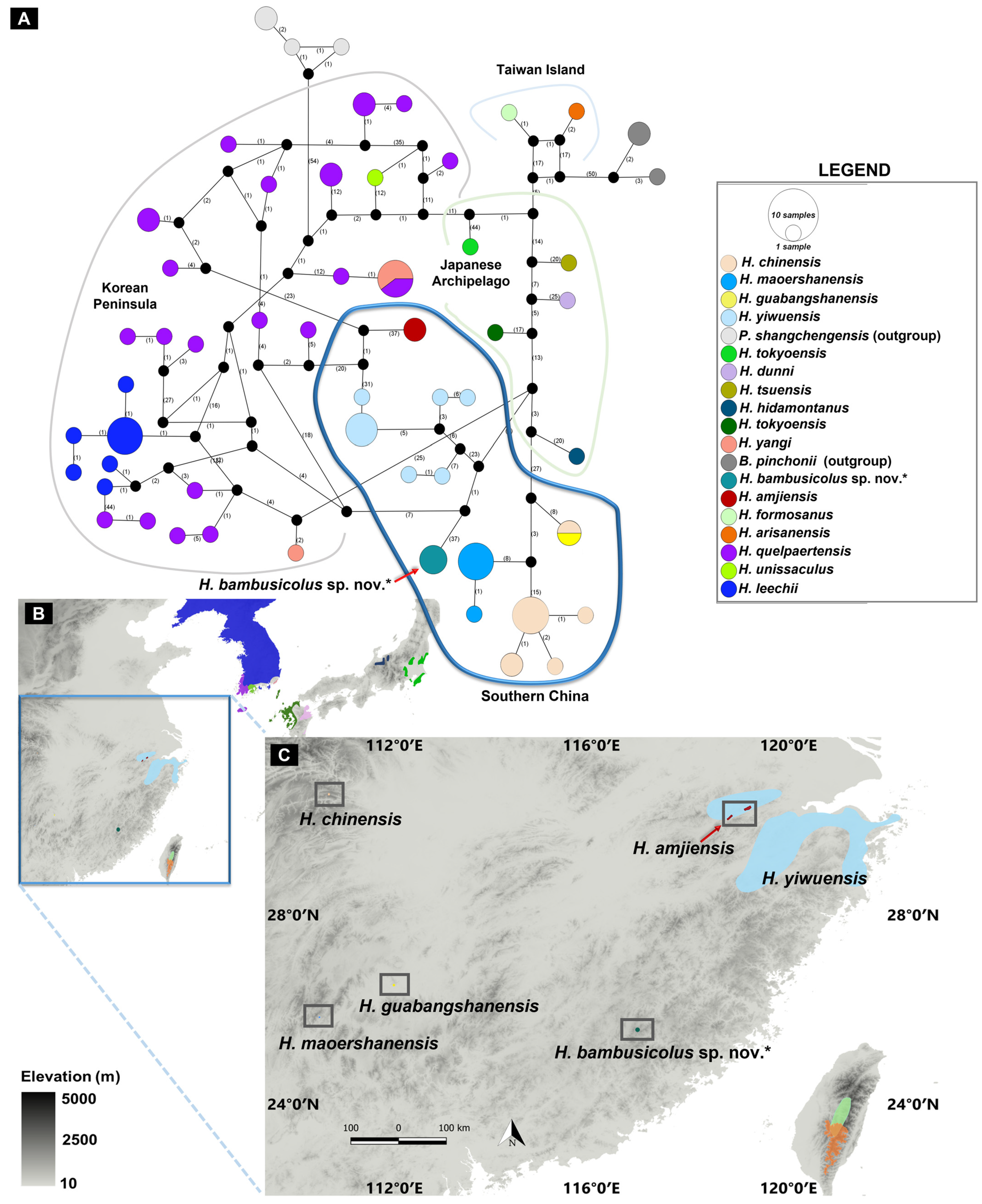
3. Results
3.1. Molecular Analyses
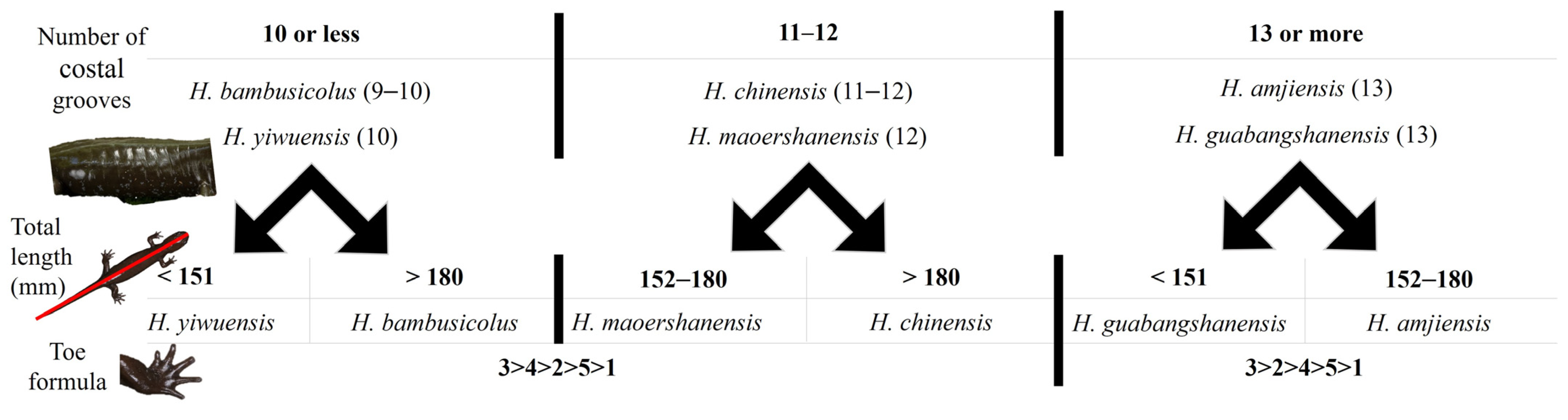
3.2. Morphometric results
- -
- The combination of 10 or fewer grooves with the following:
- -
- A total length < 151 mm assigns the individual to H. yiwuensis;
- -
- A total length > 180 mm assigns the individual to H. bambusicolus sp. nov;
- -
- The combination of 11 or 12 grooves with the following:
- -
- A total length < 180 mm assigns the individual to H. maoershanensis;
- -
- A total length > 180 mm assigns the individual to H. chinensis;
- -
- The combination of 13 or more grooves with the following:
- -
- A total length < 151 mm assigns the individual to H. guabangshanensis;
- -
- A total length > 152 mm assigns the individual to H. amjiensis.
