Wild Avian Gut Microbiome at a Small Spatial Scale: A Study from a Mediterranean Island Population of Alectoris rufa
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Sampling
2.2. DNA Extraction
2.3. 16S rRNA Gene Amplification and Sequencing
2.4. Analysis of the Sequences
2.5. Statistical Analyses
3. Results
3.1. Sequencing Outcome
3.2. Composition of the Microbial Communities
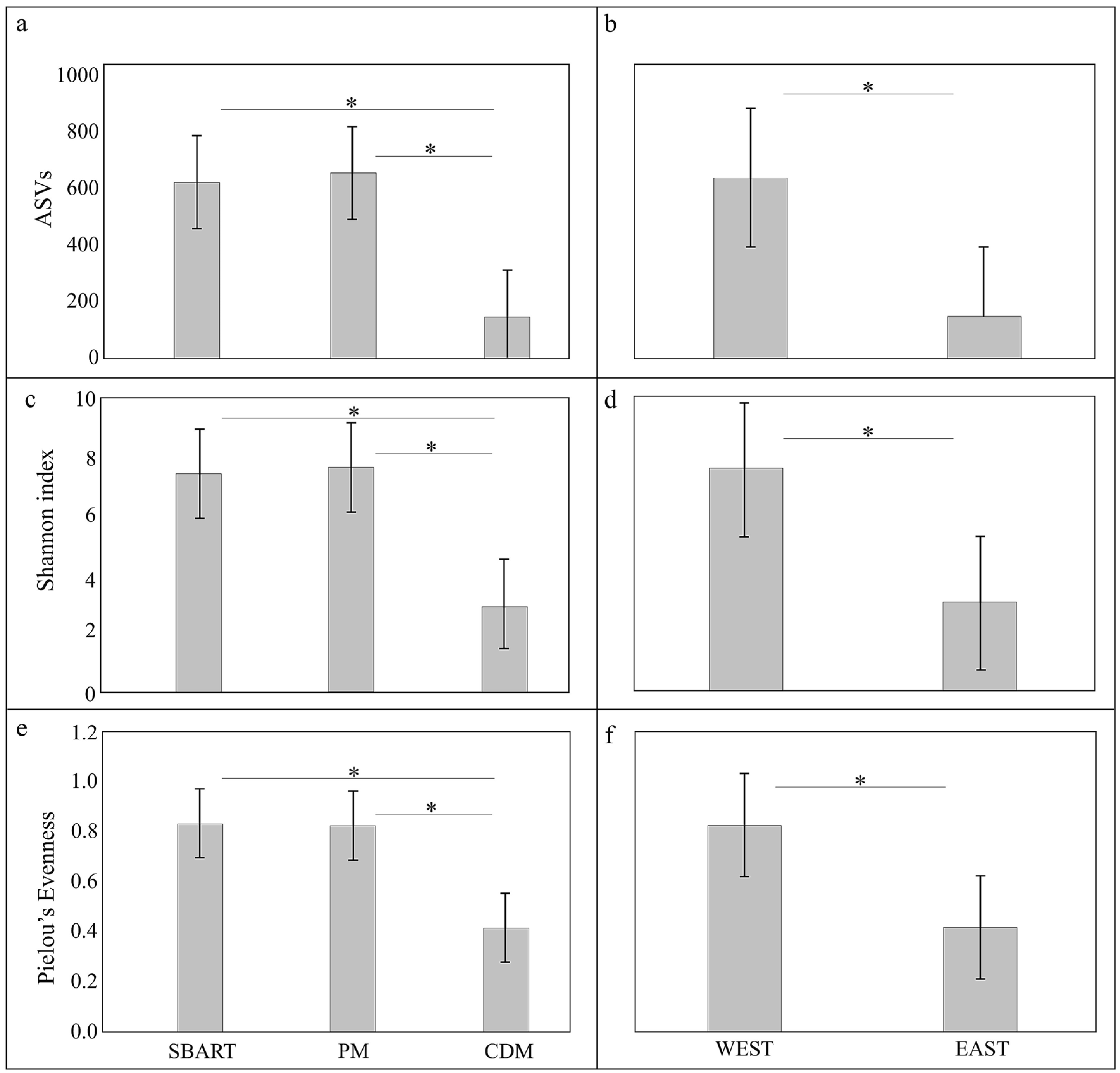
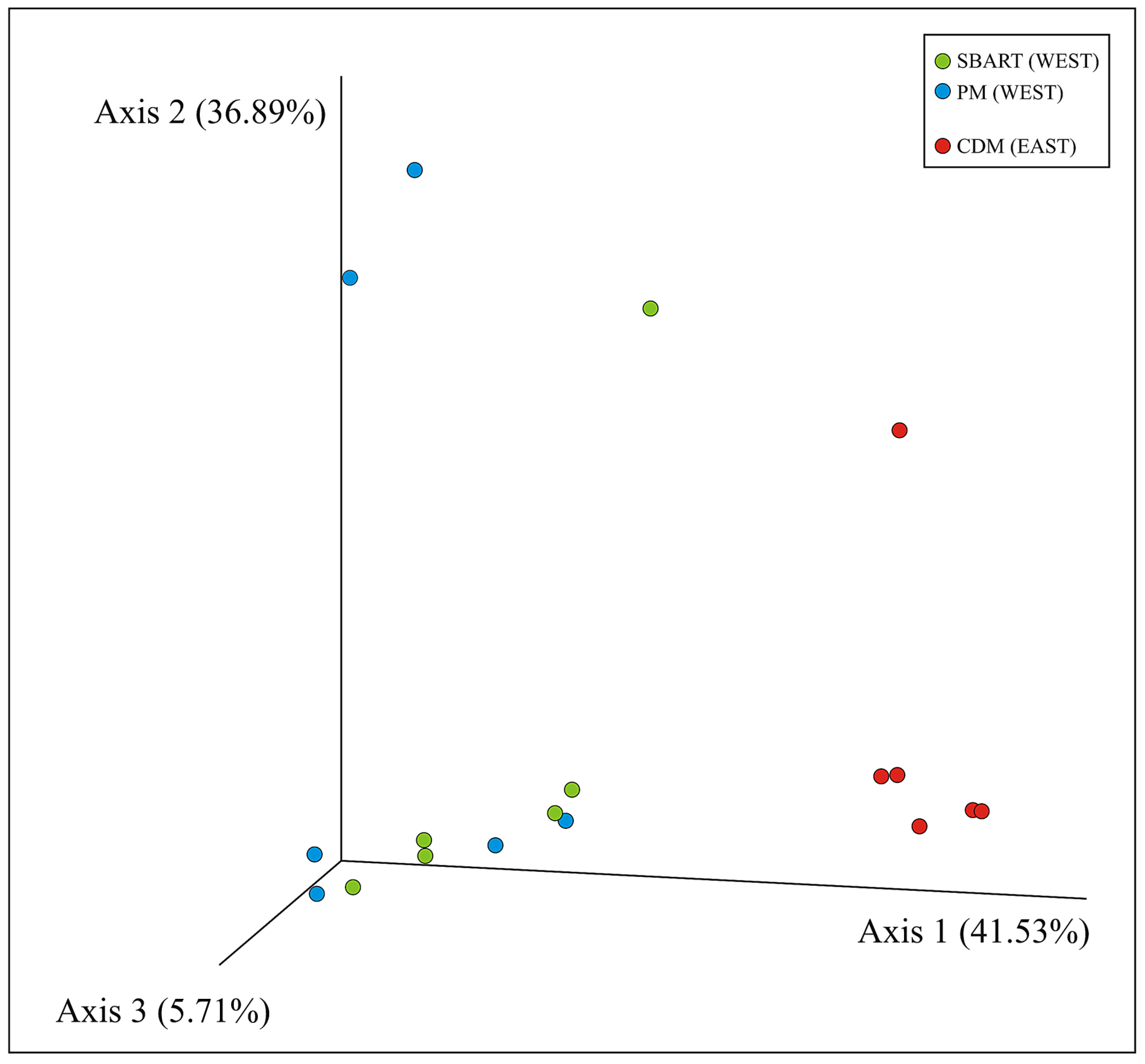
3.3. Comparative Analyses of Microbial Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woese, C.R. Default taxonomy: Ernst Mayr’s view of the microbial world. Proc. Natl. Acad. Sci. USA 1998, 95, 1043–11046. [Google Scholar] [CrossRef]
- Shropshire, J.D.; Bordenstein, S.R. Speciation by symbiosis: The microbiome and behavior. mBio 2016, 7, e01785-15. [Google Scholar] [CrossRef]
- Lee, W.J.; Hase, K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014, 10, 416–424. [Google Scholar] [CrossRef]
- Amato, K.R.; Metcalf, J.L.; Song, S.J.; Hale, V.L.; Clayton, J.; Ackermann, G.; Humphrey, G.; Niu, K.; Cui, D.; Zhao, H.; et al. Using the gut microbiota as a novel tool for examining colobine primate GI health. Glob. Ecol. Conserv. 2016, 7, 225–237. [Google Scholar] [CrossRef]
- Petrelli, S.; Buglione, M.; Rivieccio, E.; Ricca, E.; Baccigalupi, L.; Scala, G.; Fulgione, D. Reprogramming of the gut microbiota following feralization in Sus scrofa. Anim. Microbiome 2023, 5, 14. [Google Scholar] [CrossRef]
- Dehority, B.A. Rumen Microbiology; Nottingham University Press: Nottingham, UK, 2003. [Google Scholar]
- Kartzinel, T.R.; Hsing, J.C.; Musili, P.M.; Brown, B.R.P.; Pringle, R.M. Covariation of diet and gut microbiome in African megafauna. Proc. Natl. Acad. Sci. USA 2019, 116, 23588–23593. [Google Scholar] [CrossRef]
- Olsen, M.A.; Blix, A.S.; Utsi, T.H.A.; Sørmo, W.; Mathiesen, S.D. Chitinolytic bacteria in the minke whale forestomach. Can. J. Microbiol. 2000, 46, 85–94. [Google Scholar] [CrossRef][Green Version]
- Silva, A.M.; Barbosa, F.H.; Duarte, R.; Vieira, L.Q.; Arantes, R.M.; Nicoli, J.R. Effect of Bifidobacterium longum ingestion on experimental salmonellosis in mice. J. Appl. Microbiol. 2004, 97, 29–37. [Google Scholar] [CrossRef]
