Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity
Abstract
:1. Nitrogen Metabolism
1.1. Fundamentals of Nitrogen Assimilation in Prokaryotes
1.2. Nitrogen Assimilation in Gram-Negative Bacteria
1.2.1. Nitrogen Assimilation and Control in Escherichia coli
1.2.2. Nitrogen Assimilation and Control in Cyanobacteria
1.3. Nitrogen Assimilation in Gram-Positive Bacteria
1.3.1. Nitrogen Assimilation and Its Control in Bacillus subtilis
1.3.2. Nitrogen Assimilation and Its Control in Corynebacterium glutamicum
1.3.3. Nitrogen Assimilation and Its Control in Streptomyces coelicolor
1.3.4. Nitrogen Assimilation and Its Control in Mycobacterium tuberculosis
1.4. The Central Role of Glutamine Synthetases in the Bacterial Nitrogen Metabolism
2. Polyamine and monoamine Metabolism
2.1. Polyamine Metabolism in Bacteria
2.1.1. Distribution and Role of Polyamines
2.1.2. Importance of Polyamines for Intracellular Pathogens
2.1.3. Occurrence of Polyamines in Bacterial Cells
Polyamine Biosynthesis
Polyamine Uptake
2.1.4. Polyamine Assimilation in Bacteria
Polyamine Utilization in E. coli
Polyamine Utilization in P. aeruginosa
Polyamine Utilization in S. coelicolor
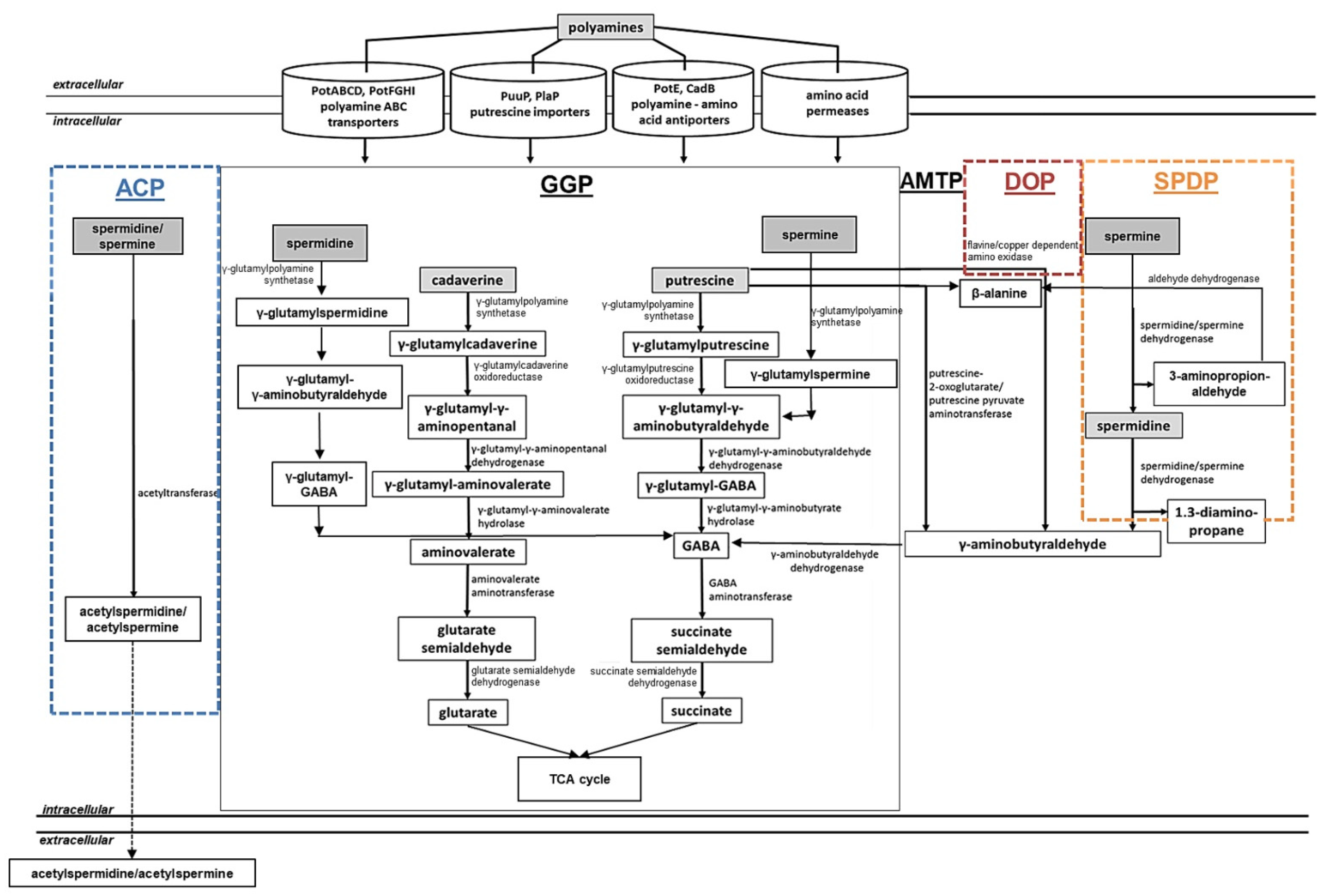
2.1.5. Regulation of Polyamine Assimilation Genes in Bacteria
2.2. Monoamine Metabolism in Bacteria
2.2.1. Distribution and Role of the Monoamine Ethanolamine
2.2.2. Ethanolamine Biosynthesis and Uptake in Bacteria
2.2.3. Ethanolamine Assimilation in Bacteria
Ethanolamine Assimilation in E. coli and S. typhimurium
Ethanolamine Assimilation in S. coelicolor and M. tuberculosis
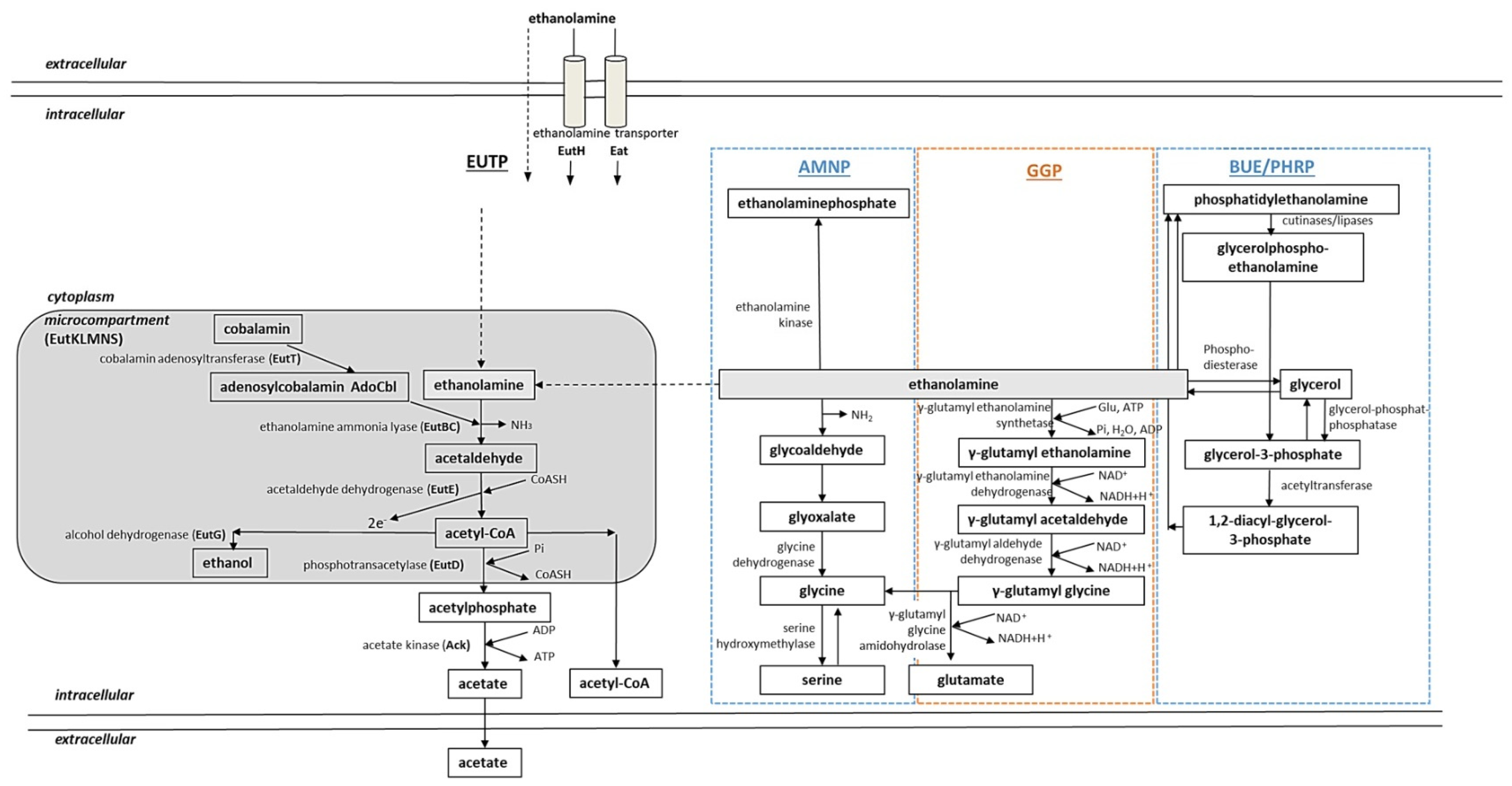
2.2.4. Regulation of Ethanolamine Assimilation Genes in Bacteria
3. Recent Advances in Drug Development Targeting Bacterial Nitrogen, Mono- and Polyamine Metabolism
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aharonowitz, Y. Nitrogen metabolite regulation of antibiotic biosynthesis. Annu. Rev. Microbiol. 1980, 34, 209–233. [Google Scholar] [PubMed]
- Arcondéguy, T.; Jack, R.; Merrick, M. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 2001, 65, 80–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, S. A Guide to the Polyamines; Oxford University Press: Oxford, UK, 1998; p. 610. [Google Scholar]
- Tyler, B. Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 1978, 47, 1127–1162. [Google Scholar] [CrossRef] [PubMed]
- Magasanik, B. Genetic control of nitrogen assimilation in bacteria. Annu. Rev. Genet. 1982, 16, 135–168. [Google Scholar] [CrossRef]
- Merrick, M.J.; Edwards, R.A. Nitrogen control in bacteria. Microbiol. Rev. 1995, 59, 604–622. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Matthews, A.; Busche, T.; Bera, A.; Wohlleben, W. Poly- and Monoamine Metabolism in Streptomyces coelicolor: The New Role of Glutamine Synthetase-Like Enzymes in the Survival under Environmental Stress. Microb. Physiol. 2021, 31, 233–247. [Google Scholar] [CrossRef]
- Sperber, A.M.; Herman, J.K. Metabolism Shapes the Cell. J. Bacteriol. 2017, 199, e00039-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, G.; Mullins, J.G.L.; Merrick, M. Membrane topology of the Mep/Amt family of ammonium transporters. Mol. Microbiol. 2000, 37, 331–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, S.; Vining, L.C. Suppression of nitrate utilization by ammonium and its relationship to chloramphenicol production in Streptomyces venezuelae. Can. J. Microbiol. 1984, 30, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Soda, K. Biochemistry and biotechnology of amino acid dehydrogenases. Adv. Biochem. Eng. Biotechnol. 1990, 42, 187. [Google Scholar]
- Moir, J.W.B.; Wood, N.J. Nitrate and nitrite transport in bacteria. Cell. Mol. Life Sci. 2001, 58, 215–224. [Google Scholar] [CrossRef]
- Fischer, M.; Alderson, J.; van Keulen, G.; White, J.; Sawers, R.G. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 2010, 156, 3166–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Kurihara, S. Polyamine Catabolism in Prokaryotes. In Polyamines: A Universal Molecular Nexus for Growth, Survival, and Specialized Metabolism; Kusano, T., Suzuki, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 47–59. [Google Scholar]
- Yuan, J.; Doucette, C.D.; Fowler, W.U.; Feng, X.J.; Piazza, M.; Rabitz, H.A.; Wingreen, N.S.; Rabinowitz, J.D. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 2009, 5, 302. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Wohlleben, W. Überlebenswichtig: Glutaminsynthetasehomologe Proteine in Streptomyceten. Biospektrum 2022, 28, 23–26. [Google Scholar] [CrossRef]
- Reitzer, L.J.; Magasanik, B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, L-alanine, and D-alanine. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, Volume 1; Neidhardt, F.C., Ingraham, J.L., Low, K.B., Magasanik, B., Schaechter, M., Umbarger, H.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1987; pp. 302–320. [Google Scholar]
- Blauwkamp, T.A.; Ninfa, A.J. Physiological role of the GlnK signal transduction protein of Escherichia coli: Survival of nitrogen starvation. Mol. Microbiol. 2002, 46, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Ninfa, A.J.; Jiang, P.; Atkinson, M.R.; Peliska, J.A. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell. Regul. 2001, 36, 31–75. [Google Scholar]
- He, L.; Soupene, E.; Kustu, S. NtrC is required for control of Klebsiella pneumoniae NifL activity. J. Bacteriol. 1997, 179, 7446–7455. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, B.M.; Stadtman, E.R. Glutamine synthetase (Escherichia coli). Methods Enzymol. 1970, 17, 910–922. [Google Scholar]
- Stadtman, E.R.; Ginsburg, A. The glutamine synthetase of Escherichia coli: Structure and control. In The Enzymes; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1974; Volume 10, pp. 755–807. [Google Scholar]
- Carroll, P.; Pashley, C.A.; Parish, T. Functional analysis of GlnE, an essential adenylyl transferase in Mycobacterium tuberculosis. J. Bacteriol. 2008, 190, 4894–4902. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Pioszak, A.A.; Ninfa, A.J. Structure/function analysis of glutamine synthetase adenylyltransferase (ATase, E.C. 2.7.7.49) of Escherichia coli. Biochemistry 2007, 46, 4117–4132. [Google Scholar] [CrossRef]
- Bolay, P.; Muro-Pastor, M.I.; Florencio, F.J.; Klähn, S. The Distinctive Regulation of Cyanobacterial Glutamine Synthetase. Life 2018, 8, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Dominguez, M.; Reyes, J.C.; Florencio, F.J. Glutamine synthetase inactivation by protein-protein interaction. Proc. Natl. Acad. Sci. USA 1999, 96, 7161–7166. [Google Scholar] [CrossRef] [Green Version]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [PubMed]
- Vazquez-Bermudez, M.F.; Herrero, A.; Flores, E. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002, 512, 71–74. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Peliska, J.A.; Ninfa, A.J. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 1998, 37, 12782–12794. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, M.I.; Florencio, F.J. Regulation of ammonium assimilation in cyanobacteria. Plant. Physiol. Biochem. 2003, 41, 595–603. [Google Scholar] [CrossRef]
- Atkinson, M.R.; Fisher, S.H. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J. Bacteriol. 1991, 173, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Schreier, H.J. Biosynthesis of glutamine and glutamate and the assimilation of ammonia in Bacillus subtilis and other Gram-positive bacteria. In Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics; Sonenshein, A.L., Hoch, J.A., Losick, R., Eds.; American Society for Microbiology: Washington, DC, USA, 1993; pp. 281–298. [Google Scholar]
- Picossi, S.; Belitsky, B.R.; Sonenshein, A.L. Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC. J. Mol. Biol. 2007, 365, 1298–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, S.H. Regulation of nitrogen metabolism in Bacillus subtilis: Vive la difference! Mol. Microbiol. 1999, 32, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, J.L.; Wray, L.V., Jr.; Beier, L.; Jarmer, H.; Saxild, H.H.; Fisher, S.H. Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter. J. Bacteriol. 2002, 184, 6060–6064. [Google Scholar] [CrossRef] [Green Version]