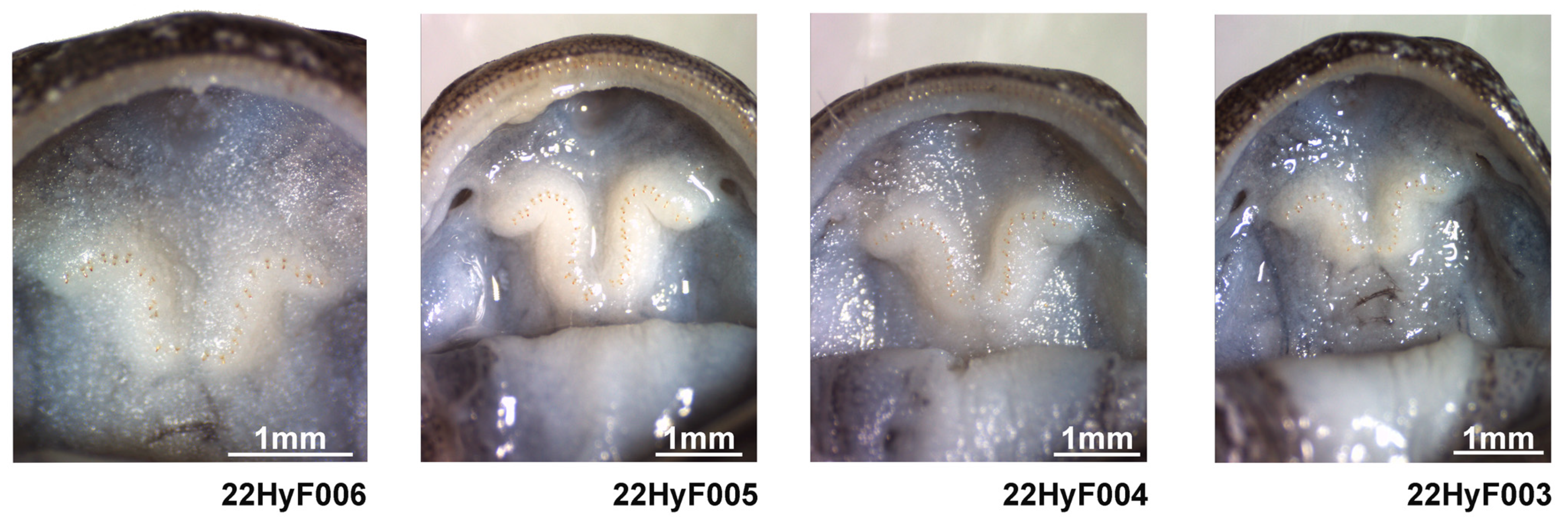
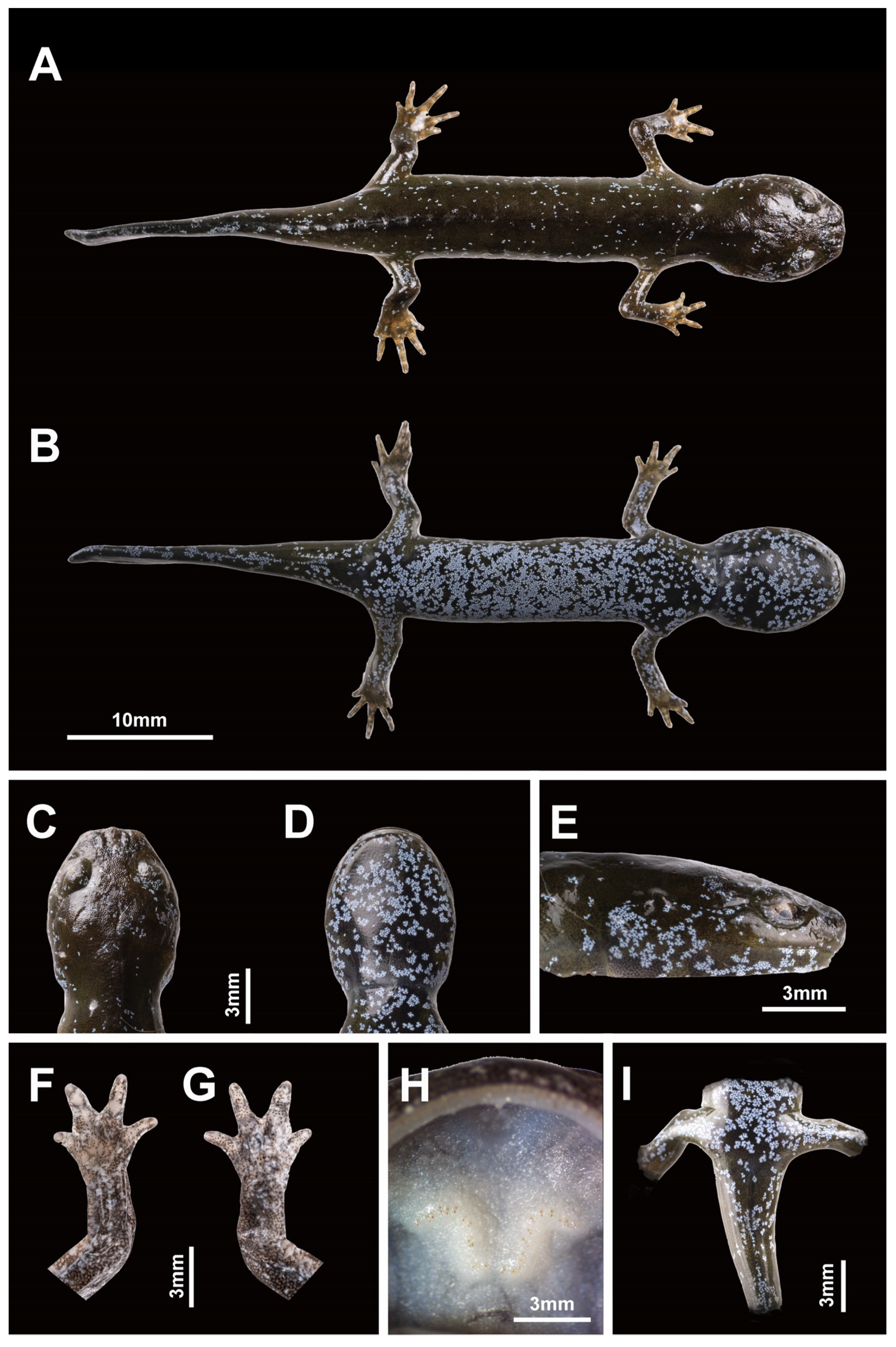
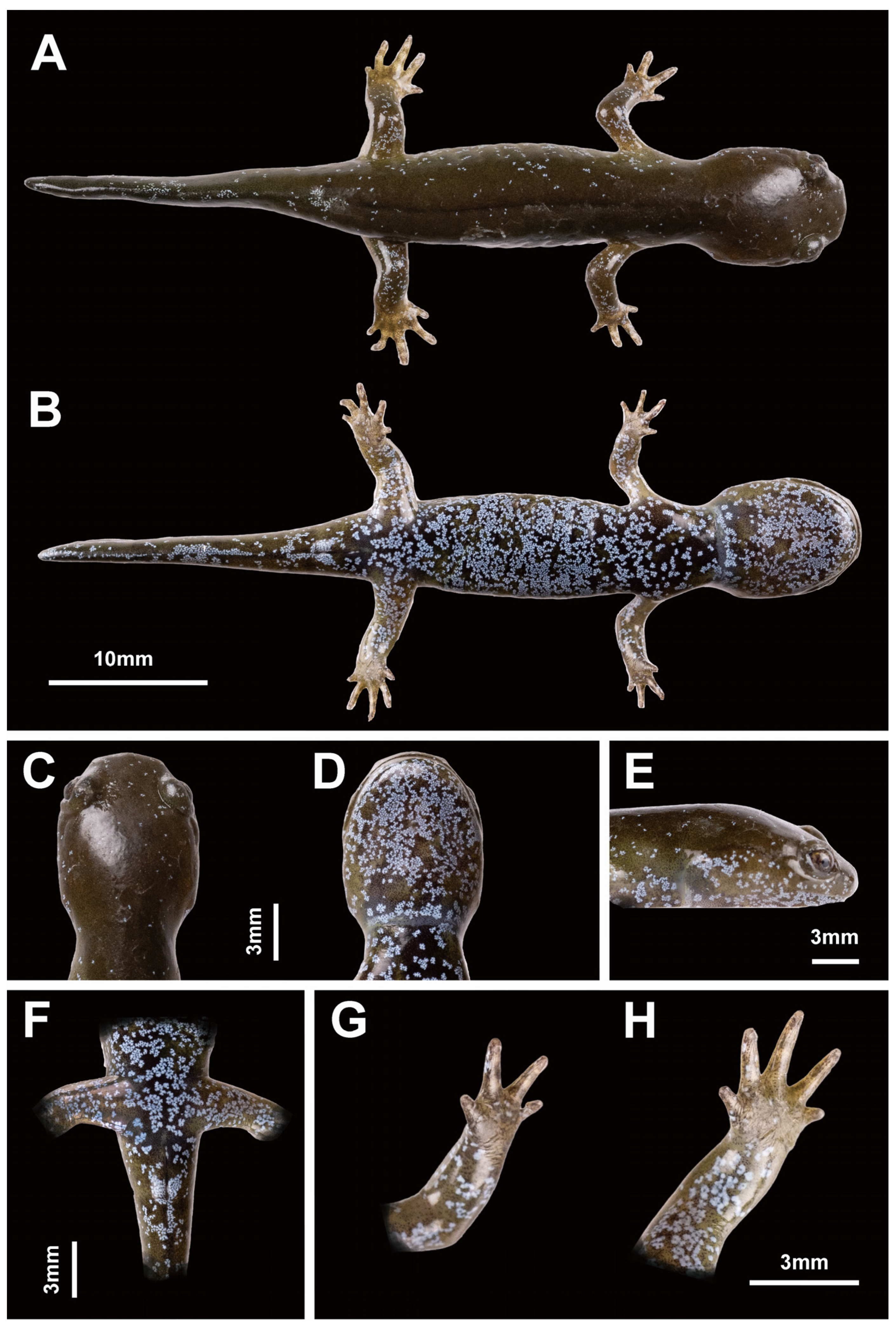
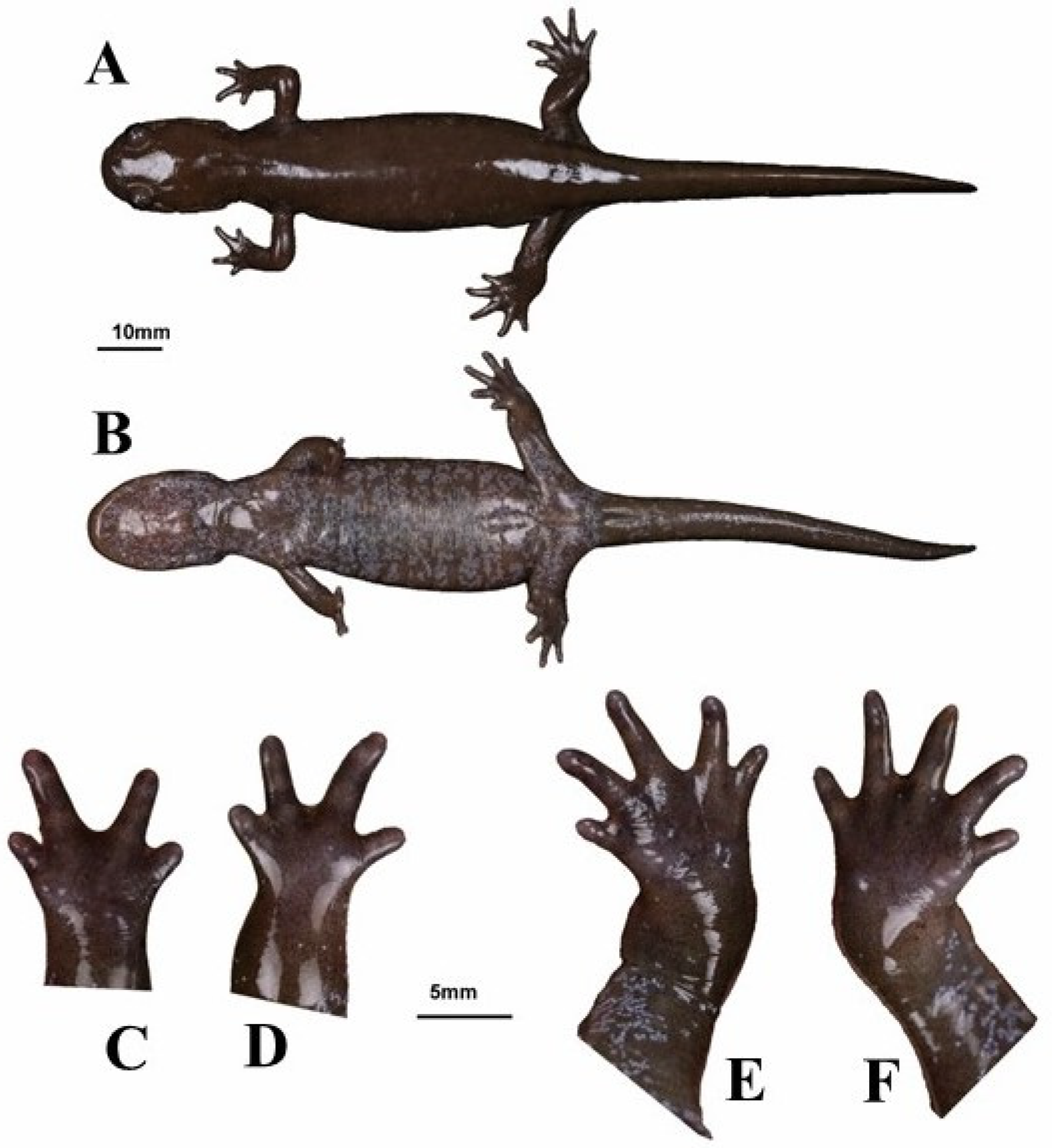
3.3. Species Description
Nomenclature History
- Holotype
- Paratypes
- Etymology
- Identity, diagnosis, and distribution
- ZooBank registration
- Nomenclatural acts
- Habitat and behavior
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Wake, D.B.; Vredenburg, V.T. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. 1), 11466–11473. [Google Scholar] [CrossRef]
- Ceballos, G.; Ehrlich, P.R.; Barnosky, A.D.; García, A.; Pringle, R.M.; Palmer, T.M. Accelerated modern human–induced species losses: Entering the sixth mass extinction. Sci. Adv. 2015, 1, e1400253. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.A.; Elliott Smith, R.E.; Lyons, S.K.; Payne, J.L. Body size downgrading of mammals over the late Quaternary. Science 2018, 360, 310–313. [Google Scholar] [CrossRef]
- Manne, L.L.; Pimm, S.L. Beyond eight forms of rarity: Which species are threatened and which will be next? Anim. Conserv. 2001, 4, 221–229. [Google Scholar] [CrossRef]
- Régnier, C.; Achaz, G.; Lambert, A.; Cowie, R.H.; Bouchet, P.; Fontaine, B. Mass extinction in poorly known taxa. Proc. Natl. Acad. Sci. USA 2015, 112, 7761–7766. [Google Scholar] [CrossRef]
- Tedesco, P.A.; Bigorne, R.; Bogan, A.E.; Giam, X.; Jézéquel, C.; Hugueny, B. Estimating how many undescribed species have gone extinct. Conserv. Biol. 2014, 28, 1360–1370. [Google Scholar] [CrossRef]
- Brodie, J.F.; Aslan, C.E.; Rogers, H.S.; Redford, K.H.; Maron, J.L.; Bronstein, J.L.; Groves, C.R. Secondary extinctions of biodiversity. Trends Ecol. Evol. 2014, 29, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Rix, M.G.; Huey, J.A.; Main, B.Y.; Waldock, J.M.; Harrison, S.E.; Comer, S.; Austin, A.D.; Harvey, M.S. Where have all the spiders gone? The decline of a poorly known invertebrate fauna in the agricultural and arid zones of southern Australia. Austral Entomol. 2017, 56, 14–22. [Google Scholar] [CrossRef]
- Rinawati, F.; Stein, K.; Lindner, A. Climate change impacts on biodiversity—The setting of a lingering global crisis. Diversity 2013, 5, 114–123. [Google Scholar] [CrossRef]
- Otto, S.P. Adaptation, speciation and extinction in the Anthropocene. Proc. R. Soc. B 2018, 285, 20182047. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, J.; Meng, W.; Lohman, D.J.; Pierce, N.E. Out of sight, out of mind: Public and research interest in insects is negatively correlated with their conservation status. Insect Conserv. Divers. 2021, 14, 700–708. [Google Scholar] [CrossRef]