- Ley, R.; Turnbaugh, P.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- West, A.G.; Waite, D.W.; Deines, P.; Bourne, D.G.; Digby, A.; McKenzie, V.J.; Taylor, M.W. The microbiome in threatened species conservation. Biol. Conserv. 2019, 229, 85–98. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Quinn, R.A.; Debelius, J.; Xu, Z.Z.; Morton, J.; Garg, N.; Kansson, J.K.; Dorrestein, P.C.; Knight, R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016, 535, 94–103. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, V.J.; Kueneman, J.G.; Harris, R.N. Probiotics as a tool for disease mitigation in wildlife: Insights from food production and medicine. Ann. N. Y. Acad. Sci. 2018, 1429, 18–30. [Google Scholar] [CrossRef]
- Kimura, N.; Yoshikane, M.; Kobayashi, A. Microflora of the bursa of Fabricius of chickens. Poult. Sci. 1986, 65, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Ley, R.E.; Gao, Z.; Pei, Z.; Ortiz-Zuazaga, H.; Pericchi, L.R.; Garcia-Amado, M.A.; Michelangeli, F.; Blaser, M.J.; Gordon, J.I.; et al. Bacterial community in the crop of the hoatzin, a neotropical folivorous flying bird. Appl. Environ. Microbiol. 2008, 74, 5905–5912. [Google Scholar] [CrossRef]
- Grond, K.; Sandercock, B.K.; Jumpponen, A.; Zeglin, L.H. The avian gut microbiota: Community, physiology and function in wild birds. J. Avian Biol. 2018, 49, e01788. [Google Scholar] [CrossRef]
- Roggenbuck, M.; Bærholm Schnell, I.; Blom, N.; Bælum, J.; Bertelsen, M.F.; Sicheritz-Pontén, T.; Sørensen, S.J.; Gilbert, M.T.P.; Graves, G.R.; Hansen, L.H. The microbiome of New World vultures. Nat. Commun. 2014, 5, 5498. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Flores, A.; Tveit, A.T.; Wright, A.-D.; Pope, P.B.; Sundset, M.A. Characterization of the cecum microbiome from wild and captive rock ptarmigans indigenous to Arctic Norway. PLoS ONE 2019, 14, e0213503. [Google Scholar] [CrossRef] [PubMed]
- Barbanera, F.; Forcina, G.; Guerrini, M.; Dini, F. Molecular phylogeny and diversity of Corsican red-legged partridge: Hybridization and management issues. J. Zool. 2011, 285, 56–65. [Google Scholar] [CrossRef]