- Kormelink, T.; Koenders, E.; Hagemeijer, Y.; Overmars, L.; Siezen, R.J.; de Vos, W.M.; Francke, C. Comparative genome analysis of central nitrogen metabolism and its control by GlnR in the class bacilli. BMC Genom. 2012, 13, 191. [Google Scholar] [CrossRef] [Green Version]
- Meier-Wagner, J.; Nolden, L.; Jakoby, M.; Siewe, R.; Krämer, R.; Burkovski, A. Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: Role of Amt and AmtB. Microbiology 2001, 147, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siewe, R.M.; Weil, B.; Burkovski, A.; Eikmanns, B.J.; Eikmanns, M.; Krämer, R. Functional and genetic characterization of the (methyl) ammonium uptake carrier of Corynebacterium glutamicum. J. Biol. Chem. 1996, 271, 5398–5403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, B.; Kuspert, M.; Ansorge, D.; Kramer, R.; Burkovski, A. Dissection of ammonium uptake systems in Corynebacterium glutamicum: Mechanism of action and energetics of AmtA and AmtB. J. Bacteriol. 2008, 190, 2611–2614. [Google Scholar] [CrossRef] [Green Version]
- Amon, J.; Titgemeyer, F.; Burkovski, A. Common patterns—Unique features: Nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol. Rev. 2010, 34, 588–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckers, G.; Strosser, J.; Hildebrandt, U.; Kalinowski, J.; Farwick, M.; Kramer, R.; Burkovski, A. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: Mechanism and characterization of the AmtR regulon. Mol. Microbiol. 2005, 58, 580–595. [Google Scholar] [CrossRef]
- Jakoby, M.; Krämer, R.; Burkovski, A. Nitrogen regulation in Corynebacterium glutamicum: Isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiol. Lett. 1999, 173, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolden, L.; Beckers, G.; Mockel, B.; Pfefferle, W.; Nampoothiri, K.M.; Kramer, R.; Burkovski, A. Urease of Corynebacterium glutamicum: Organization of corresponding genes and investigation of activity. FEMS Microbiol. Lett. 2000, 189, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Burkovski, A. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: An example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 2003, 27, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Harper, C.; Hayward, D.; Wiid, I.; van Helden, P. Regulation of nitrogen metabolism in Mycobacterium tuberculosis: A comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor. IUBMB Life 2008, 60, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Forty years of genetics with Streptomyces: From in vivo through in vitro to in silico. Microbiology 1999, 145, 2183–2202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchanan, R.E. Studies in the nomenclature and classification of the bacteria. II. The Primary Sub-divisions of the Schizomycetes. J. Bacteriol. 1917, 2, 155–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, F.; Bibb, M.J. Codon usage in the G+C-rich Streptomyces genome. Gene 1992, 113, 55–65. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A.; Chater, K.F.; Bibb, M.J. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 1995, 28, 65–102. [Google Scholar] [PubMed]
- Hodgson, D.A. Primary metabolism and its control in Streptomycetes. Adv. Microbial. Physiol. 2000, 42, 47–238. [Google Scholar]
- McCarthy, A.J.; Williams, S.T. Actinomycetes as agents of biodegradation in the environment—A review. Gene 1992, 115, 189–192. [Google Scholar] [CrossRef]
- Hopwood, D.A. Soil to genomics: The Streptomyces chromosome. Annu. Rev. Genet. 2006, 40, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karandikar, A.; Sharples, G.P.; Hobbs, G. Differentiation of Streptomyces coelicolor A3(2) under nitrate limited conditions. Microbiology 1997, 143, 3581–3590. [Google Scholar] [CrossRef] [Green Version]
- Wray, L.V., Jr.; Atkinson, M.R.; Fisher, S.H. Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2). J. Bacteriol. 1991, 173, 7351–7360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wray, L.V., Jr.; Fisher, S.H. The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene 1993, 130, 145–150. [Google Scholar] [CrossRef]
- Reuther, J.; Wohlleben, W. Nitrogen Metabolism in Streptomyces coelicolor: Transcriptional and Post-Translational Regulation. J. Mol. Microbiol. Biotechnol. 2007, 12, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Tiffert, Y.; Supra, P.; Wurm, R.; Wohlleben, W.; Wagner, R.; Reuther, J. The Streptomyces coelicolor GlnR regulon: Identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 2008, 67, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Tiffert, Y.; Franz-Wachtel, M.; Fladerer, C.; Nordheim, A.; Reuther, J.; Wohlleben, W.; Mast, Y. Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 2011, 89, 1149–1159. [Google Scholar] [CrossRef]
- Fink, D.; Falke, D.; Wohlleben, W.; Engels, A. Nitrogen metabolism in Streptomyces coelicolor A3(2): Modification of glutamine synthetase I by an adenylyltransferase. Microbiology 1999, 145, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Weissschuh, N.; Reuther, J.; Wohlleben, W.; Engels, A. Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 2002, 46, 331–347. [Google Scholar] [CrossRef]
- Pullan, S.T.; Chandra, G.; Bibb, M.J.; Merrick, M. Genomewide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genom. 2011, 12, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, R.; Franz-Wachtel, M.; Tiffert, Y.; Heberer, M.; Meky, M.; Ahmed, Y.; Matthews, A.; Krysenko, S.; Jakobi, M.; Hinder, M.; et al. Post-translational Serine/Threonine Phosphorylation and Lysine Acetylation: A Novel Regulatory Aspect of the Global Nitrogen Response Regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 2016, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhao, G.-P. GlnR positively regulates nasA transcription in Streptomyces coelicolor. Biochem. Biophys. Res. Commun. 2009, 386, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Reuther, J.; Bera, A.; Wohlleben, W.; Mast, Y. A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 2012, 158, 1172–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissschuh, N.; Fink, D.; Vierling, S.; Bibb, M.J.; Wohlleben, W.; Engels, A. Transcriptional analysis of the gene for glutamine synthetase II and two upstream genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 2000, 264, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Hindra-Mulder, D.; Yin, C.; Elliot, M.A. Crp is a global regulator of antibiotic production in Streptomyces. mBio 2012, 3, e00407-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-García, A.; Sola-Landa, A.; Apel, K.; Santos-Beneit, F.; Martín, J.F. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: Direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 2009, 37, 3230–3242. [Google Scholar] [CrossRef] [PubMed]
- Sola-Landa, A.; Rodríguez-García, A.; Amin, R.; Wohlleben, W.; Martín, J.F. Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res. 2013, 41, 1767–1782. [Google Scholar] [CrossRef] [Green Version]