- Sodhi, N.S.; Brook, B.W.; Bradshaw, C.J. Causes and consequences of species extinctions. In The Princeton Guide to Ecology; Levin, S.A., Carpenter, S.R., Godfray, H.C.J., Kinzig, A.P., Loreau, M., Losos, J.B., Walker, B., Wilcove, D.S., Eds.; Princeton University Press: Princeton, NJ, USA, 2009. [Google Scholar]
- Lindenmayer, D.B.; Fischer, J. Habitat Fragmentation and Landscape Change: An Ecological and Conservation Synthesis; Island Press: Washington, DC, USA, 2013. [Google Scholar]
- Wang, K.; Ren, J.; Chen, H.; Lyu, Z.; Guo, X.; Jiang, K.; Chen, J.; Li, J.; Guo, P.; Wang, Y.; et al. The updated checklists of amphibians and reptiles of China. Biodivers. Sci. 2020, 28, 189–218. [Google Scholar]
- Moura, M.R.; Jetzt, W. Shortfalls and opportunities in terrestrial vertebrate species discovery. Nat. Ecol. Evol. 2021, 5, 631–639. [Google Scholar] [CrossRef]
- Button, S.; Borzée, A. An integrative synthesis to global amphibian conservation priorities. Glob. Chang. Biol. 2021, 27, 4516–4529. [Google Scholar] [CrossRef] [PubMed]
- Borzée, A.; Min, M.-S. Disentangling the impact of speciation, sympatry and island effect on the morphology of seven Hynobius sp. salamanders. Animals 2021, 11, 187. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Misawa, Y.; Yoshikawa, N.; Nishikawa, K. Taxonomic reappraisal of Hynobius tokyoensis, with description of a new species from northeastern Honshu, Japan (Amphibia: Caudata). Zootaxa 2022, 5168, 207–221. [Google Scholar] [CrossRef]
- Sugawara, H.; Naito, J.I.; Iwata, T.; Nagano, M. Molecular phylogenetic and morphological problems of the Aki Salamander Hynobius akiensis: Description of two new species from Chugoku, Japan. Bull. Kanagawa Prefect. Mus. 2022, 2022, 35–46. [Google Scholar]
- Sugawara, H.; Fujitani, T.; Seguchi, S.; Sawahata, T.; Nagano, M. Taxonomic re-examination of the Yamato Salamander Hynobius vandenburghi: Description of a new species from Central Honshu, Japan. Bull. Kanagawa Prefect. Mus. 2022, 51, 47–59. [Google Scholar]
- Lai, J.S.; Lue, K.Y. Two new Hynobius (Caudata: Hynobiidae) salamanders from Taiwan. Herpetologica 2008, 64, 63–80. [Google Scholar] [CrossRef]
- Günther, A.C.L.G. Third contribution to our knowledge of reptiles and fishes from the upper Yangtze-Kiang. Ann. Mag. Nat. Hist. 1889, 6, 218–229. [Google Scholar] [CrossRef]
- Gu, H.-q. A new species of Hynobius-Hynobius amjiensis. In Animal Science Research: A Symposium Issued to Celebrate the 90th Birthday of the Professor Mangven Ly Chang; Qian, Y., Zhao, E.-M., Zhao, K.-t., Eds.; Chinese Forestry Press: Beijing, China, 1992; pp. 39–43. [Google Scholar]
