Modulation of In Vitro Macrophage Responses via Primary and Secondary Bile Acids in Dogs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biochemical Reagents
2.2. Cell Culture Medium
2.3. Canine Macrophage Cell Lines
2.4. Generation of Monocyte-Derived Macrophages
2.5. Bile Acid Treatment of Macrophage Cultures
2.6. Flow Cytometry for Assessment of Macrophage Expression of the Bile Acid Receptor TGR5
2.7. ELISA Assays for Canine Cytokines
2.8. Assessment of TGR5 Expression via Fluorescence Microscopy
2.9. MTT Assay
2.10. RNA Sequencing and Analysis Pipeline
2.11. Statistical Analysis
3. Results
3.1. Impact of Primary and Secondary BAs on Cytokine Secretion via LPS-Activated Canine MH588 Macrophages
3.2. Impact of Primary and Secondary BAs on Cytokine Secretion via LPS-Activated Primary Monocyte-Derived Macrophages
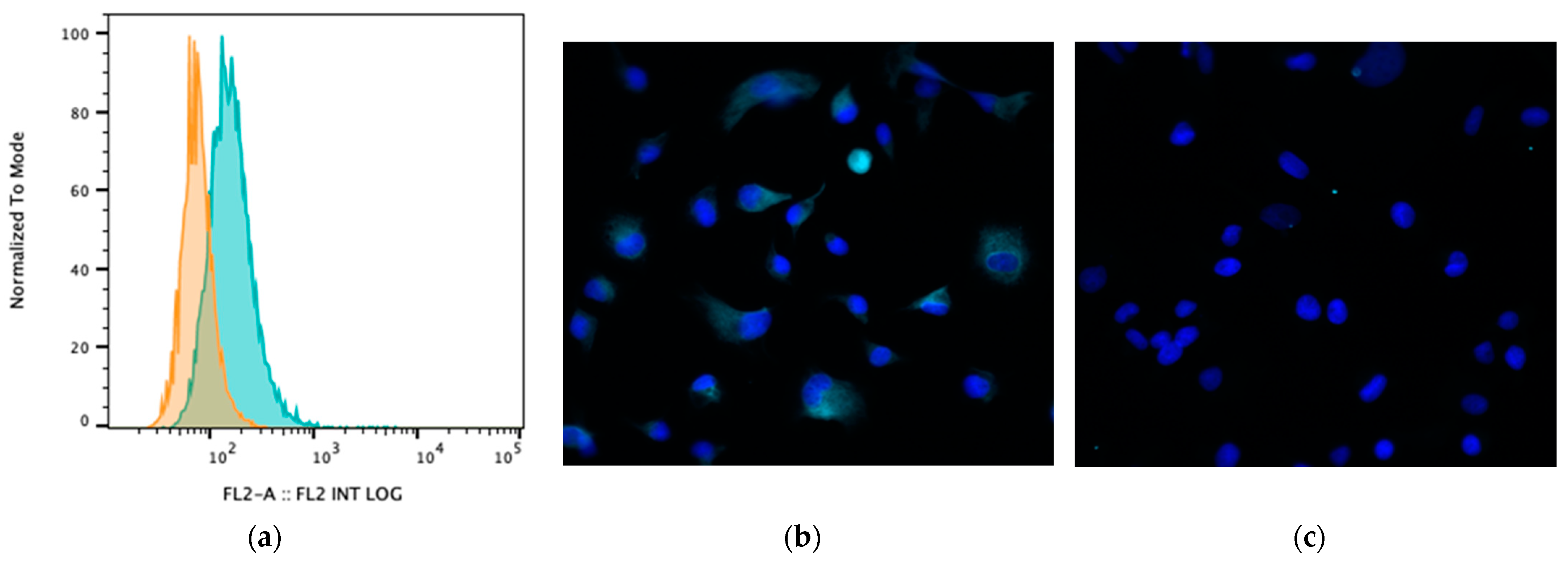
3.3. Impact of BA Exposure on TGR5 Expression
3.4. Macrophage Transcriptomic Responses to BAs
4. Discussion
4.1. Potential Impact of the New Findings
4.2. Study Potential Weaknesses and How to Address Them
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hegyi, P.; Maléth, J.; Walters, J.R.; Hofmann, A.F.; Keely, S.J. Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev. 2018, 98, 1983–2023. [Google Scholar] [CrossRef] [PubMed]
- Giaretta, P.R.; Suchodolski, J.S.; Blick, A.K.; Steiner, J.M.; Lidbury, J.A.; Rech, R.R. Distribution of bile acid receptor TGR5 in the gastrointestinal tract of dogs. Histol. Histopathol. 2019, 34, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, R.; Takayama, T.; Yoneno, K.; Kamada, N.; Kitazume, M.T.; Higuchi, H.; Matsuoka, K.; Watanabe, M.; Itoh, H.; Kanai, T.; et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology 2012, 136, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Kim, M.; Yim, M. FXR/TGR5 mediates inflammasome activation and host resistance to bacterial infection. Biochem. Biophys. Rep. 2021, 27, 101051. [Google Scholar] [CrossRef]
- Duboc, H.; Tache, Y.; Hofmann, A.F. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig. Liver Dis. 2014, 46, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef]
- Keitel, V.; Donner, M.; Winandy, S.; Kubitz, R.; Haussinger, D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 2008, 372, 78–84. [Google Scholar] [CrossRef]
- Yoneno, K.; Hisamatsu, T.; Shimamura, K.; Kamada, N.; Ichikawa, R.; Kitazume, M.T.; Mori, M.; Uo, M.; Namikawa, Y.; Matsuoka, K.; et al. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology 2013, 139, 19–29. [Google Scholar] [CrossRef]
- Wammers, M.; Schupp, A.K.; Bode, J.G.; Ehlting, C.; Wolf, S.; Deenen, R.; Köhrer, K.; Häussinger, D.; Graf, D. Reprogramming of pro-inflammatory human macrophages to an anti-inflammatory phenotype by bile acids. Sci. Rep. 2018, 8, 255. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef]
- Peleman, C.; Camilleri, M.; Busciglio, I.; Burton, D.; Donato, L.; Zinsmeister, A.R. Colonic Transit and Bile Acid Synthesis or Excretion in Patients with Irritable Bowel Syndrome-Diarrhea without Bile Acid Malabsorption. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2017, 15, 720–727.e721. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Martins, R.; Sullivan, M.C.; Friedman, E.S.; Misic, A.M.; El-Fahmawi, A.; De Martinis, E.C.P.; O’Brien, K.; Chen, Y.; Bradley, C.; et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome 2019, 7, 126. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Honneffer, J.B.; Jergens, A.E.; Jonika, M.M.; Toresson, L.; Lawrence, Y.A.; Webb, C.B.; Hill, S.; Lidbury, J.A.; Steiner, J.M.; et al. Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid-responsive chronic inflammatory enteropathy. J. Vet. Intern. Med. 2019, 33, 1295–1305. [Google Scholar] [CrossRef]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020, 27, 659–670.e655. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Manchester, A.C.; Webb, C.B.; Blake, A.B.; Sarwar, F.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Long-term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J. Vet. Intern. Med. 2019, 33, 2605–2617. [Google Scholar] [CrossRef]
- Pilla, R.; Gaschen, F.P.; Barr, J.W.; Olson, E.; Honneffer, J.; Guard, B.C.; Blake, A.B.; Villanueva, D.; Khattab, M.R.; AlShawaqfeh, M.K.; et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J. Vet. Intern. Med. 2020, 34, 1853–1866. [Google Scholar] [CrossRef]
- Xenoulis, P.G.; Palculict, B.; Allenspach, K.; Steiner, J.M.; Van House, A.M.; Suchodolski, J.S. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 2008, 66, 579–589. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Markel, M.E.; Garcia-Mazcorro, J.F.; Unterer, S.; Heilmann, R.M.; Dowd, S.E.; Kachroo, P.; Ivanov, I.; Minamoto, Y.; Dillman, E.M.; et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e51907. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Takamine, F.; Imamura, T. Isolation and characterization of bile acid 7-dehydroxylating bacteria from human feces. Microbiol. Immunol. 1995, 39, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Marion, S.; Studer, N.; Desharnais, L.; Menin, L.; Escrig, S.; Meibom, A.; Hapfelmeier, S.; Bernier-Latmani, R. In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 2019, 10, 481–503. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.; Bee, A.; Bell, S.; Gilmore, W.; Mee, A.; Morris, R.; Carter, S.D. Immunological and inflammatory characterisation of three canine cell lines: K1, K6 and DH82. Vet. Immunol. Immunopathol. 2000, 75, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Harrus, S.; Waner, T.; Friedmann-Morvinski, D.; Fishman, Z.; Bark, H.; Harmelin, A. Down-regulation of MHC class II receptors of DH82 cells, following infection with Ehrlichia canis. Vet. Immunol. Immunopathol. 2003, 96, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Bin Park, W.; Kim, S.; Kyung, S.M.; Lee, E.S.; Lee, Y.J.; Yoo, H.S. Gene expression of Toll-like receptors, cytokines and a nuclear factor and cytokine secretion in DH82 canine macrophage cells infected with Brucella canis. Vet. Immunol. Immunopathol. 2023, 260, 110607. [Google Scholar] [CrossRef]
- Chow, L.; Soontararak, S.; Wheat, W.; Ammons, D.; Dow, S. Canine polarized macrophages express distinct functional and transcriptomic profiles. Front. Vet. Sci. 2022, 9, 988981. [Google Scholar] [CrossRef]
- van Faassen, A. Bile acids, neutral steroids, and bacteria in feces as affected by a mixed, lacto-ovovegetarian, and a vegan diet. Am. J. Clin. Nutr. 1987, 46, 962–967. [Google Scholar] [CrossRef]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Chen, J.; Rao, J.N.; Zou, T.; Liu, L.; Marasa, B.S.; Xiao, L.; Zeng, X.; Turner, D.J.; Wang, J.Y. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G568–G576. [Google Scholar] [CrossRef]
