The Effect of Dietary Synbiotics in Actively Racing Standardbred Horses Receiving Trimethoprim/Sulfadiazine
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design
- Antibiotic only (AB; Trimethoprim/Sulfadiazine, 75 mg/kg BW administered by oral syringe once per day on days 1–10).
- Synbiotic only (PBP; PROBIOPlusTM (Selected Bioproducts Inc., Guelph, ON, Canada), 30 mg/kg BW administered as a feed top-dress once per day (per manufacturer directions) on days 1–28).
- Synbiotic + Antibiotic (PBP + AB).
- Unsupplemented control (CO).
2.3. Sampling
2.4. DNA Extraction and Illumina Sequencing
2.5. Bioinformatics
- Baseline period: days −1, 0, and 1 (prior to treatment)
- Early period: day 2 (commencement of treatment)
- Mid period: days 9, 10, and 11 (end of AB treatment)
- Late period: days 28, 29, and 30 (end of PBP treatment)
2.6. Fecal pH and Dry Matter Measurements
2.7. Synbiotic Culture
2.8. Statistics
3. Results
3.1. Microbiome Analysis
3.2. Beta Diversity
3.3. Alpha Diversity
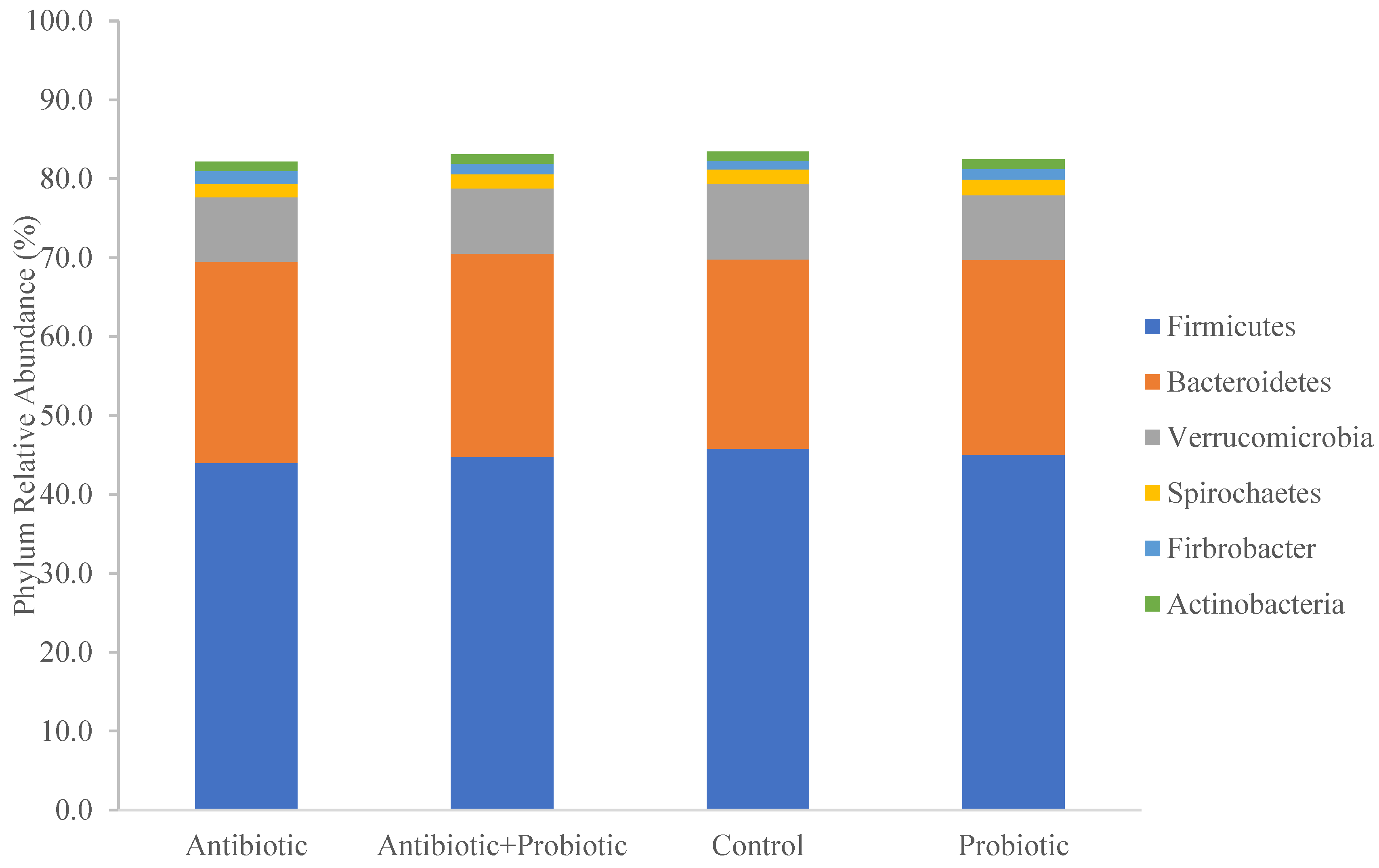
3.4. Taxonomy
3.5. LEfSe Analysis
3.6. Fecal pH and Dry Matter
3.7. Synbiotic Culture
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gustafson, R.H.; Bowen, R.E. Antibiotic use in animal agriculture. J. Appl. Microbiol. 1997, 83, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Khusro, A.; Aarti, C.; Buendía-Rodriguez, G.; Arasu, M.V.; Al-Dhabi, N.A.; Barbabosa-Pliego, A. Adverse Effect of Antibiotics Administration on Horse Health: An Overview. J. Equine Vet. Sci. 2021, 97, 103339. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, A. Antibiotic-associated diarrhoea in horses. Equine Vet. Educ. 2002, 14, 186. [Google Scholar] [CrossRef]
- Båverud, V.; Gustafsson, A.; Franklin, A.; Lindholm, A.; Gunnarsson, A. Clostridium difficile associated with acute colitis in mature horses treated with antibiotics. Equine Vet. J. 1997, 29, 279–284. [Google Scholar] [CrossRef]
- World Health Organization. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Available online: https://www.fao.org/3/a0512e/a0512e.pdf (accessed on 13 July 2023).
- Schoster, A. Probiotic Use in Equine Gastrointestinal Disease. Vet. Clin. N. Am. Equine Pract. 2018, 34, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Quigley, E.M.M. Prebiotics and Probiotics in Digestive Health. Clin. Gastroenterol. Hepatol. 2019, 17, 333–344. [Google Scholar] [CrossRef]
- Vanderhoof, J.A.; Whitney, D.B.; Antonson, D.L.; Hanner, T.L.; Lupo, J.V.; Young, R.J. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J. Pediatr. 1999, 135, 564–568. [Google Scholar] [CrossRef]