- Shen, Y.-h.; Deng, X.-j.; Wang, B. A new hynobiid species, Hynobius guabangshanensis, from Hunan Province, China (Amphiba: Hynobiidae). Acta Zool. Sin. 2004, 50, 209–215. [Google Scholar]
- Zhou, F.; Jiang, A.-W.; Jiang, D.-b. A new species of the genus Hynobius from Guangxi Zhuang Autonomous Region, China (Caudata, Hynobiidae). Acta Zootaxonomica Sin. 2006, 31, 670–674. [Google Scholar]
- Cai, C.-m. A survey of tailed amphibians for Zhejiang, with description of a new species of Hynobius. Acta Herpetol. Sin. 1985, 4, 109–114. [Google Scholar]
- Hu, S.; Fei, L.; Ye, C. Investigation report of amphibians in Fujian. In Research Material of Amphibians and Reptiles; Sichuan Institute of Biology: Chengdu, China, 1978; Volume 4, pp. 22–29. [Google Scholar]
- Fei, L.; Hu, S.; Ye, C.; Huang, Y. Fauna Sinica: Amphibia; Science Press: Beijing, China, 2006; Volume 1. [Google Scholar]
- IUCN. IUCN Policy Statement on Research Involving Species at Risk of Extinction; International Union for the Conservation of Nature: Gland, Switzerland, 1989. [Google Scholar]
- Kusano, T.; Miyashita, K. Dispersal of the salamander, Hynobius nebulosus tokyoensis. J. Herpetol. 1984, 18, 349–353. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Palumbi, S. Nucleic acids II: The polymerase chain reaction. In Molecular Systematics; Hillis, D., Moritz, C., Mable, B., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- Rassmann, K. Evolutionary age of the Galapagos iguanas predates the age of the present Galapagos Islands. Mol. Phylogenet. Evol. 1997, 7, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Xia, Y.; Gu, H.F.; Peng, R.; Chen, Q.; Zheng, Y.C.; Murphy, R.W.; Zeng, X.M. COI is better than 16S rRNA for DNA barcoding Asiatic salamanders (Amphibia: Caudata: Hynobiidae). Mol. Ecol. Resour. 2012, 12, 48–56. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System.; Open Source Geospatial Foundation Project. Available online: http://qgis.osgeo.org (accessed on 3 May 2023).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Chen, H.; Bu, R.; Ning, M.; Yang, B.; Wu, Z.; Huang, H. Sexual Dimorphism in the Chinese Endemic Species Hynobius maoershanensis (Urodela: Hynobiidae). Animals 2022, 12, 1712. [Google Scholar] [CrossRef]
- Chen, C.; Yang, J.; Wu, Y.; Fan, Z.; Lu, W.; Chen, S.; Yu, L. The breeding ecology of a critically endangered salamander, Hynobius amjiensis (Caudata: Hynobiidae), Endemic to Eastern China. Asian Herpetol. Res. 2016, 7, 54–59. [Google Scholar] [CrossRef]