- Barbanera, F.; Forcina, G.; Cappello, A.; Guerrini, M.; van Grouw, H.; Aebischer, N.J. Introductions over introductions: The genomic adulteration of an early genetically valuable alien species in the United Kingdom. Biol. Invasions 2015, 17, 409–422. [Google Scholar] [CrossRef]
- Barbanera, F. On the origins and history of the red-legged partridge (Alectoris rufa) from Elba Island (Tuscan Archipelago, Italy). Atti Soc. Toscana Sci. Nat. Mem. Serie B 2021, 128, 45–55. [Google Scholar] [CrossRef]
- Tanini, D.; Guerrini, M.; Vannini, C.; Barbanera, F. Unexpected genetic integrity boosts hope for the conservation of the red-legged partridge (Alectoris rufa, Galliformes) in Italy. Zoology 2022, 155, 126056. [Google Scholar] [CrossRef] [PubMed]
- Chiatante, G.; Meriggi, A.; Giustini, D.; Baldaccini, N.E. Density and habitat requirements of red-legged partridge on Elba Island (Tuscan Archipelago, Italy). Ital. J. Zool. 2013, 80, 402–411. [Google Scholar] [CrossRef]
- Foggi, B.; Cartei, L.; Pignotti, L.; Signorini, M.A.; Viciani, D. Il paesaggio vegetale dell’Isola d’Elba (Arcipelago toscano): Studio fitosociologico e cartografico. Fitosociologia 2006, 43, 3–94. (In Italian) [Google Scholar]
- Carta, A.; Taboada, T.; Müller, J.V. Diachronic analysis using aerial photographs across fifty years reveals significant land use and vegetation changes on a Mediterranean island. Appl. Geogr. 2018, 98, 78–86. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 72–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Werner, J.J.; Koren, O.; Hugenholtz, P.; DeSantis, T.Z.; Walters, W.A.; Caporaso, J.G.; Angenent, L.T.; Knight, R.; Ley, R.E. Impact of training sets on classification of high-throughput bacterial 16S rRNA gene surveys. ISME J. 2012, 6, 94–103. [Google Scholar] [CrossRef]
- Hird, S.M.; Sánchez, C.; Carstens, B.C.; Brumfield, R.T. Comparative Gut microbiota of 59 Neotropical Bird Species. Front. Microbiol. 2015, 6, 1403. [Google Scholar] [CrossRef]
- Schmiedová, L.; Tomášek, O.; Pinkasová, H.; Albrecht, T.; Kreisinger, J. Variation in diet composition and its relation to gut microbiota in a passerine bird. Sci. Rep. 2022, 12, 3787. [Google Scholar] [CrossRef] [PubMed]