- McFarland, L.V.; Surawicz, C.M.; Greenberg, R.N.; Elmer, G.W.; Moyer, K.A.; Melcher, S.A.; Bowen, K.E.; Cox, J.L. Prevention of b-lactam-associated diarrhea by Saccharomyces boulardii compared with placebo. Am. J. Gastroenterol. 1995, 90, 439–448. [Google Scholar] [PubMed]
- Jouany, J.P.; Medina, B.; Bertin, G.; Julliand, V. Effect of live yeast culture supplementation on hindgut microbial communities and their polysaccharidase and glycoside hydrolase activities in horses fed a high-fiber or high-starch diet. J. Anim. Sci. 2009, 87, 2844–2852. [Google Scholar] [CrossRef] [Green Version]
- Swyers, K.L.; Burk, A.O.; Hartsock, T.G.; Ungerfeld, E.M.; Shelton, J.L. Effects of direct-fed microbial supplementation on digestibility and fermentation end-products in horses fed low-and high-starch concentrates. J. Anim. Sci. 2008, 86, 2596–2608. [Google Scholar] [CrossRef]
- MacNicol, J.L.; Renwick, S.; Ganobis, C.M.; Allen-Vercoe, E.; Weese, J.S.; Pearson, W. The influence of a probiotic/prebiotic supplement on microbial and metabolic parameters of equine cecal fluid or fecal slurry in vitro. J. Anim. Sci. 2023, 101, skad034. [Google Scholar] [CrossRef] [PubMed]
- John, J.; Roediger, K.; Schroedl, W.; Aldaher, N.; Vervuert, I. Development of intestinal microflora and occurrence of diarrhoea in sucking foals: Effects of Bacillus cereus var. toyoi supplementation. BMC Vet. Res. 2015, 11, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2015, 1, e0009-15. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacNicol, J.L.; Renwick, S.; Ganobis, C.M.; Allen-Vercoe, E.; Weese, J.S.; Pearson, W. A Comparison of Methods to Maintain the Equine Cecal Microbial Environment In Vitro Utilizing Cecal and Fecal Material. Animals 2022, 12, 2009. [Google Scholar] [CrossRef]
- Gihring, T.M.; Green, S.J.; Schadt, C.W. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ. Microbiol. 2012, 14, 285–290. [Google Scholar] [CrossRef]
- Hydock, K.L.; Nissley, S.G.; Staniar, W.B. A standard protocol for fecal pH measurements in the horse. Prof. Anim. Sci. 2014, 30, 643–648. [Google Scholar] [CrossRef]
- Yang, L.; Bajinka, O.; Jarju, P.O.; Tan, Y.; Taal, A.M.; Ozdemir, G. The varying effects of antibiotics on gut microbiota. AMB Express 2021, 11, 116. [Google Scholar] [CrossRef]
- Bajinka, O.; Tan, Y.; Abdelhalim, K.A.; Özdemir, G.; Qiu, X. Extrinsic factors influencing gut microbes, the immediate consequences and restoring eubiosis. AMB Express 2020, 10, 130. [Google Scholar] [CrossRef]