- Perez-Redondo, R.; Rodriguez-Garcia, A.; Botas, A.; Santamarta, I.; Martin, J.F.; Liras, P. ArgR of Streptomyces coelicolor is a versatile regulator. PLoS ONE 2012, 7, e32697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Beneit, F.; Rodriguez-Garcia, A.; Martin, J.F. Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiol. Res. 2012, 167, 532–535. [Google Scholar] [CrossRef]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wezel, G.P.; McDowall, K.J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 2011, 28, 1311–1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Mast, Y.; Wang, J.; Zhang, W.; Zhao, G.; Wohlleben, W.; Lu, Y.; Jiang, W. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol. 2013, 87, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Hou, Y.; Zhang, H.; Chu, Y.; Xia, H.; Tian, Y. Regulatory genes and their roles for improvement of antibiotic biosynthesis in Streptomyces. Biotech 2017, 7, 250. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel, E.; Herbig, A.; Battke, F.; Amin, R.; Nentwich, M.; Nieselt, K.; Ellingsen, T.E.; Wentzel, A.; Hodgson, D.A.; Wohlleben, W.; et al. The PII protein GlnK is a pleiotropic regulator for morphological differentiation and secondary metabolism in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2011, 92, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Wohlleben, W.; Bera, A.; Mast, Y.; Stegmann, E. Regulation of Secondary Metabolites of Actinobacteria. In Biology and Biotechnology of Actinobacteria; Wink, J., Mohammadipanah, F., Hamedi, J., Eds.; Springer International Publishing AG: Berlin/Heidelberg, Germany, 2017; p. 395. [Google Scholar]
- Hesketh, A.; Fink, D.; Gust, B.; Rexer, H.U.; Scheel, B.; Chater, K.; Wohlleben, W.; Engels, A. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 2002, 46, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Tullius, M.V.; Harth, G.; Horwitz, M.A. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 2003, 71, 3927–3936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agapova, A.; Serafini, A.; Petridis, M.; Hunt, D.M.; Garza-Garcia, A.; Sohaskey, C.D.; Sório de Carvalho, L.P. Flexible nitrogen utilisation by the metabolic generalist pathogen Mycobacterium tuberculosis. eLife 2019, 8, e41129. [Google Scholar] [CrossRef] [PubMed]
- Gouzy, A.; Poquet, Y.; Neyrolles, O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 2014, 12, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Harth, G.; Maslesa-Galic, S.; Tullius, M.V.; Horwitz, M.A. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 2005, 58, 1157–1172. [Google Scholar] [CrossRef]
- Jenkins, V.A.; Robertson, B.D.; Williams, K.J. Aspartate D48 is essential for the GlnR-mediated transcriptional response to nitrogen limitation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2012, 330, 38–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, K.J.; Jenkins, V.A.; Barton, G.R.; Bryant, W.A.; Krishnan, N.; Robertson, B.D. Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol. Microbiol. 2015, 97, 1142–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesole, G.; Gissi, C.; Lanave, C.; Saccone, C. Glutamine synthetase gene evolution in bacteria. Mol. Biol. Evol. 1995, 12, 189–197. [Google Scholar] [PubMed] [Green Version]
- Reitzer, L.; Schneider, B.L. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 2001, 65, 422–444. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Sun, Y.; Zhi, Y.; Wei, X.; Luo, Y.; Mao, L.; Zhou, P. Identification and characterization of the nitrate assimilation genes in the isolate of Streptomyces griseorubens JSD-1. Microb. Cell Factories 2014, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Kumada, Y.; Benson, D.R.; Hillemann, D.; Hosted, T.J.; Rochefort, D.A.; Thompson, C.J.; Wohlleben, W.; Tateno, Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. USA 1993, 90, 3009–3013. [Google Scholar] [CrossRef] [Green Version]
- Mathis, R.; Gamas, P.; Meyer, Y.; Cullimore, J.V. The presence of GSI-like genes in higher plants: Support for the paralogous evolution of GSI and GSII genes. J. Mol. Evol. 2000, 50, 116–122. [Google Scholar] [CrossRef]
- Wyatt, K.; White, H.E.; Wang, L.; Bateman, O.A.; Slingsby, C.; Orlova, E.V.; Wistow, G. Lengsin is a survivor of an ancient family of class I glutamine synthetases re-engineered by evolution for a role in the vertebrate lens. Structure 2006, 14, 1823–1834. [Google Scholar] [CrossRef] [Green Version]
- Behrmann, I.; Hillemann, D.; Pühler, A.; Strauch, E.; Wohlleben, W. Overexpression of a Streptomyces viridochromogenes gene (glnII) encoding a glutamine synthetase similar to those of eucaryotes confers resistance against the antibiotic phosphinothricyl-alanyl-alanine. J. Bacteriol. 1990, 172, 5326–5334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S.L.; Young, J.P. The glutamine synthetases of rhizobia: Phylogenetics and evolutionary implications. Mol. Biol. Evol. 2000, 17, 309–319. [Google Scholar] [PubMed] [Green Version]
- Reyes, J.C.; Florencio, F.J. A mutant lacking the glutamine synthetase gene (glnA) is impaired in the regulation of the nitrate assimilation system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1994, 176, 7516–7523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, S.; Isu, S.; Kaneko, G.; Yamada, H.; Hara, T.; Itoh, Y.; Watabe, S. The occurrence of eukaryotic type III glutamine synthetase in the marine diatom Chaetoceros compressum. Mar. Genom. 2009, 2, 103–111. [Google Scholar] [CrossRef]
- Chiurazzi, M.; Meza, R.; Lara, M.; Lahm, A.; Defez, R.; Iaccarino, M.; Espín, G. The Rhizobium leguminosarum biovar phaseoli glnT gene, encoding glutamine synthetase III. Gene 1992, 119, 1–8. [Google Scholar] [CrossRef]
- Shatters, R.G.; Liu, Y.; Kahn, M.L. Isolation and characterization of a novel glutamine synthetase from Rhizobium meliloti. J. Biol. Chem. 1993, 268, 469–475. [Google Scholar] [CrossRef]
- Rossbach, S.; Schell, J.; Bruijn, F.J. Cloning and analysis of Agrobacterium tumefaciens C58 loci involved in glutamine biosynthesis: Neither the glnA (GSI) nor the glnII (GSII) gene plays a special role in virulence. Mol. Gen. Genet. 1988, 212, 38–47. [Google Scholar] [CrossRef]
- Eisenberg, D.; Gill, H.S.; Pfluegl, G.M.; Rotstein, S.H. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 2000, 1477, 122–145. [Google Scholar] [CrossRef] [Green Version]
- Robertson, D.L.; Alberte, R.S. Isolation and characterization of glutamine synthetase from the marine diatom Skeletonema costatum. Plant. Physiol. 1996, 111, 1169–1175. [Google Scholar] [CrossRef] [Green Version]
- Unno, H.; Uchida, T.; Sugawara, H.; Kurisu, G.; Sugiyama, T.; Yamaya, T.; Sakakibara, H.; Hase, T.; Kusunoki, M. Atomic structure of plant glutamine synthetase: A key enzyme for plant productivity. J. Biol. Chem. 2006, 281, 29287–29296. [Google Scholar] [CrossRef] [Green Version]
- Seabra, A.R.; Carvalho, H.; Pereira, P.J. Crystallization and preliminary crystallographic characterization of glutamine synthetase from Medicago truncatula. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 1309–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krajewski, W.W.; Collins, R.; Holmberg-Schiavone, L.; Jones, T.A.; Karlberg, T.; Mowbray, S.L. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J. Mol. Biol. 2008, 375, 217. [Google Scholar] [CrossRef] [Green Version]
- van Rooyen, J.M.; Abratt, V.R.; Belrhali, H.; Sewell, T. Crystal structure of Type III glutamine synthetase: Surprising reversal of the inter-ring interface. Structure 2011, 19, 471–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaw, S.H.; Pan, C.; Eisenberg, D. Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine. Proc. Natl. Acad. Sci. USA 1993, 90, 4996–5000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, H.S.; Eisenberg, D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 2001, 40, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.K.; Park, Y.W.; Jang, Y.Y.; Lee, J.Y. Structural Analysis of Glutamine Synthetase from Helicobacter pylori. Sci. Rep. 2018, 8, 11657. [Google Scholar] [CrossRef] [Green Version]
- Murray, D.S.; Chinnam, N.; Tonthat, N.K.; Whitfill, T.; Wray, L.V., Jr.; Fisher, S.H.; Schumacher, M.A. Structures of the Bacillus subtilis glutamine synthetase dodecamer reveal large intersubunit catalytic conformational changes linked to a unique feedback inhibition mechanism. J. Biol. Chem. 2013, 288, 35801–35811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, H.S.; Pfluegl, G.M.; Eisenberg, D. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry 2002, 41, 9863–9872. [Google Scholar] [CrossRef] [PubMed]
- Wray, L.V.; Fisher, S.H. Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene 1988, 71, 247–256. [Google Scholar] [CrossRef]