- Allan, B.M.; Nimmo, D.G.; Ierodiaconou, D.; Wal, J.V.D.; Koh, L.P.; Ritchie, E.G. Futurecasting ecological research: The rise of technoecology. Ecosphere 2018, 9, e02163. [Google Scholar] [CrossRef]
- Fernando, M.d.F.L. 3D print so more scholars can access unique specimens. Nature 2018, 563, 7731. [Google Scholar]
- Iwasawa, H.; Yamashita, K. Normal stages of development of a hynobiid salamander, Hynobius nigrescens Stejneger. Jpn. J. Herpetol. 1991, 14, 39–62. [Google Scholar] [CrossRef]
- Prasad, V.K.; Chuang, M.F.; Das, A.; Ramesh, K.; Yi, Y.; Dinesh, K.P.; Borzée, A. Coexisting good neighbours: Acoustic and calling microhabitat niche partitioning in two elusive syntopic species of balloon frogs, Uperodon systoma and U. globulosus (Anura: Microhylidae) and potential of individual vocal signatures. BMC Zool. 2022, 7, 27. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World: An Online Reference; Version 6.1; Electronic Database; American Museum of Natural History: New York, NY, USA, 2022. [Google Scholar]
- Kuzmin, S.L.; Dunayev, E.A. On the problem of the type territory of the Turkestanian Salamander (Hynobius turkestanicus Nikolsky, 1909). Adv. Amphib. Res. Former Sov. Union 2000, 5, 243–250. [Google Scholar]
- Min, M.-S.; Baek, H.; Song, J.-Y.; Chang, M.; Poyarkov Jr, N. A new species of salamander of the genus Hynobius (Amphibia, Caudata, Hynobiidae) from South Korea. Zootaxa 2016, 4169, 475–503. [Google Scholar] [CrossRef]
- Xiong, J.L.; Chen, Q.; Zeng, X.M.; Zhao, E.M.; Qing, L.Y. Karyotypic, morphological, and molecular evidence for Hynobius yunanicus as a synonym of Pachyhynobius shangchengensis (Urodela: Hynobiidae). J. Herpetol. 2007, 41, 664–671. [Google Scholar] [CrossRef]
- Nishikawa, K.; Jiang, J.P.; Matsui, M.; Mo, Y.M.; Chen, X.H.; Kim, J.B.; Tominaga, A.; Yoshikawa, N. Invalidity of Hynobius yunanicus and molecular phylogeny of Hynobius salamander from continental China (Urodela, Hynobiidae). Zootaxa 2010, 2426, 65–67. [Google Scholar] [CrossRef]
- Jin, Y. The Demi-Gods and Semi-Devils; Joint Publishing Press: Beijing, China, 1994. [Google Scholar]
- Heo, K.; Shin, Y.; Messenger, K.R. Hynobius notialis (Southern Korean Salamander). Behavior. Herpetol. Rev. 2022, 53, 642. [Google Scholar]
- Fauth, J.E.; Resetarits, W.J. Biting in the salamander Siren intermedia intermedia: Courtship component or agonistic behavior? J. Herpetol. 1999, 33, 493–496. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, Y.-Q.; Zhou, H.; Liu, Y.-F.; Wang, X.-L.; Papenfuss, T.J.; Wake, D.B.; Qu, L.-H. Phylogeny, evolution, and biogeography of Asiatic salamanders (Hynobiidae). Proc. Natl. Acad. Sci. USA 2006, 103, 7360–7365. [Google Scholar] [CrossRef]