- Lucas López, R.; Grande Burgos, M.J.; Gálvez, A.; Pérez Pulido, R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: A state of the science review. APMIS 2017, 125, 3–10. [Google Scholar] [CrossRef]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Weese, J.S.; Holcombe, S.J.; Embertson, R.M.; Kurtz, K.A.; Roessner, H.A.; Jalali, M.; Wismer, S.E. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 2015, 47, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Stewart, H.L.; Southwood, L.L.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Pitta, D. Differences in the equine faecal microbiota between horses presenting to a tertiary referral hospital for colic compared with an elective surgical procedure. Equine Vet. J. 2019, 51, 336–342. [Google Scholar] [CrossRef]
- Theelen, M.J.P.; Luiken, R.E.C.; Wagenaar, J.A.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Rossen, J.W.A.; Schaafstra, F.J.W.C.; van Doorn, D.A.; Zomer, A.L. Longitudinal study of the short- and long-term effects of hospitalisation and oral trimethoprim-sulfadiazine administration on the equine faecal microbiome and resistome. Microbiome 2023, 11, 33. [Google Scholar] [CrossRef]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stämpfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the Fecal Microbiota of Healthy Horses and Horses with Colitis by High Throughput Sequencing of the V3-V5 Region of the 16S rRNA Gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.C.; Silva, G.; Ramos, R.V.; Staempfli, H.R.; Arroyo, L.G.; Kim, P.; Weese, J.S. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, N.A.; Walker, A.W.; Berry, S.H.; Duncan, S.H.; Farquarson, F.M.; Louis, P.; Thomson, J.M.; UK IBD Genetics Consortium; Satsangi, J.; Flint, H.J.; et al. The Impact of Different DNA Extraction Kits and Laboratories upon the Assessment of Human Gut Microbiota Composition by 16S rRNA Gene Sequencing. PLoS ONE 2014, 9, e88982. [Google Scholar] [CrossRef]
- Lim, M.Y.; Park, Y.S.; Kim, J.H.; Nam, Y.D. Evaluation of fecal DNA extraction protocols for human gut microbiome studies. BMC Microbiol. 2020, 20, 212. [Google Scholar] [CrossRef]
- Wang, Z.; Zolnik, C.P.; Qiu, Y.; Usyk, M.; Wang, T.; Strickler, H.D.; Isasi, C.R.; Kaplan, R.C.; Kurland, I.J.; Qi, Q.; et al. Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies. Front. Cell. Infect. Microbiol. 2018, 8, 301. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems 2016, 1, e00021-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, C.; Pilla, R.; Chaffin, K.; Lidbury, J.; Steiner, J.; Suchodolski, J. Alterations in the fecal microbiome and metabolome of horses with antimicrobial-associated diarrhea compared to antibiotic-treated and non-treated healthy case controls. Animals 2021, 11, 1807. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; de Vuyst, L. Inulin-type fructan degradation capacity of clostridium cluster IV and XlVa butyrate- producing colon bacteria and their associated metabolic outcomes. Benef. Microbes 2017, 8, 473–490. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Livanos, A.E.; Snider, E.J.; Whittier, S.; Chong, D.H.; Wang, T.C.; Abrams, J.A.; Freedberg, D.E. Rapid gastrointestinal loss of Clostridial Clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS ONE 2018, 13, e0200322. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.P.; McCormick, C.A.; Suen, G. Fibrobacter communities in the gastrointestinal tracts of diverse hindgut-fermenting herbivores are distinct from those of the rumen. Environ. Microbiol. 2017, 19, 3768. [Google Scholar] [CrossRef]
- Holdeman, L.V.; Moore, W.E.C. New genus, Coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int. J. Syst. Bacteriol. 1974, 24, 260–277. [Google Scholar] [CrossRef] [Green Version]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021, 12, 1943. [Google Scholar] [CrossRef]
- Collinet, A.; Grimm, P.; Julliand, S.; Julliand, V. Multidimensional Approach for Investigating the Effects of an Antibiotic–Probiotic Combination on the Equine Hindgut Ecosystem and Microbial Fibrolysis. Front. Microbiol. 2021, 25, 646294. [Google Scholar] [CrossRef]
- O’ Donnell, M.M.; Harris, H.M.; Jeffery, I.B.; Claesson, M.J.; Younge, B.; O’Toole, P.W.; Ross, R.P. The core faecal bacterial microbiome of Irish Thoroughbred racehorses. Lett. Appl. Microbiol. 2013, 57, 492–501. [Google Scholar] [CrossRef]
- Cai, S.; Li, J.; Hu, F.Z.; Zhang, K.; Luo, Y.; Janto, B.; Boissy, R.; Ehrlich, G.; Dong, X. Cellulosilyticum ruminicola, a Newly Described Rumen Bacterium That Possesses Redundant Fibrolytic-Protein-Encoding Genes and Degrades Lignocellulose with Multiple Carbohydrate- Borne Fibrolytic Enzymes. Appl. Environ. Microbiol. 2010, 76, 3818. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, D.; McBride, S.M. The impact of pH on clostridioides difficile sporulation and physiology. Appl. Environ. Microbiol. 2020, 86, e02706-19. [Google Scholar] [CrossRef]
- Blackmore, T.M.; Dugdale, A.; Argo, C.M.; Curtis, G.; Pinloche, E.; Harris, P.A.; Worgan, H.J.; Girdwood, S.E.; Dougal, K.; Newbold, C.J.; et al. Strong Stability and Host Specific Bacterial Community in Faeces of Ponies. PLoS ONE 2013, 8, e75079. [Google Scholar] [CrossRef] [Green Version]
- Garber, A.; Hastie, P.; Murray, J.A. Factors Influencing Equine Gut Microbiota: Current Knowledge. J. Equine Vet. Sci. 2020, 88, 102943. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S. Evaluation of deficiencies in labeling of commercial probiotics. Can. Vet. J. 2003, 44, 982–983. [Google Scholar]
- Weese, J.S.; Martin, H. Assessment of commercial probiotic bacterial contents and label accuracy. Can. Vet. J. 2011, 52, 43–46. [Google Scholar] [PubMed]