- Hillemann, D.; Dammann, T.; Hillemann, A.; Wohlleben, W. Genetic and biochemical characterization of the two glutamine synthetases GSI and GSII of the phosphinothricyl-alanyl-alanine producer, Streptomyces viridochromeogenes Tu494. J. Gen. Microbiol. 1993, 139, 1773–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayward, D.; van Helden, P.D.; Wiid, I.J. Glutamine synthetase sequence evolution in the mycobacteria and their use as molecular markers for Actinobacteria speciation. BMC Evol. Biol. 2009, 9, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rexer, H.U.; Schaberle, T.; Wohlleben, W.; Engels, A. Investigation of the functional properties and regulation of three glutamine synthetase like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 2006, 186, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Okoniewski, N.; Kulik, A.; Matthews, A.; Grimpo, J.; Wohlleben, W.; Bera, A. Gamma-Glutamylpolyamine Synthetase GlnA3 is involved in the first step of polyamine degradation pathway in Streptomyces coelicolor M145. Front. Microbiol. 2017, 8, 726. [Google Scholar] [CrossRef] [Green Version]
- Krysenko, S.; Matthews, A.; Okoniewski, N.; Kulik, A.; Girbas, M.G.; Tsypik, O.; Meyners, C.S.; Hausch, F.; Wohlleben, W.; Bera, A. Initial metabolic step of a novel ethanolamine utilization pathway and its regulation in Streptomyces coelicolor M145. mBio 2019, 10, e00326-19. [Google Scholar] [CrossRef] [Green Version]
- Krysenko, S.; Okoniewski, N.; Nentwich, M.; Matthews, A.; Bäuerle, M.; Zinser, A.; Busche, T.; Kulik, A.; Gursch, S.; Kemeny, A.; et al. A Second Gamma-Glutamylpolyamine Synthetase, GlnA2, Is Involved in Polyamine Catabolism in Streptomyces coelicolor. Int. J. Mol. Sci. 2022, 23, 3752. [Google Scholar] [CrossRef] [PubMed]
- Yatin, M. Polyamines in living organisms. Mol. Cell. Biol. 2002, 1, 57–67. [Google Scholar]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018, 293, 18693–18701. [Google Scholar] [CrossRef] [Green Version]
- Oshima, T.; Hamasaki, N.; Uzawa, T. Biochemical Properties of Unusual Polyamines Found in an Extreme Thermophile, Thermus thermophiles. In Progress in Polyamine Research; Zappia, V., Pegg, A.E., Eds.; Springer: Boston, MA, USA, 1988; pp. 633–642. [Google Scholar]
- Hamana, K.; Matsuzaki, S. Distribution of polyamines in actinomycetes. FEMS Microbiol. Lett. 1987, 41, 1574–6968. [Google Scholar] [CrossRef]
- Fukuda, W.; Yamori, Y.; Hamakawa, M.; Osaki, M.; Fukuda, M.; Hidese, R.; Kanesaki, Y.; Okamoto-Kainuma, A.; Kato, S.; Fujiwara, S. Genes regulated by branched-chain polyamine in the hyperthermophilic archaeon Thermococcus kodakarensis. Amino Acids 2020, 52, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Young, C.C.; Chen, L.F. Polyamines in humic acid and their effect on radical growth of lettuce seedlings. Plant Soil 1997, 195, 143–149. [Google Scholar] [CrossRef]
- Lasbury, M.E.; Merali, S.; Durant, P.J.; Tschang, D.; Ray, C.A.; Lee, C.H. Polyamine-mediated apoptosis of alveolar macrophages during Pneumocystis pneumonia. J. Biol. Chem. 2007, 282, 11009–11020. [Google Scholar] [CrossRef] [Green Version]
- Broshtilova, V.; Lozanov, V.; Miteva, L. Comparative analysis of polyamine metabolism in benign and neoplastic keratinocytic proliferations. Acta Derm. Venereol. 2012, 21, 3–5. [Google Scholar]
- Tabor, C.W.; Tabor, H. Polyamines in microorganisms. Microbiol. Rev. 1985, 49, 81–99. [Google Scholar] [CrossRef]
- Grasemann, H.; Shehnaz, D.; Enomoto, M.; Leadley, M.; Belik, J.; Ratjen, F. L-Ornithine Derived Polyamines in Cystic Fibrosis Airways. PLoS ONE 2012, 7, e46618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishna, S.; Guarino, L.; Cohen, S.S. Polyamines of Anacystis nidulans and metabolism of exogenous spermidine and spermine. J. Bacteriol. 1978, 134, 744–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, A.; Sahara, J.; Kawai, G.; Yamamoto, K.; Ishihama, A.; Uemura, T.; Igarashi, K.; Kashiwagi, K.; Terui, Y. Cytotoxic Mechanism of Excess Polyamines Functions through Translational Repression of Specific Proteins Encoded by Polyamine Modulon. Int. J. Mol. Sci. 2020, 21, 2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, K.; Kashiwagi, K. Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 2018, 293, 18702–18709. [Google Scholar] [CrossRef] [Green Version]
- Hanfrey, C.; Sommer, S.; Mayer, M.J.; Burtin, D.; Michael, A.J. Arabidopsis polyamine biosynthesis: Absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 2001, 27, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, Y.; Itoh, Y. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 2003, 149, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Rosenthal, S.M. The acetylation of polyamines in Escherichia coli. J. Biol. Chem. 1960, 235, 776–782. [Google Scholar] [CrossRef]
- Shah, P.; Swiatlo, E. Immunization with polyamine transport protein PotD protects mice against systemic infection with Streptococcus pneumoniae. Infect. Immun. 2006, 74, 5888–5892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, H.J.; Kim, E.J.; Lee, J.K. Physiological polyamines: Simple primordial stress molecules. J. Cell. Mol. Med. 2007, 11, 685–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrell, M.; Hanfrey, C.C.; Murray, E.J.; Stanley-Wall, N.R.; Michael, A.J. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J. Biol. Chem. 2010, 285, 39224–39238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrell, M.; Hanfrey, C.C.; Kinch, L.N.; Elliot, K.A.; Michael, J.A. Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 2012, 86, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.N.; Wortham, B.W.; Lines, J.L.; Fetherston, J.D.; Perry, R.D.; Oliveira, M.A. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 2006, 188, 2355–2363. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Sperandio, V.; Frantz, D.E.; Longgood, J.; Camilli, A.; Phillips, M.A.; Michael, A.J. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 2009, 284, 9899–9907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, R.; Hanfrey, C.C.; Elliott, K.A.; McCloskey, D.E.; Wang, X.; Kanugula, S.; Pegg, A.E.; Michael, A.J. Independent evolutionary origins of functional polyamine biosynthetic enzyme fusions catalysing de novo diamine to triamine formation. Mol. Microbiol. 2011, 81, 1109–1124. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, M.K.; Tabor, C.W.; Tabor, H. Polyamines are not required for aerobic growth of Escherichia coli: Preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 2009, 191, 5549–5552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain ATyagi, A.K. Role of polyamines in the synthesis of RNA in mycobacteria. Mol. Cell. Biochem. 1987, 78, 3–8. [Google Scholar]
- Huang, S.C.; Panagiotidis, C.A.; Canellakis, E.S. Transcriptional effects of polyamines on ribosomal proteins and on polyamine-synthesizing enzymes in Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 3464–3468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, N.K.; Shankar, S.; Tyagi, A.K. Polyamines exert regulatory control on mycobacterial transcription: A study using RNA polymerase from Mycobacterium phlei. Biochem. Mol. Biol. Int. 1995, 35, 1189–1198. [Google Scholar]
- Algranati, I.D.; Echandi, G.; Garcia-Patrone, M.; Gonzalez, N.S.; Goldemberg, S.H. Polyamines, equilibrium between ribosomal particles and protein synthesis in bacteria. Arch. Biol. Med. Exp. 1976, 10, 49–60. [Google Scholar] [PubMed]
- Nastri, H.G.; Fastame, I.G.; Algranati, I.D. Polyamines modulatestreptomycin-induced mistranslation in Escherichia coli. Biochim. Biophys. Acta 1993, 1216, 455–459. [Google Scholar] [CrossRef]
- Higashi, K.; Kashiwagi, K.; Taniguchi, S.; Terui, Y.; Yamamoto, K.; Ishihama, A.; Igarashi, K. Enhancement of C1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli. J. Biol. Chem. 2006, 281, 9527–9537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terui, Y.; Akiyama, M.; Sakamoto, A.; Tomitori, H.; Yamamoto, K.; Ishihama, A.; Igarashi, K.; Kashiwagi, K. Increase in cell viability by polyamines through stimulation of the synthesis of ppGpp regulatory protein and omega protein of RNA polymerase in Escherichia coli. Int. J. Biochem. Cell. Biol. 2012, 44, 412–422. [Google Scholar] [CrossRef]