- Lu, B.; Zheng, Y.; Murphy, R.W.; Zeng, X. Coalescence patterns of endemic Tibetan species of stream salamanders (Hynobiidae: Batrachuperus). Mol. Ecol. 2012, 21, 3308–3324. [Google Scholar] [CrossRef]
- Li, J.; Fu, C.; Lei, G. Biogeographical consequences of Cenozoic tectonic events within East Asian margins: A case study of Hynobius biogeography. PLoS ONE 2011, 6, e21506. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Zhu, H.; Li, P.; Yang, B.; Ma, J. Phylogeny and biogeography of South Chinese brown frogs (Ranidae, Anura). PLoS ONE 2017, 12, e0175113. [Google Scholar] [CrossRef]
- Jacques, F.M.; Shi, G.; Su, T.; Zhou, Z. A tropical forest of the middle Miocene of Fujian (SE China) reveals Sino-Indian biogeographic affinities. Rev. Palaeobot. Palynol. 2015, 216, 76–91. [Google Scholar] [CrossRef]
- Che, R.; Sun, Y.; Wang, R.; Xu, T. Transcriptomic analysis of endangered Chinese salamander: Identification of immune, sex and reproduction-related genes and genetic markers. PLoS ONE 2014, 9, e87940. [Google Scholar]
- IUCN Standards and Petitions Committee. Guidelines for Using the IUCN Red List Categories and Criteria; Version 14; Prepared by the Standards and Petitions Committee; IUCN: Gland, Switzerland, 2019. [Google Scholar]
- Shin, Y.; Min, M.S.; Borzée, A. Driven to the edge: Species distribution modeling of a Clawed Salamander (Hynobiidae: Onychodactylus koreanus) predicts range shifts and drastic decrease of suitable habitats in response to climate change. Ecol. Evol. 2021, 11, 14669–14688. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M.; Lannoo, M.J.; Lesbarrères, D.; Muths, E. Amphibian population declines: 30 years of progress in confronting a complex problem. Herpetologica 2020, 76, 97–100. [Google Scholar] [CrossRef]
- Moor, H.; Bergamini, A.; Vorburger, C.; Holderegger, R.; Bühler, C.; Egger, S.; Schmidt, B.R. Bending the curve: Simple but massive conservation action leads to landscape-scale recovery of amphibians. Proc. Natl. Acad. Sci. USA 2022, 119, e2123070119. [Google Scholar] [CrossRef]
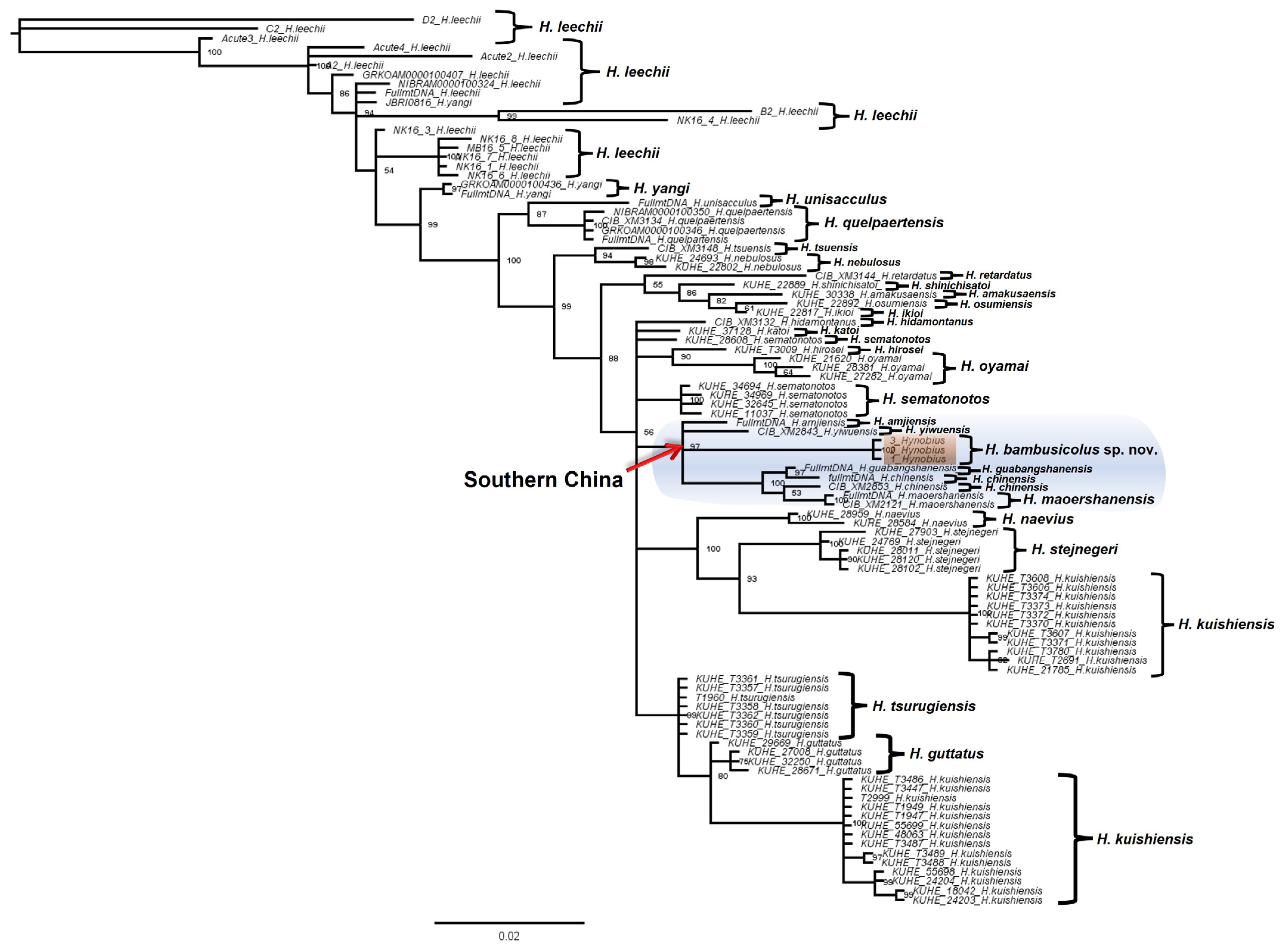
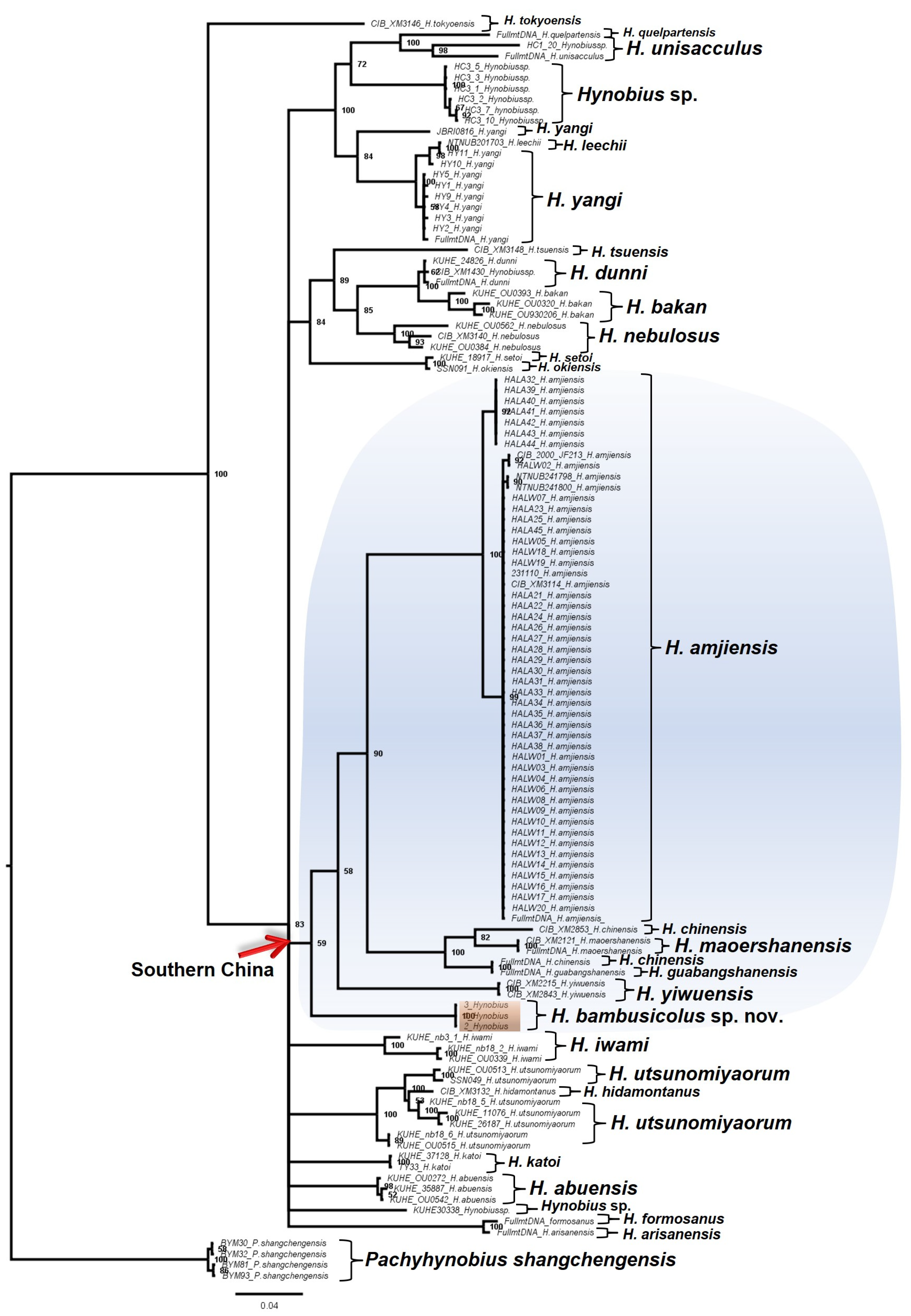
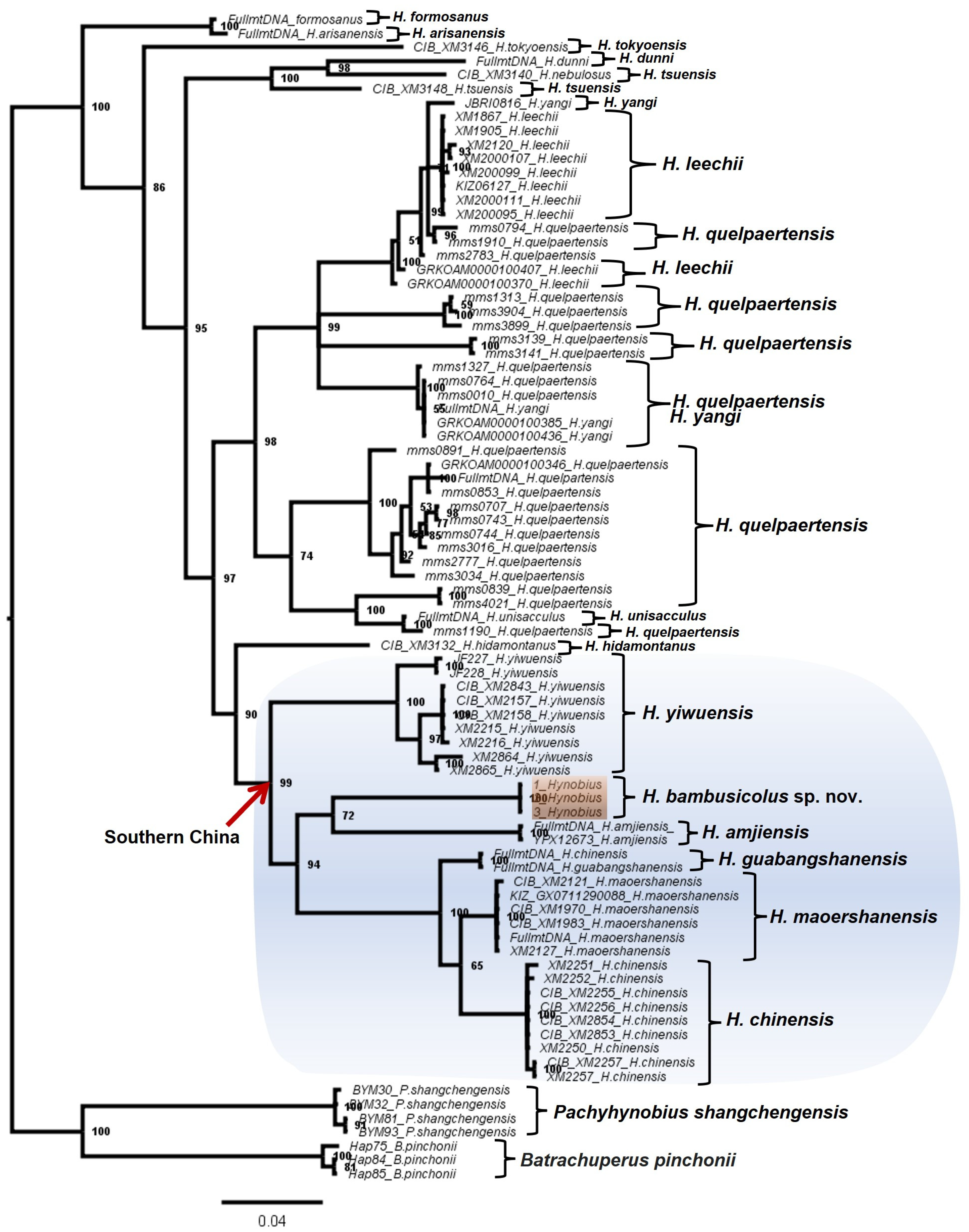
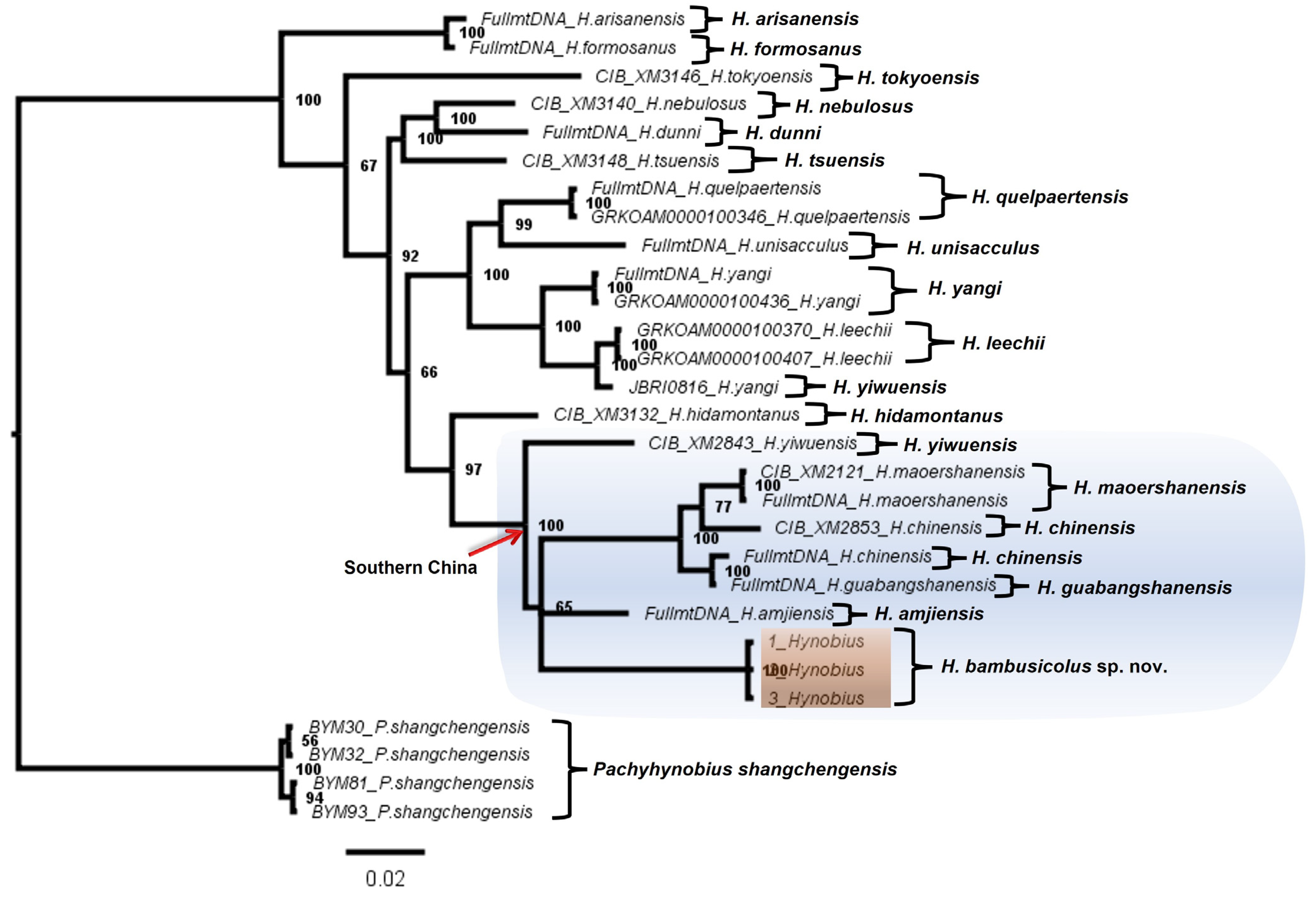



| Fragment | Primer | 5′-3′ | Length | Source | PCR Condition |
|---|---|---|---|---|---|