- Weese, J.S. Microbiologic evaluation of commercial probiotics. J. Am. Vet. Med. Assoc. 2002, 220, 794–797. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]



| Horse | Sex | Age (yr) | Height (hh) | Weight (kg) | Treatment | |||
|---|---|---|---|---|---|---|---|---|
| Trial 1 | Trial 2 | Trial 3 | Trial 4 | |||||
| 1 | G | 3 | 15.2 | 485 | CO | × | × | × |
| 2 | M | 2 | 15.3 | 567 | CO | PBP + AB | AB | PBP |
| 3 | M | 2 | 15.3 | 540 | PBP | CO | PBP + AB | AB |
| 4 | G | 2 | 15.3 | 485 | PBP | CO | AB | PBP + AB |
| 5 | S | 2 | 15.3 | 460 | AB | PBP | CO | PBP + AB |
| 6 | G | 7 | 15.3 | 522 | AB | PBP | × | × |
| 7 | G | 6 | 15.2 | 472 | PBP + AB | AB | × | × |
| 8 | G | 4 | 16.2 | 540 | PBP + AB | × | × | × |
| 9 | G | 3 | 15.3 | 512 | × | PBP + AB | × | × |
| 10 | S | 2 | 15.1 | 512 | × | AB | PBP | CO |
| 11 | G | 2 | 15.1 | 460 | × | × | PBP | × |
| 12 | M | 3 | 16.2 | 540 | × | × | PBP + AB | × |
| 13 | M | 3 | 15.2 | 526 | × | × | CO | × |
| 14 | G | 3 | 15.3 | 472 | × | × | × | AB |
| 15 | M | 3 | 14.2 | 460 | × | × | × | CON |
| 16 | G | 3 | 14.3 | 485 | × | × | × | PBP |
| Trt | Sampling Period | p | ||||
|---|---|---|---|---|---|---|
| Baseline | Early | Mid | Late | trt | 0.409 | |
| CO | 0.6004 ± 0.005 | 0.6143 ± 0.007 | 0.5980 ± 0.005 | 0.5993 ± 0.005 | period | 0.054 |
| AB | 0.5982 ± 0.005 | 0.6047 ± 0.007 | 0.6075 ± 0.005 | 0.5993 ± 0.005 | trt × period | 0.043 |
| PBP | 0.5998 ± 0.005 | 0.5871 ± 0.007 | 0.6068 ± 0.005 | 0.6013 ± 0.005 | ||
| PBP + AB | 0.5996 ± 0.005 | 0.6089 ± 0.007 | 0.6090 ± 0.005 | 0.6000 ± 0.005 | ||
| Trt | Sampling Period | p | ||||
|---|---|---|---|---|---|---|
| Baseline | Early | Mid | Late | trt | 0.098 | |
| CO | 10.83 ± 0.2 | 11.62 ± 0.4 a | 10.32 ± 0.2 | 10.33 ± 0.2 | period | 0.041 |
| AB | 10.32 ± 0.2 | 10.99 ± 0.3 ab | 10.78 ± 0.2 | 10.41 ± 0.2 | trt × period | 0.007 |
| PBP | 10.32 ± 0.2 | 9.95 ± 0.3 b | 10.64 ± 0.2 | 10.44 ± 0.2 | ||
| PBP + AB | 10.60 ± 0.2 | 10.79 ± 0.3 ab | 10.87 ± 0.2 | 10.31 ± 0.2 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagounova, M.; MacNicol, J.L.; Weese, J.S.; Pearson, W. The Effect of Dietary Synbiotics in Actively Racing Standardbred Horses Receiving Trimethoprim/Sulfadiazine. Animals 2023, 13, 2344. https://doi.org/10.3390/ani13142344
Lagounova M, MacNicol JL, Weese JS, Pearson W. The Effect of Dietary Synbiotics in Actively Racing Standardbred Horses Receiving Trimethoprim/Sulfadiazine. Animals. 2023; 13(14):2344. https://doi.org/10.3390/ani13142344
Chicago/Turabian StyleLagounova, Maria, Jennifer L. MacNicol, J. Scott Weese, and Wendy Pearson. 2023. "The Effect of Dietary Synbiotics in Actively Racing Standardbred Horses Receiving Trimethoprim/Sulfadiazine" Animals 13, no. 14: 2344. https://doi.org/10.3390/ani13142344