- Griffiths, G.L.; Sigel, S.P.; Payne, S.M.; Neilands, J.B. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 1984, 259, 383–385. [Google Scholar] [CrossRef]
- Brickman, T.J.; Armstrong, S.K. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: Putrescine is a precursor of alcaligin. J. Bacteriol. 1996, 178, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oves-Costales, D.; Kadi, N.; Fogg, M.J.; Song, L.; Wilson, K.S.; Challis, G.L. Petrobactin biosynthesis: AsbB catalyzes condensation of spermidine with N8-citryl-spermidine and its N1-(3,4-dihydroxybenzoyl) derivative. Chem. Commun. 2008, 34, 4034–4036. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Kashiwagi, K.; Shigemasa, A.; Taniguchi, S.; Yamamoto, K.; Makinoshima, H.; Ishihama, A.; Igarashi, K. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 2004, 279, 46008–46013. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, K.; Kashiwagi, K. Polyamine Modulon in Escherichia coli: Genes involved in the stimulation of cell growth by polyamines. J. Biochem. 2006, 139, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Wortham, B.W.; Patel, C.N.; Oliveira, M.A. Polyamines in bacteria: Pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 2007, 603, 106–115. [Google Scholar]
- Nesse, L.L.; Berg, K.; Vestby, L.K. Effects of norspermidine and spermidine on biofilm formation by potentially pathogenic Escherichia coli and Salmonella enterica wild-type strains. Appl. Environ. Microbiol. 2015, 81, 2226–2232. [Google Scholar] [CrossRef] [Green Version]
- Jung, I.L.; Kim, I.G. Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: Polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem. Biophys. Res. Commun. 2003, 301, 915–922. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Nesterova, L.Y. Polyamines as modulators of gene expression under oxidative stress in Escherichia coli. Biochemistry 2003, 68, 850–856. [Google Scholar]
- Oh, T.J.; Kim, I.G. The expression of Escherichia coli SOS genes recA and uvrA is inducible by polyamines. Biochem. Biophys. Res. Commun. 1999, 264, 584–589. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Tabor, H. Polyamines are critical for the induction of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2013, 288, 33559–33570. [Google Scholar] [CrossRef] [Green Version]
- Schneider, B.L.; Ruback, S.; Kiupakis, A.K.; Kasbarian, H.; Pybus, C.; Reitzer, L. The Escherichia coli gabDTPC operon: Specific γ-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 2002, 184, 6976–6986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nastri, H.G.; Algranati, I.D. Effect of polyamines on plasmid-mediated kanamycin resistance and kanamycin phosphotransferase gene expression in Escherichia coli. Cell. Mol. Biol. 1996, 42, 711–717. [Google Scholar] [PubMed]
- Kwon, D.H.; Lu, C.D. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2006, 50, 1615–1622. [Google Scholar] [CrossRef] [Green Version]
- Tkachenko, A.G.; Pozhidaeva, O.N.; Shumkov, M.S. Role of polyamines in formation of multiple antibiotic resistance of Escherichia coli under stress conditions. Biochemistry 2006, 71, 1042–1049. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Lee, E.; Dartois, V. Polyamines inhibit porin-mediated fluoroquinolone uptake in mycobacteria. PLoS ONE 2013, 8, e65806. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Bogaert, P.; van Hengel, J.; Guérin, C.J.; Berx, G.; Movahedi, K.; Van den Bergh, R.; Pereira-Fernandes, A.; Geuns, J.M.; Pircher, H.; et al. Alternatively activated macrophages engage in homotypic and heterotypic interactions through IL-4 and polyamine-induced E-cadherin/catenin complexes. Blood 2009, 114, 4664–4674. [Google Scholar] [CrossRef] [PubMed]
- Muraille, E.; Leo, O.; Moser, M. TH1/TH2 paradigm extended: Macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front. Immunol. 2014, 5, 603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hesse, M.; Modolell, M.; La Flamme, A.C.; Schito, M.; Fuentes, J.M.; Cheever, A.W.; Pearce, E.J.; Wynn, T.A. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: Granulomatous pathology is shaped by the pattern of L-arginine metabolism. J. Immunol. 2001, 167, 6533–6544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cano, V.; March, C.; Insua, J.L.; Aguiló, N.; Llobet, E.; Moranta, D.; Regueiro, V.; Brennan, G.P.; Millán-Lou, M.I.; Martín, C.; et al. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol. 2015, 17, 1537–1560. [Google Scholar]
- Monack, D.M.; Bouley, D.M.; Falkow, S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. Exp. Med. 2004, 199, 231–241. [Google Scholar] [CrossRef]
- Kerrinnes, T.; Winter, M.G.; Young, B.M.; Diaz-Ochoa, V.E.; Winter, S.E.; Tsolis, R.M. Utilization of host polyamines in alternatively activated macrophages promotes chronic infection by Brucella abortus. Infect. Immun. 2018, 86, e00458-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sycz, G.; Di Venanzio, G.; Distel, J.S.; Sartorio, M.G.; Le, N.H.; Scott, N.E.; Beatty, W.L.; Feldman, M.F. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLoS Pathog. 2021, 17, e1009802. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.K.; Thomas, S.M.; Olive, A.J.; Abramovitch, R.B. Macrophage Infection Models for Mycobacterium tuberculosis. In Methods in Molecular Biology; Clifton, N.J., Ed.; Humana Press: Totowa, NJ, USA, 2021; pp. 167–182. [Google Scholar]
- Gevrekci, A.Ö. The roles of polyamines in microorganisms. World. J. Microbiol. Biotechnol. 2017, 33, 204. [Google Scholar] [CrossRef] [PubMed]
- Hamana, K.; Tanaka, T.; Hosoya, R.; Niitsu, M.; Itoh, T. Cellular polyamines of the acidophilic, thermophilic and thermoacidophilic archaebacteria, Acidilobus, Ferroplasma, Pyrobaculum, Pyrococcus, Staphylothermus, Thermococcus, Thermodiscus and Vulcanisaeta. J. Gen. Appl. Microbiol. 2003, 49, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Nakada, Y.; Itoh, Y. Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2002, 184, 3377–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wargnies, B.; Lauwers, N.; Stalon, V. Structure and properties of the putrescine carbamoyltransferase of Streptococcus faecalis. Eur. J. Biochem. 1979, 101, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Potter, A.J.; Paton, J.C. Spermidine biosynthesis and transport modulate pneumococcal autolysis. J. Bacteriol. 2014, 196, 3556–3561. [Google Scholar] [CrossRef] [Green Version]
- Tomar, P.C.; Lakra, N.; Mishra, S.N. Cadaverine: A lysine catabolite involved in plant growth and development. Plant Signal. Behav. 2013, 8, e25850. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.; Wendisch, V.F. Biotechnological production of polyamines by Bacteria: Recent achievements and future perspectives. Appl. Microbiol. Biotechnol. 2011, 91, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Oda, S.; Kato, K.; Kim, H.G.; Koyanagi, T.; Kumagai, H.; Suzuki, H. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 2005, 280, 4602–4608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, S.; Tsuboi, Y.; Oda, S.; Kim, H.G.; Kumagai, H.; Suzuki, H. The putrescine importer PuuP of Escherichia coli K-12. J. Bacteriol. 2009, 191, 2776–2782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashiwagi, K.; Miyamoto, S.; Suzuki, F.; Kobayashi, H.; Igarashi, K. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 4529–4533. [Google Scholar] [CrossRef] [Green Version]
- Pistocchi, R.; Kashiwagi, K.; Miyamoto, S.; Nukui, E.; Sadakata, Y.; Kobayashi, H.; Igarashi, K. Characteristics of the operon for a putrescine transport system that maps at 19 min on the Escherichia coli chromosome. J. Biol. Chem. 1993, 268, 146–152. [Google Scholar] [CrossRef]