| mtDNA—COI | LCO1490 | GGTCAACAAATCATAAAGATATTG | 680 bp | [31] | 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 53 °C for 30 s and 72 °C for 40 s, and final elongation at 72 °C for 7 min |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | 680 bp | [31] | ||
| 16S rRNA | L02510 | CGCCTGTTTATCAAAAACAT | 500 bp | [32] | 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 53 °C for 30 s and 72 °C for 40 s, and final elongation at 72 °C for 7 min |
| H03063 | CTCCGGTTTGAACTCAGATC | 500 bp | [33] | ||
| mtDNA-Cytb (in-house design) | CytbHy-F1 | TGTAGACCTCCCAACCCCC | 780 bp | This study | 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55–60 °C for 30 s and 72 °C for 60 s, and final elongation at 72 °C for 10 min |
| CytbHy-R1 | CGTAGGCGAATAAGAAATACCACT | 780 bp | This study |
| Mitochondrial Gene Fragment | Type | Partition’s Strategy | Best Sequence Substitution Model |
|---|---|---|---|
| 16S rRNA | Non-coding ribosomal | 1–528 bp | HKY + I + G |
| Cytb | Protein-coding | Exon by three codons’ position (1–630 bp; 2–630 bp; 3–630 bp) | GTR + I + G |
| COI | Protein-coding | Exon by three codons’ position (1–567 bp; 2–567 bp; 3–567 bp) | GTR + I + G |
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | H. bambusicolus sp. nov. | N/A | |||||||||||||||||
| 2 | P. shangchengensis | 0.166 | |||||||||||||||||
| 3 | H. maoershanensis | 0.113 | 0.170 | ||||||||||||||||
| 4 | H. yiwuensis | 0.102 | 0.157 | 0.106 | |||||||||||||||
| 5 | H. chinensis | 0.106 | 0.166 | 0.035 # | 0.096 | ||||||||||||||
| 6 | H. hidamontanus | 0.101 | 0.146 | 0.093 | 0.090 | 0.091 | |||||||||||||
| 7 | H. nebulosus | 0.119 | 0.149 | 0.106 | 0.100 | 0.106 | 0.083 | ||||||||||||
| 8 | H. tokyoensis | 0.138 | 0.173 | 0.132 | 0.128 | 0.126 | 0.118 | 0.126 | |||||||||||
| 9 | H. tsuensis | 0.115 | 0.147 | 0.106 | 0.096 | 0.101 | 0.086 | 0.074 | 0.126 | ||||||||||
| 10 | H. amjiensis | 0.098 * | 0.161 | 0.092 | 0.096 | 0.090 | 0.095 | 0.102 | 0.132 | 0.099 | |||||||||
| 11 | H. arisanensis | 0.118 | 0.145 | 0.121 | 0.110 | 0.121 | 0.109 | 0.104 | 0.121 | 0.098 | 0.122 | ||||||||
| 12 | H. dunni | 0.122 | 0.156 | 0.109 | 0.107 | 0.106 | 0.091 | 0.060 | 0.121 | 0.077 | 0.099 | 0.105 | |||||||
| 13 | H. formosanus | 0.115 | 0.144 | 0.121 | 0.106 | 0.122 | 0.108 | 0.104 | 0.117 | 0.100 | 0.123 | 0.008 # | 0.104 | ||||||
| 14 | H. guabangshanensis | 0.106 | 0.161 | 0.032 # | 0.093 | 0.021 # | 0.083 | 0.103 | 0.127 | 0.098 | 0.091 | 0.117 | 0.106 | 0.118 | |||||
| 15 | H. quelpaertensis | 0.114 | 0.146 | 0.105 | 0.099 | 0.100 | 0.085 | 0.092 | 0.123 | 0.088 | 0.095 | 0.111 | 0.094 | 0.107 | 0.095 | ||||