- Leaphart, C.L.; Cavallo, J.; Gribar, S.C.; Cetin, S.; Li, J.; Branca, M.F.; Dubowski, T.D.; Sodhi, C.P.; Hackam, D.J. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J. Immunol. 2007, 179, 4808–4820. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, D.K.; Borcherding, D.C.; Chandra, L.; Jergens, A.E.; Atherly, T.; Bourgois-Mochel, A.; Ellinwood, N.M.; Snella, E.; Severin, A.J.; Martin, M.; et al. Differential Transcriptomic Profiles Following Stimulation with Lipopolysaccharide in Intestinal Organoids from Dogs with Inflammatory Bowel Disease and Intestinal Mast Cell Tumor. Cancers 2022, 14, 3525. [Google Scholar] [CrossRef] [PubMed]
- Costa-Pereira, C.; Moreira, M.L.; Soares, R.P.; Marteleto, B.H.; Ribeiro, V.M.; Franca-Dias, M.H.; Cardoso, L.M.; Viana, K.F.; Giunchetti, R.C.; Martins-Filho, O.A.; et al. One-year timeline kinetics of cytokine-mediated cellular immunity in dogs vaccinated against visceral leishmaniasis. BMC Vet. Res. 2015, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Soontararak, S.; Chow, L.; Johnson, V.; Coy, J.; Webb, C.; Wennogle, S.; Dow, S. Humoral immune responses against gut bacteria in dogs with inflammatory bowel disease. PLoS ONE 2019, 14, e0220522. [Google Scholar] [CrossRef] [PubMed]
- Wheat, W.; Chow, L.; Kuzmik, A.; Soontararak, S.; Kurihara, J.; Lappin, M.; Dow, S. Local immune and microbiological responses to mucosal administration of a Liposome-TLR agonist immunotherapeutic in dogs. BMC Vet. Res. 2019, 15, 330. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 6. [Google Scholar] [CrossRef]
- Regan, D.P.; Chow, L.; Das, S.; Haines, L.; Palmer, E.; Kurihara, J.N.; Coy, J.W.; Mathias, A.; Thamm, D.H.; Gustafson, D.L.; et al. Losartan Blocks Osteosarcoma-Elicited Monocyte Recruitment, and Combined With the Kinase Inhibitor Toceranib, Exerts Significant Clinical Benefit in Canine Metastatic Osteosarcoma. Clin. Cancer Res. 2022, 28, 662–676. [Google Scholar] [CrossRef]
- Pezzanite, L.M.; Chow, L.; Griffenhagen, G.M.; Bass, L.; Goodrich, L.R.; Impastato, R.; Dow, S. Distinct differences in immunological properties of equine orthobiologics revealed by functional and transcriptomic analysis using an activated macrophage readout system. Front. Vet. Sci. 2023, 10, 1109473. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Haselow, K.; Bode, J.G.; Wammers, M.; Ehlting, C.; Keitel, V.; Kleinebrecht, L.; Schupp, A.K.; Haussinger, D.; Graf, D. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J. Leukoc. Biol. 2013, 94, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Wilke, V.L.; Nettleton, D.; Wymore, M.J.; Gallup, J.M.; Demirkale, C.Y.; Ackermann, M.R.; Tuggle, C.K.; Ramer-Tait, A.E.; Wannemuehler, M.J.; Jergens, A.E. Gene expression in intestinal mucosal biopsy specimens obtained from dogs with chronic enteropathy. Am. J. Vet. Res. 2012, 73, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Taman, H.; Fenton, C.G.; Hensel, I.V.; Anderssen, E.; Florholmen, J.; Paulssen, R.H. Transcriptomic Landscape of Treatment-Naive Ulcerative Colitis. J. Crohn’s Colitis 2018, 12, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Greve, J.W.; Gouma, D.J.; Buurman, W.A. Bile acids inhibit endotoxin-induced release of tumor necrosis factor by monocytes: An in vitro study. Hepatology 1989, 10, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Barroso, A.; Mahler, J.V.; Fonseca-Castro, P.H.; Quintana, F.J. The aryl hydrocarbon receptor and the gut-brain axis. Cell Mol. Immunol. 2021, 18, 259–268. [Google Scholar] [CrossRef]
- Sandhu, N.; Rana, S.; Meena, K. Nuclear receptor subfamily 5 group A member 2 (NR5A2): Role in health and diseases. Mol. Biol. Rep. 2021, 48, 8155–8170. [Google Scholar] [CrossRef]