- Kurihara, S.; Suzuki, H.; Oshida, M.; Benno, Y. A novel putrescine importer required for type pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J. Biol. Chem. 2011, 286, 10185–10192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soksawatmaekhin, W.; Kuraishi, A.; Sakata, K.; Kashiwagi, K.; Igarashi, K. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 2004, 51, 1401–1412. [Google Scholar] [CrossRef]
- Furuchi, T.; Kashiwagi, K.; Kobayashi, H.; Igarashi, K. Characteristics of the gene for a spermidine and putrescine transport-system that maps at 15-min on the Escherichia coli chromosome. J. Biol. Chem. 1991, 266, 20928–20933. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. Polyamine transport in bacteria and yeast. Biochem. J. 1999, 344, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Ishigure, H.; Demizu, R.; Uemura, T.; Nishino, K.; Yamaguchi, A.; Kashiwagi, K.; Igarashi, K. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008, 190, 872–878. [Google Scholar] [CrossRef] [Green Version]
- Karatan, E.; Duncan, T.R.; Watnick, P.I. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 2005, 187, 7434–7443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGinnis, M.W.; Parker, Z.M.; Walter, N.E.; Rutkovsky, A.C.; Cartaya-Marin, C.; Karatan, E. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 2009, 299, 166–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zijnge, V.; Kieselbach, T.; Oscarsson, J. Proteomics of protein secretion by Aggregatibacter actinomycetemcomitans. PLoS ONE 2012, 7, e41662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, P.; Swiatlo, E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Sakai, Y.; Suzuki, H.; Muth, A.; Phanstiel, O., IV; Rather, P.N. Putrescine importer PlaP contributes to swarming motility and urothelial cell invasion in Proteus mirabilis. J. Biol. Chem. 2013, 288, 15668–15676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Maezato, Y.; Kim, S.H.; Kurihara, S.; Liang, J.; Michael, A.J. Polyamine-independent growth and biofilm formation, and functional spermidine/spermine N-acetyltransferases in Staphylococcus aureus and Enterococcus faecalis. Mol. Microbiol. 2019, 111, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; He, W.; Lu, C.-D. Functional characterization of seven Glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and Alanine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2011, 193, 3923–3930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Li, C.; Zhang, J.; Lu, C.-D. γ-glutamyl Spermine Synthetase PauA2 as a potential target of antibiotic development against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2012, 56, 5309–5314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, G.S.; Spontak, J.S.; Klapper, D.G.; Richardson, A.R. Arginine catabolic mobile element encoded speG abrogates the unique hyper-sensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 2011, 82, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, A.Q.; Schneider, J.; Wendisch, V.F. Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J. Biotechnol. 2015, 201, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Oda, S.; Kumagai, H.; Suzuki, H. γ-Glutamyl-γ-aminobutyrate hydrolase in the putrescine utilization pathway of Escherichia coli K-12. FEMS Microbiol. Lett. 2006, 256, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Oda, S.; Tsuboi, Y.; Kim, H.G.; Oshida, M.; Kumagai, H.; Suzuki, H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 2008, 283, 19981–19990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, B.L.; Reitzer, L. Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J. Bacteriol. 2012, 194, 4080–4088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, S.; Kato, K.; Asada, K.; Kumagai, H.; Suzuki, H. A putrescine-inducible pathway comprising PuuE-YneI in which γ-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 2010, 192, 4582–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsonova, N.N.; Smirnov, S.V.; Altman, I.B.; Ptitsyn, L.R. Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 2003, 3, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaibe, E.; Metzer, E.; Halpern, Y.S. Metabolic pathway for the utilization of Larginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J. Bacteriol. 1985, 163, 933–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaibe, E.; Metzer, E.; Halpern, Y.S. Control of utilization of L-arginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J. Bacteriol. 1985, 163, 938–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forouhar, F.; Lee, I.S.; Vujcic, J.; Vujcic, S.; Shen, J.; Vorobiev, S.M.; Xiao, R.; Acton, T.B.; Montelione, G.T.; Porter, C.W.; et al. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1- spermidine/spermine acetyltransferase. J. Biol. Chem. 2005, 280, 40328–40336. [Google Scholar] [CrossRef] [Green Version]
- Foster, A.; Barnes, N.; Speight, R.; Keane, M.A. Genomic organisation, activity and distribution analysis of the microbial putrescine oxidase degradation pathway. Syst. Appl. Microbiol. 2013, 36, 457–466. [Google Scholar] [CrossRef]
- Campilongo, R.; Di Martino, M.L.; Marcocci, L.; Pietrangeli, P.; Leuzzi, A.; Grossi, M.; Casalino, M.; Nicoletti, M.; Micheli, G.; Colonna, B.; et al. Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: Looking into the gap between commensal E. coli and harmful Shigella. PLoS ONE 2014, 9, e106589. [Google Scholar] [CrossRef]
- Fukuchi, J.; Kashiwagi, K.; Takio, K.; Igarashi, K. Properties and structure of spermidine acetyltransferase in Escherichia coli. J. Biol. Chem. 1994, 269, 22581–22585. [Google Scholar] [CrossRef]
- Bollinger, J.M., Jr.; Kwon, D.S.; Huisman, G.W.; Kolter, R.; Walsh, C.T. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J. Biol. Chem. 1995, 270, 14031–14041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, D.S.; Lin, C.H.; Chen, S.; Coward, J.K.; Walsh, C.T.; Bollinger, J.M., Jr. Dissection of glutathionylspermidine synthetase/amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J. Biol. Chem. 1997, 272, 2429–2436. [Google Scholar] [CrossRef] [Green Version]
- Cardona-Cardona, Y.V.; Regla, I.; Juárez-Díaz, J.A.; Carrillo-Campos, J.; López-Ortiz, M.; Aguilera-Cruz, A.; Mújica-Jiménez, C.; Muñoz-Clares, R.A. The critical role of the aldehyde dehydrogenase PauC in spermine, spermidine, and diaminopropane toxicity in Pseudomonas aeruginosa: Its possible use as a drug target. FEBS J. 2022, 289, 2685–2705. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.D.; Itoh, Y.; Nakada, Y.; Jiang, Y. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2002, 184, 3765–3773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, H.T.; Li, J.Y.; Peng, Y.C.; Lu, C.D. Molecular characterization of PauR and its role in control of putrescine and cadaverine catabolism through the γ-glutamylation pathway in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2013, 195, 3906–3913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Lu, C.-D. Characterization of Staphylococcus aureus responses to spermine stress. Curr. Microbiol. 2014, 69, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Che, S.; Liang, Y.; Chen, Y.; Wu, W.; Liu, R.; Zhang, Q.; Bartlam, M. Structure of Pseudomonas aeruginosa spermidine dehydrogenase: A polyamine oxidase with a novel heme-binding fold. FEBS J. 2022, 289, 1911–1928. [Google Scholar] [CrossRef] [PubMed]
- Kakegawa, T.; Guo, Y.; Chiba, Y.; Miyazaki, T.; Nakamura, M.; Hirose, S.; Canellakis, Z.N.; Igarashi, K. Effect of acetylpolyamines on in vitro protein synthesis and on the growth of a polyamine-requiring mutant of Escherichia coli. J. Biochem. 1991, 109, 627–631. [Google Scholar] [CrossRef]