- Carey, H.V.; Walters, W.A.; Knight, R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R33–R42. [Google Scholar] [CrossRef] [PubMed]
- Maurice, C.F.; Knowles, S.C.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.; Jiang, Y.; et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. mBio 2015, 6, e00022-15. [Google Scholar] [CrossRef] [PubMed]
- Turjeman, S.; Pekarsky, S.; Corl, A.; Kamath, P.L.; Getz, W.M.; Bowie, R.C.K.; Markin, Y.; Nathan, R. Comparing invasive and noninvasive faecal sampling in wildlife microbiome studies: A case study on wild common cranes. Mol. Ecol. Res. 2023, 23, 359–367. [Google Scholar] [CrossRef]
- Forcina, G.; Guerrini, M.; Barbanera, F. Non-native and hybrid in a changing environment: Conservation perspectives for the last Italian red-legged partridge (Alectoris rufa) population with long natural history. Zoology 2020, 138, 125740. [Google Scholar] [CrossRef]
- Suzuki, T.A.; Martins, F.M.; Nachman, M.W. Altitudinal variation of the gut microbiota in wild house mice. Mol. Ecol. 2018, 9, 2378–2390. [Google Scholar] [CrossRef]
- Spanò, S. La Pernice Rossa; Edizioni Il Piviere: Gavi, Italy, 2010. (In Italian) [Google Scholar]
- Wilkinson, N.; Hughes, R.J.; Aspden, W.J.; Chapman, J.; Moore, R.J.; Stanley, D. The gastrointestinal tract microbiota of the Japanese quail, Coturnix japonica. Appl. Microbiol. Biotechnol. 2016, 100, 4201–4209. [Google Scholar] [CrossRef]
- Sekelja, M.; Rud, I.; Knutsen, S.H.; Denstadli, V.; Westereng, B.; Næs, T.; Rudi, K. Abrupt Temporal Fluctuations in the Chicken Fecal Microbiota Are Explained by Its Gastrointestinal Origin. Appl. Environ. Microbiol. 2012, 78, 2941–2948. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Tan, C.L.; Niu, K.; Yang, Y.; Cui, D.; Zhao, H.; Knight, R.; Amato, K.R. Effects of field conditions on fecal microbiota. J. Microbiol. Methods 2016, 130, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Steury, R.A.; Currey, M.C.; Cresko, W.A.; Bohannan, B.J.M. Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback. Genes 2019, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Fan, Y.; Zhang, Z.; Shen, X.; Li, X.; Liang, X.; Bi, R.; Wu, Y.; Zhai, J.; Dai, J.; et al. Covariation of the Fecal Microbiome with Diet in Nonpasserine Birds. mSphere 2021, 6, e00308-21. [Google Scholar] [CrossRef]