- Cassmann, E.; White, R.; Atherly, T.; Wang, C.; Sun, Y.; Khoda, S.; Mosher, C.; Ackermann, M.; Jergens, A. Alterations of the Ileal and Colonic Mucosal Microbiota in Canine Chronic Enteropathies. PLoS ONE 2016, 11, e0147321. [Google Scholar] [CrossRef]
- Walker, H.K. Serum metabolomic profiles in dogs with chronic enteropathy. J. Vet. Intern. Med. 2022, 36, 1752–1759. [Google Scholar] [CrossRef]
- Yu, J. Serum proteome of dogs with chronic enteropathy. J. Vet. Intern. Med. 2023, 37, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, Y.M. Treatment with Hydrolyzed Diet Supplemented with Prebiotics and Glycosaminoglycans Alters Lipid Metabolism in Canine Inflammatory Bowel Disease. Front. Vet. Sci. 2022, 7, 451. [Google Scholar] [CrossRef] [PubMed]
- Dandrieux, J.R.S.; Mansfield, C.S. Chronic Enteropathy in Canines: Prevalence, Impact and Management Strategies. Vet. Med. 2019, 10, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Manchester, A.C.; Gagne, J.W.; Lappin, M.; CHow, L.; Dow, S. Efficacy of an elemental diet in achieving clinical remission in dogs with chronic enteropathy. J. Vet. Intern. Med. 2023, 37, 2322–2333. [Google Scholar] [CrossRef]
- Hall, E.J. Antibiotic-responsive diarrhea in small animals. Vet. Clin. N. Am. Small Anim. Pract. 2011, 41, 273–286. [Google Scholar] [CrossRef]
- Dandrieux, J.R. Inflammatory bowel disease versus chronic enteropathy in dogs: Are they one and the same? J. Small Anim. Pract. 2016, 57, 589–599. [Google Scholar] [CrossRef]





| Gene ID | Description | Log2 (Ratio) (LPS + CA vs LPS + LCA) |
|---|---|---|
| CCL8 | C-C motif chemokine 8 | 4.13 |
| RSAD2 | Radical S-adenosyl methionine domain-containing protein 2 | 4.10 |
| GAP43 | Neuromodulin | 3.96 |
| IL1B | Interleukin-1 beta | 3.37 |
| IL1A | Interleukin-1 alpha | 3.31 |
| ENSCAFG00000016245 | Adhesion G protein-coupled receptor E2 | 3.10 |
| CXCL6 | C-X-C motif chemokine 6 | 2.75 |
| TBX3 | T-box transcription factor TBX3 | 2.69 |
| SERPINB2 | Plasminogen activator inhibitor 2 | 2.62 |
| TIFAB | TRAF-interacting protein with FHA domain-containing protein B | 2.61 |
| SELENOM | Selenoprotein M | 2.61 |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | 2.45 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | −6.34 |
| PGF | Placenta growth factor | −5.26 |
| SLA | Src-like-adapter | −5.18 |
| FCMR | Fas apoptotic inhibitory molecule 3 | −4.61 |
| CFAP45 | Cilia- and flagella-associated protein 45 | −4.52 |
| CRLF1 | Cytokine receptor-like factor 1 | −4.47 |
| SLC1A7 | Excitatory amino acid transporter 5 | −4.40 |
| KEL | Kell blood group glycoprotein | −4.31 |
| SAMD11 | Sterile alpha motif domain-containing protein 11 | −4.26 |
| KCNMA1 | Calcium-activated potassium channel subunit alpha-1 | −4.25 |
| TMEM59L | Transmembrane protein 59-like | −3.76 |
| SELPLG | P-selectin glycoprotein ligand 1 | −3.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manchester, A.C.; Chow, L.; Wheat, W.; Dow, S. Modulation of In Vitro Macrophage Responses via Primary and Secondary Bile Acids in Dogs. Animals 2023, 13, 3714. https://doi.org/10.3390/ani13233714
Manchester AC, Chow L, Wheat W, Dow S. Modulation of In Vitro Macrophage Responses via Primary and Secondary Bile Acids in Dogs. Animals. 2023; 13(23):3714. https://doi.org/10.3390/ani13233714
Chicago/Turabian StyleManchester, Alison C., Lyndah Chow, William Wheat, and Steven Dow. 2023. "Modulation of In Vitro Macrophage Responses via Primary and Secondary Bile Acids in Dogs" Animals 13, no. 23: 3714. https://doi.org/10.3390/ani13233714