- Bai, F.; Xu, H.; Zhang, Q.; Qi, X.; Mou, R.; Bai, G.; Qiao, M. Functional characterization of pfm in protein secretion and lung infection of Pseudomonas aeruginosa. Can. J. Microbiol. 2011, 57, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J.; Larussa, S.J.; Dana, A.; Smith, H.; Xu, A.; Ryan, C.; Uhlemann, A.C.; Boundy, S.; Goldberg, J.; Narechania, A.; et al. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 2013, 4, e00889-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmer, D.P.; Soupene, E.; Lee, H.L.; Wendisch, V.F.; Khodursky, A.B.; Peter, B.J.; Bender, R.A.; Kustu, S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 2000, 97, 14674–14679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciag, A.; Peano, C.; Pietrelli, A.; Egli, T.; De Bellis, G.; Landini, P. In vitro transcription profiling of the σ S subunit of bacterial RNA polymerase: Redefinition of the σ S regulon and identification of σ S-specific promoter sequence elements. Nucleic Acids Res. 2011, 39, 5338–5355. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Liu, M.; Burgess, R.R. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 2010, 38, 1273–1283. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, N.; Kurihara, S.; Kitahara, Y.; Asada, K.; Kato, K.; Suzuki, H. Mechanism for regulation of the putrescine utilization pathway by the transcription factor PuuR in Escherichia coli K-12. J. Bacteriol. 2012, 194, 3437–3447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, J.D.; Scott, C.; Tang, Y.; Poole, R.K.; Green, J. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 2006, 281, 27806–27815. [Google Scholar] [CrossRef] [Green Version]
- Randle, C.L.; Albro, P.W.; Dittmer, J.C. The phosphoglyceride composition of gram-negative bacteria and the changes in composition during growth. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1969, 187, 214–220. [Google Scholar] [CrossRef]
- White, D.A. The phospholipids composition of mammalian tissues. In Form and Function of Phospholipids, 2nd ed.; Ansel, G.B., Hawthorne, J.N., Dawson, R.M.C., Eds.; Elsevier: Amsterdam, The Netherlands, 1973; pp. 441–483. [Google Scholar]
- Knaak, J.B.; Leung, H.-W.; Stott, W.T.; Busch, J.; Bilsky, J. Toxicology of mono-, di-, and triethanolamine. Rev. Environ. Contam. Toxicol. 1997, 149, 1–86. [Google Scholar] [PubMed]
- Vance, J.E.; Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 2012, 1831, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Bingham, E.; Cohrssen, B.; Powell, C.H. Patty’s Toxicology, 5th ed.; John Wiley & Sons: New York, NY, USA, 2001; Volume 1–9, p. 782. [Google Scholar]
- Anderson, J.C.; Kendall, M.M. Location, location, location. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression. Microb. Cell 2016, 3, 89–91. [Google Scholar] [CrossRef]
- Kofoid, E.; Rappleye, C.; Stojiljkovic, I.; Roth, J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 1999, 181, 5317–5329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, T.J.; Ehrmann, M.; Boos, W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 1983, 258, 5428–5432. [Google Scholar] [CrossRef]
- Proulx, P.; Fung, C.K. Metabolism of phosphoglycerides in E. coli. IV. The positional specificity and properties of phospholipase A. Can. J. Biochem. 1969, 47, 1125–1128. [Google Scholar]
- Nandedkar, A.K. Report on the utilization of ethanolamine-1-14C by Mycobacterium 607. Biochem. Med. 1974, 11, 67–70. [Google Scholar] [CrossRef]
- Tsoy, O.; Ravcheev, D.; Mushegian, A. Comparative genomics of ethanolamine utilization. J. Bacteriol. 2009, 191, 7157–7164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garsin, D.A. Ethanolamine: A Signal to Commence a Host-Associated Lifestyle? mBio 2012, 1, e00066-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlt, J.A. Tools and strategies for discovering novel enzymes and metabolic pathways. Perspect. Sci. 2016, 9, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Korbel, J.O.; Doerks, T.; Jensen, L.J.; Perez-Iratxeta, C.; Kaczanowski, S.; Hooper, S.D.; Andrade, M.A.; Bork, P. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005, 3, e134. [Google Scholar] [CrossRef] [Green Version]
- Cronan, C.E., Jr.; Rock, C.O. Biosynthesis of membrane lipids. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Ed.; ASM Press: Washington, DC, USA, 1996; pp. 612–636. [Google Scholar]
- Gibellini, F.; Smith, T.K. The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. [Google Scholar] [CrossRef]
- Penrod, J.T.; Mace, C.C.; Roth, J.R. A pH-sensitive function and phenotype: Evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J. Bacteriol. 2004, 186, 6885–6890. [Google Scholar] [CrossRef] [Green Version]
- Stojiljkovic, I.; Baumler, A.J.; Heffron, F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the cchA, cchB, eutE, eutJ, eutG, eutH gene cluster. J. Bacteriol. 1995, 177, 1357–1366. [Google Scholar] [CrossRef] [Green Version]
- Garsin, D.A. Ethanolamine Utilization in Bacterial Pathogens: Roles and Regulation. Nat. Rev. Microbiol. 2010, 8, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, G.W.; Chang, J.T. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 1975, 254, 150–151. [Google Scholar] [CrossRef]
- Del Papa, M.F.; Perego, M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 2008, 190, 7147–7156. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, C.M.; Scarlett, F.A.; Turner, J.M. Ethanolamine catabolism by bacteria, including Escherichia coli. Biochem. Soc. Trans. 1976, 4, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, C.M.; Turner, J.M. Microbial metabolism of amino alcohols. Formation of coenzyme B12-dependent ethanolamine ammonia-lyase and its concerted induction in Escherichia coli. Biochem. J. 1978, 176, 751–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatri, N.; Khatri, I.; Subramanian, S.; Raychaudhuri, S. Ethanolamine utilization in Vibrio alginolyticus. Biol. Direct 2012, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman, D.S.; Wagner, M.J. Enzyme systems in the mycobacteria. XIII. Glycine dehydrogenase and the glyoxylic acid cycle. Biochim. Biophys. Acta 1962, 65, 297–306. [Google Scholar] [CrossRef]
- Shukla, S.D.; Turner, J.M. Biosynthetic utilization of ethanolamine for lipid synthesis by bacteria. Biochem. J. 1980, 186, 13–19. [Google Scholar] [CrossRef] [Green Version]
- San Francisco, B.; Zhang, X.; Whalen, K.; Gerlt, K. A novel pathway for bacterial ethanolamine metabolism. FASEB J. 2015, 29, 573.45. [Google Scholar] [CrossRef]
- Roof, D.M.; Roth, J.R. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 1988, 170, 3855–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheppard, D.E.; Penrod, J.T.; Bobik, T.; Kofoid, E.; Roth, J.R. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 2004, 186, 7635–7644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaval, K.G.; Garsin, D.A. Ethanolamine Utilization in Bacteria. mBio 2018, 3, e00172-12. [Google Scholar] [CrossRef] [Green Version]
- Yeates, T.O.; Kerfeld, C.A.; Heinhorst, S.; Cannon, G.C.; Shively, J.M. Proteinbased organelles in bacteria: Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 2008, 6, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Brinsmade, S.R.; Escalante-Semerena, J.C. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 2004, 186, 1890–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penrod, J.T.; Roth, J.R. Conserving a volatile metabolite: A role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 2006, 188, 2865–2874. [Google Scholar] [CrossRef] [Green Version]
- Bradbeer, C. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J. Biol. Chem. 1965, 240, 4675–4681. [Google Scholar] [CrossRef]