- Angelakis, E.; Raoult, D. The increase of Lactobacillus species in the gut flora of newborn broiler chicks and ducks is associated with weight gain. PLoS ONE 2010, 5, e10463. [Google Scholar] [CrossRef]
- Awad, W.A.; Ghareeb, K.; Böhm, J. Effect of addition of a probiotic micro-organism to broiler diet on intestinal mucosal architecture and electrophysiological parameters. J. Anim. Physiol. Anim. Nutr. 2010, 94, 486–494. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Castillo-Carranza, S.A.; Guard, B.; Gomez-Vazquez, J.P.; Dowd, S.E.; Brigthsmith, D.J. Comprehensive Molecular Characterization of Bacterial Communities in Feces of Pet Birds Using 16S Marker Sequencing. Microb. Ecol. 2017, 73, 224–235. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the Gastrointestinal Tract Microbiota Correlated with Improved Growth and Feed Conversion: Challenges Presented for the Identification of Performance Enhancing Probiotic Bacteria. Front. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef]
- Hauffe, H.C.; Barelli, C. Conserve the germs: The gut microbiota and adaptive potential. Conserv. Genet. 2019, 20, 19–27. [Google Scholar] [CrossRef]
- Chong, R.; Grueber, C.E.; Fox, S.; Wise, P.; Barrs, V.R.; Hogg, C.J.; Belov, K. Looking like the locals—Gut microbiome changes post-release in an endangered species. Anim. Microbiome 2019, 1, 8. [Google Scholar] [CrossRef]
- Quiroga-González, C.; Cardenas, L.A.C.; Ramírez, M.; Reyes, A.; González, C.; Stevenson, P.R. Monitoring the variation in the gut microbiota of captive woolly monkeys related to changes in diet during a reintroduction process. Sci. Rep. 2021, 11, 6522. [Google Scholar] [CrossRef]
- Eliades, S.J.; Brown, J.C.; Colston, T.J.; Fisher, R.N.; Niukula, J.B.; Gray, K.; Vadada, J.; Rasalato, S.; Siler, C.D. Gut microbial ecology of the Critically Endangered Fijian crested iguana (Brachylophus vitiensis): Effects of captivity status and host reintroduction on endogenous microbiomes. Ecol. Evol. 2021, 11, 4731–4743. [Google Scholar] [CrossRef]