| 16 | H. unisacculus | 0.116 | 0.141 | 0.105 | 0.096 | 0.102 | 0.095 | 0.093 | 0.120 | 0.092 | 0.096 | 0.113 | 0.096 | 0.110 | 0.102 | 0.062 | |||
| 17 | H. yangi | 0.104 | 0.147 | 0.114 | 0.101 | 0.107 | 0.083 | 0.088 | 0.122 | 0.088 | 0.099 | 0.105 | 0.096 | 0.104 | 0.108 | 0.075 | 0.073 | ||
| 18 | H. leechii | 0.100 | 0.148 | 0.107 | 0.099 | 0.104 | 0.081 | 0.089 | 0.121 | 0.089 | 0.098 | 0.105 | 0.097 | 0.104 | 0.107 | 0.066 | 0.070 | 0.036 | N/A |
| Age | Adults | Juveniles | ||||
|---|---|---|---|---|---|---|
| Type | Wild | Wild | Paratype | Paratype | Paratype | Holotype |
| ID | 22HyF001 | 22HyF002 | 22HyF003 | 22HyF004 | 22HyF005 | 22HyF006 |
| TOL | 191.57 | 196.94 | 48.06 | 48.38 | 50.43 | 55.11 |
| SVL | 136.36 | 137.81 | 28.39 | 27.72 | 31.32 | 34.05 |
| TL | 55.21 | 59.13 | 19.57 | 18.74 | 20.32 | 21.73 |
| HL | 22.78 | 21.86 | 7.27 | 8.48 | 8.25 | 10.32 |
| HW | 18.57 | 15.84 | 6.22 | 6.80 | 6.82 | 7.82 |
| IOD | 6.19 | 5.25 | 2.67 | 2.92 | 2.83 | 3.44 |
| IND | 6.29 | 5.77 | 3.16 | 3.29 | 2.72 | 3.16 |
| BW | 17.75 | 16.08 | 5.24 | 6.24 | 6.83 | 7.09 |
| AG | 52.07 | 51.06 | 16.46 | 16.14 | 17.95 | 21.04 |
| FOL | 21.13 | 20.77 | 6.49 | 6.73 | 5.89 | 8.08 |
| HIL | 25.53 | 23.08 | 6.38 | 6.30 | 8.73 | 7.77 |
| COS | 9 | 10 | 9 | 9 | 10 | 9 |
| SL | 19.78 | 20.82 | 4.72 | 5.47 | 4.44 | 5.37 |
| TH | 10.12 | 10.52 | 3.26 | 4.02 | 3.06 | 3.49 |
| TW | 11.03 | 10.90 | 2.21 | 2.29 | 2.83 | 2.48 |
| HH | 10.50 | 10.99 | 3.81 | 3.91 | 3.92 | 3.70 |
| DE | 5.31 | 4.64 | 2.44 | 2.31 | 2.80 | 2.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Othman, S.N.; Qiu, Z.; Lu, Y.; Prasad, V.K.; Dong, Y.; Lu, C.-H.; Borzée, A. An Isolated and Deeply Divergent Hynobius Species from Fujian, China. Animals 2023, 13, 1661. https://doi.org/10.3390/ani13101661
Wang Z, Othman SN, Qiu Z, Lu Y, Prasad VK, Dong Y, Lu C-H, Borzée A. An Isolated and Deeply Divergent Hynobius Species from Fujian, China. Animals. 2023; 13(10):1661. https://doi.org/10.3390/ani13101661
Chicago/Turabian StyleWang, Zhenqi, Siti N. Othman, Zhixin Qiu, Yiqiu Lu, Vishal Kumar Prasad, Yuran Dong, Chang-Hu Lu, and Amaël Borzée. 2023. "An Isolated and Deeply Divergent Hynobius Species from Fujian, China" Animals 13, no. 10: 1661. https://doi.org/10.3390/ani13101661
APA StyleWang, Z., Othman, S. N., Qiu, Z., Lu, Y., Prasad, V. K., Dong, Y., Lu, C.-H., & Borzée, A. (2023). An Isolated and Deeply Divergent Hynobius Species from Fujian, China. Animals, 13(10), 1661. https://doi.org/10.3390/ani13101661