- Chattopadhyay, B.; Forcina, G.; Garg, K.M.; Irestedt, M.; Guerrini, M.; Barbanera, F.; Rheindt, F.E. Novel genome reveals susceptibility of popular gamebird, the red-legged partridge (Alectoris rufa, Phasianidae), to climate change. Genomics 2021, 113, 3430–3438. [Google Scholar] [CrossRef]
- García, J.T.; Pérez-Rodríguez, L.; Calero-Riestra, M.; Sánchez-Barbudo, I.; Viñuela, J.; Casas, F. Sexual differences in blood parasite infections, circulating carotenoids and body condition in free-living red-legged partridges. J. Zool. 2023, 320, 260–270. [Google Scholar] [CrossRef]
- Ren, T.; Boutin, S.; Humphries, M.M.; Dantzer, B.; Gorrel, J.C.; Coltman, D.W.; McAdam, A.G.; Wu, M. Seasonal, spatial, and maternal effects on gut microbiome in wild red squirrels. Microbiome 2017, 5, 163. [Google Scholar] [CrossRef]
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786. [Google Scholar] [CrossRef]
- Song, S.J.; Sanders, J.G.; Delsuc, F. Comparative Analyses of Vertebrate Gut Microbiomes Reveal Convergence between Birds and Bats. mBio 2020, 11, e02901-19. [Google Scholar] [CrossRef]
- Trevelline, B.K.; Khol, K.D. The gut microbiome influences host diet selection behavior. Proc. Natl. Acad. Sci. USA 2020, 119, 2117537119. [Google Scholar] [CrossRef]
- Fleischer, R.; Risely, A.; Hoeck, P.E.A.; Keller, L.F.; Sommer, S. Mechanisms governing avian phylosymbiosis: Genetic dissimilarity based on neutral and MHC regions exhibits little relationship with gut microbiome distributions of Galápagos mockingbirds. Ecol. Evol. 2020, 10, 13345–13354. [Google Scholar] [CrossRef]
- Bodawatta, K.H.; Koane, B.; Maiah, G.; Sam, K.; Poulsen, M.; Jønsson, K.A. Species-specific but not phylosymbiotic gut microbiomes of New Guinean passerine birds are shaped by diet and flight-associated gut modifications. Proc. R. Soc. B Biol. Sci. 2021, 288, 20210446. [Google Scholar] [CrossRef]
- Díaz-Sánchez, S.; Höfle, U.; Villanúa, D.; Gortázar, C. Health Monitoring and Disease Control in Red-Legged Partridges. In The Future of the Red-Legged Partridge. Science, Hunting and Conservation; Casas, F., García, J.T., Eds.; Springer Nature: Basel, Switzerland, 2022. [Google Scholar] [CrossRef]
- Forcina, G.; Tang, Q.; Cros, E.; Guerrini, M.; Rheindt, F.E.; Barbanera, F. Genome wide markers redeem the identity of a heavily managed and doomed-too-early gamebird. Proc. R. Soc. B Biol. Sci. 2021, 288, 20210285. [Google Scholar] [CrossRef]
- Lavretsky, P.; Mohl, J.E.; Söderquist, P.; Kraus, R.H.S.; Schummer, M.L.; Brown, J.I. The meaning of wild: Genetic and adaptive consequences from large-scale releases of domestic mallards. Commun. Biol. 2023, 6, 819. [Google Scholar] [CrossRef]
- Baratti, M.; Ammannati, M.; Magnelli, C.; Dessì-Fulgheri, F. Introgression of chukar genes into a reintroduced red-legged partridge (Alectoris rufa) population in central Italy. Anim. Genet. 2004, 36, 29–35. [Google Scholar] [CrossRef]
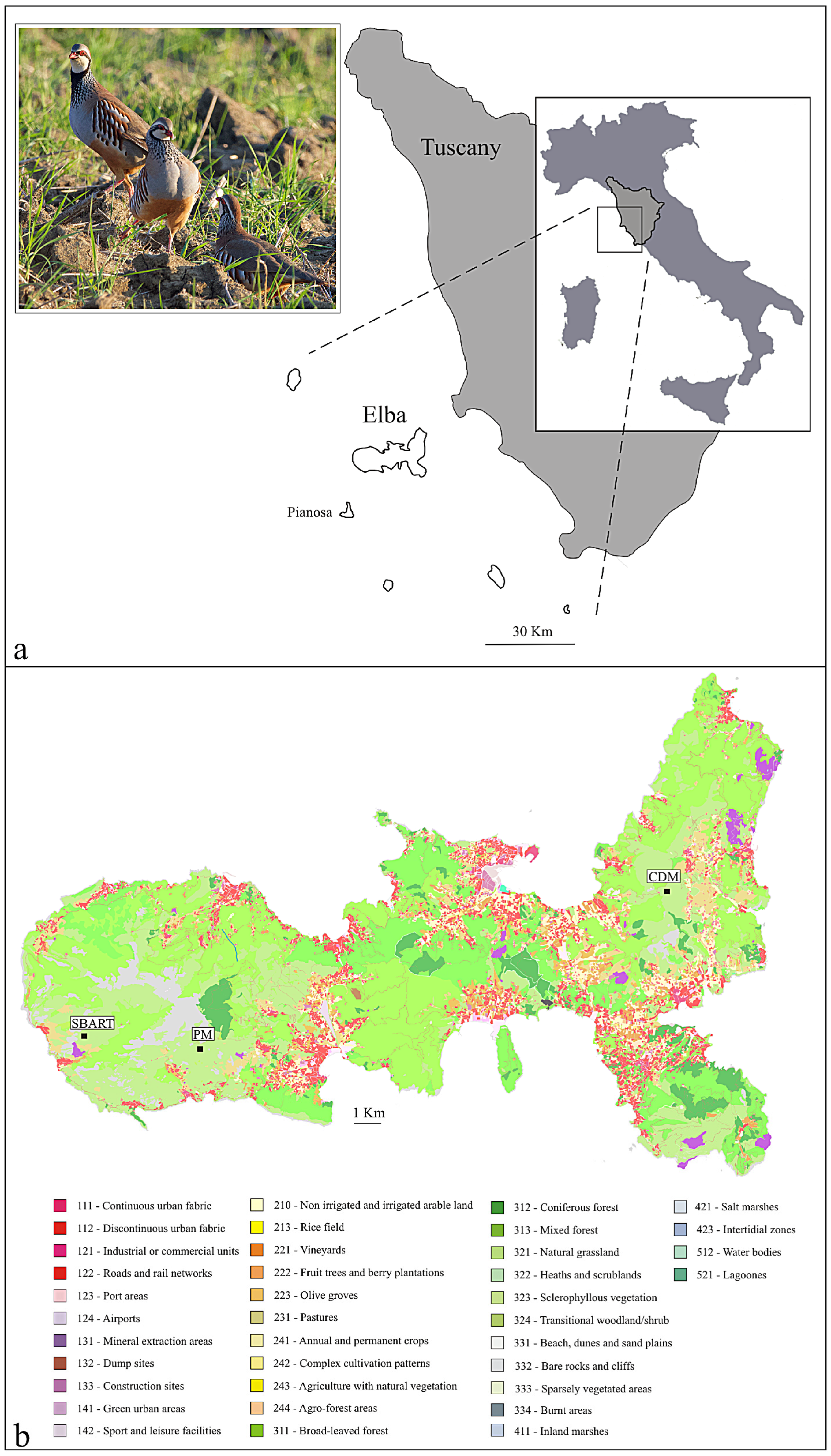

| Sample | Locality | Subpopulation | Date | Elevation |
|---|---|---|---|---|
| SBART 13 | San Bartolomeo | WEST | 17 February 2019 | 402 m |
| SBART 14 | ||||
| SBART 19 | ||||
| SBART 20 | ||||
| SBART 21 | ||||
| SBART 22 | ||||
| PM 26 | Pietra Murata | WEST | 8 February 2020 | 547 m |
| PM 41 | ||||
| PM 43 | ||||
| PM 44 | ||||
| PM 45 | ||||
| PM 46 | ||||
| CDM 3A | Cima del Monte | EAST | 15 December 2018 | 428 m |
| CDM 3B | ||||
| CDM 3C | ||||
| CDM 3MIX | ||||
| CDM 4A | ||||
| CDM 4B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrini, M.; Tanini, D.; Vannini, C.; Barbanera, F. Wild Avian Gut Microbiome at a Small Spatial Scale: A Study from a Mediterranean Island Population of Alectoris rufa. Animals 2023, 13, 3341. https://doi.org/10.3390/ani13213341
Guerrini M, Tanini D, Vannini C, Barbanera F. Wild Avian Gut Microbiome at a Small Spatial Scale: A Study from a Mediterranean Island Population of Alectoris rufa. Animals. 2023; 13(21):3341. https://doi.org/10.3390/ani13213341
Chicago/Turabian StyleGuerrini, Monica, Dalia Tanini, Claudia Vannini, and Filippo Barbanera. 2023. "Wild Avian Gut Microbiome at a Small Spatial Scale: A Study from a Mediterranean Island Population of Alectoris rufa" Animals 13, no. 21: 3341. https://doi.org/10.3390/ani13213341
APA StyleGuerrini, M., Tanini, D., Vannini, C., & Barbanera, F. (2023). Wild Avian Gut Microbiome at a Small Spatial Scale: A Study from a Mediterranean Island Population of Alectoris rufa. Animals, 13(21), 3341. https://doi.org/10.3390/ani13213341