- Bologna, F.P.; Campos-Bermudez, V.A.; Saavedra, D.D.; Andreo, C.S.; Drincovich, M.F. Characterization of Escherichia coli EutD: A phosphotransacetylase of the ethanolamine operon. J. Microbiol. 2010, 48, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.W.; Turner, J.M. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J. Gen. Microbiol. 1984, 130, 299–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinsmade, S.R.; Paldon, T.; Escalante-Semerena, J.C. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 2005, 187, 8039–8046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starai, V.J.; Garrity, J.; Escalante-Semerena, J.C. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology 2005, 151, 3793–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barisch, C.; Soldati, T. Mycobacterium marinum Degrades Both Triacylglycerols and Phospholipids from Its Dictyostelium Host to Synthesise Its Own Triacylglycerols and Generate Lipid Inclusions. PLoS Pathog. 2017, 13, e1006095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrouy-Maumus, G.; Biswas, T.; Hunt, D.M.; Kelly, G.; Tsodikov, O.V.; Sório de Carvalho, L.P. Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2013, 110, 11320–11325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandedkar, A.K.N. Biosynthesis of phosphatidyl ethanolamine in Mycobacterium 607. Biochem. Med. 1975, 12, 116–122. [Google Scholar] [CrossRef]
- Kaval, K.G.; Gebbie, M.; Goodson, J.R.; Cruz, M.R.; Winkler, W.C.; Garsin, D.A. Ethanolamine Utilization and Bacterial Microcompartment Formation Are Subject to Carbon Catabolite Repression. J. Bacteriol. 2019, 201, e00703-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roof, D.M.; Roth, J.R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 1992, 174, 6634–6643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luzader, D.H.; Clark, D.E.; Gonyar, L.A.; Kendall, M.M. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2013, 195, 4947–4953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, K.A.; Ramesh, A.; Stearns, J.E.; Bourgogne, A.; Reyes-Jara, A.; Winkler, W.C.; Garsin, D.A. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 2009, 106, 4435–4440. [Google Scholar] [CrossRef] [Green Version]
- Mowbray, S.L.; Kathiravan, M.K.; Pandey, A.A.; Odell, L.R. Inhibition of glutamine synthetase: A potential drug target in Mycobacterium tuberculosis. Molecules 2014, 19, 13161–13176. [Google Scholar] [CrossRef] [Green Version]
- Gallant, J.L.; Viljoen, A.J.; van Helden, P.D.; Wiid, I.J. Glutamate Dehydrogenase Is Required by Mycobacterium bovis BCG for Resistance to Cellular Stress. PLoS ONE 2016, 11, e0147706. [Google Scholar] [CrossRef] [PubMed]
- Somani, R.R.; Rai, P.R.; Kandpile, P.S. Ornithine Decarboxylase Inhibition: A Strategy to Combat Various Diseases. Mini Rev. Med. Chem. 2018, 18, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Carter, N.S.; Kawasaki, Y.; Nahata, S.S.; Elikaee, S.; Rajab, S.; Salam, L.; Alabdulal, M.Y.; Broessel, K.K.; Foroghi, F.; Abbas, A.; et al. Polyamine Metabolism in Leishmania Parasites: A Promising Therapeutic Target. Med. Sci. 2022, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Shack, L.A.; Lee, J.H.; Lim, J.; Eoh, H.; Swiatlo, E.; Phanstiel, O., IV; Nanduri, B. Difluoromethylornithine (DFMO) and AMXT 1501 inhibit capsule biosynthesis in pneumococci. Sci. Rep. 2022, 12, 11804. [Google Scholar] [CrossRef] [PubMed]
- Reigada, C.; Sayé, M.; Phanstiel, O., IV; Valera-Vera, E.; Miranda, M.R.; Pereira, C.A. Identification of Trypanosoma cruzi Polyamine Transport Inhibitors by Computational Drug Repurposing. Front. Med. (Lausanne) 2019, 6, 256. [Google Scholar] [CrossRef] [Green Version]
- Shah, P.; Nanduri, B.; Swiatlo, E.; Ma, Y.; Pendarvis, K. Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology 2011, 157, 504–515. [Google Scholar] [CrossRef] [Green Version]
- Jelsbak, L.; Thomsen, L.E.; Wallrodt, I.; Jensen, P.R.; Olsen, J.E. Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS ONE 2012, 7, e36149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rifkin, M.R.; Strobos, C.A.; Fairlamb, A.H. Specificity of ethanolamine transport and its further metabolism in Trypanosoma brucei. J. Biol. Chem. 1995, 270, 16160–16166. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.; Ullman, B. Parasite Polyamines as Pharmaceutical Targets. Curr. Pharm. Des. 2017, 23, 3325–3341. [Google Scholar] [CrossRef]
- Mann, S.; Frasca, K.; Scherrer, S.; Henao-Martínez, A.F.; Newman, S.; Ramanan, P.; Suarez, J.A. A Review of Leishmaniasis: Current Knowledge and Future Directions. Curr. Trop. Med. Rep. 2021, 8, 121–132. [Google Scholar] [CrossRef]
- Thiriot, J.D.; Martinez-Martinez, Y.B.; Endsley, J.J.; Torres, A.G. Hacking the host: Exploitation of macrophage polarization by intracellular bacterial pathogens. Pathog. Dis. 2020, 78, ftaa009. [Google Scholar] [CrossRef] [Green Version]
- Perdeh, J.; Berioso, B.; Love, Q.; LoGiudice, N.; Le, T.L.; Harrelson, J.P.; Roberts, S.C. Critical functions of the polyamine putrescine for proliferation and viability of Leishmania donovani parasites. Amino Acids 2020, 52, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boitz, J.M.; Yates, P.A.; Kline, C.; Gaur, U.; Wilson, M.E.; Ullman, B.; Roberts, S.C. Leishmania donovani ornithine decarboxylase is indispensable for parasite survival in the mammalian host. Infect. Immun. 2009, 77, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Samal, K.; Zhao, P.; Kendzicky, A.; Yco, L.P.; McClung, H.; Gerner, E.; Burns, M.; Bachmann, A.S.; Sholler, G. AMXT-1501, a novel polyamine transport inhibitor, synergizes with DFMO in inhibiting neuroblastoma cell proliferation by targeting both ornithine decarboxylase and polyamine transport. Int. J. Cancer 2013, 133, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Muth, A.; Kamel, J.; Kaur, N.; Shicora, A.C.; Ayene, I.S.; Gilmour, S.K.; Phanstiel, O., IV. Development of polyamine transport ligands with improved metabolic stability and selectivity against specific human cancers. J. Med. Chem. 2013, 56, 5819–5828. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Yamada, C.; Kato, K. Gamma-glutamyl compounds and their enzymatic production using bacterial gamma-glutamyltranspeptidase. Amino Acids 2007, 32, 333–340. [Google Scholar] [CrossRef]
- Kruh, N.A.; Troudt, J.; Izzo, A.; Prenni, J.; Dobos, K.M. Portrait of a pathogen: The Mycobacterium tuberculosis proteome in vivo. PLoS ONE 2010, 5, e13938. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Annotated Function | Homologue in E. coli | Homologue in P. aeruginosa | Homologue in S. coelicolor |
|---|---|---|---|
| Polyamine ABC transporter ATP-binding protein PotA-like | PotA (b1126)/YdcT (b1441) | PAO603/PAO326 | SCO3453 |
| Polyamine ABC transporter ATP-binding protein PotC-like | PotC (b1124)/YdcV (b1443) | PAO324/PotC (PA3609) | SCO3454 |
| Polyamine ABC transporter protein | PotB (b1125)/YdcU (b1442) | PotB (PA0205)/PA3252 | SCO3455 |
| Polyamine ABC transporter protein—substrate binding protein | YnjB (b1754) | PA0203 | SCO3456 |
| Amino acid/polyamine permease | PuuP (b1296)/PlaP (b2014) | PA5510 | SCO5057 |
| Lysine/ornithine decarboxylase-like enzyme | - | - | SCO5651 |
| Pyruvate-polyamine aminotransferase | PatA (b3073) | SpuC (PA0299) | SCO5655 |
| Lrp/AsnC family transcriptional regulator | - | - | SCO5656 |
| γ-aminobutyraldehyde or γ-glutamyl-γ-amino-butyraldehyde dehydrogenase | PatD (b1444)/PuuC (b1300) | BetB (PA5373)/PAO219 | SCO5657 |
| Polyamine-binding lipoprotein | PotF (b0854) | SpuD (PA0300) | SCO5658 |
| γ-aminobutyraldehyde dehydrogenase or 4-guanidino-butyraldehyde dehydrogenase | PatD (b1444) PuuC (b1300) | PauC/KauB (PA5312) | SCO5666 |
| Polyamine ABC transporter substrate-binding protein | PotF (b0854) | SpuE (PA0301) | SCO5667 |
| Polyamine ABC transporter substrate-binding protein | PotG (b0855) | SpuF (PA0302) | SCO5668 |
| Polyamine ABC-transporter integral membrane protein | PotH (b0856) | SpuG (PA0303) | SCO5669 |
| Polyamine ABC-transporter integral membrane protein | PotI (b0857) | SpuH (PA0304) | SCO5670 |
| γ-glutamyl-polyamine oxidoreductase | PuuB (b1301) | PauB3 (PA2776) | SCO5671 |
| γ-aminobutyrate aminotransferase gabT-like or puuE-like | GabT (b2662)/PuuE (b1302) | GabT (PA266) | SCO5676 |
| Succinate-semialdehyde dehydrogenase gabD-like | GabD (b2661) | GabD (PA0265) | SCO5679 |
| Amino acids/polyamine permease | PuuP (b1296) | PA5510/PAO322 | SCO5977 |
| Hydrolase | - | - | SCO6960 |
| Amidohydrolase | - | - | SCO6961 |
| γ-glutamyl-polyamine synthetase | PuuA (b1297) | PauA7 (PA5508)/SpuI (PA0296) | SCO6962 |
| Annotated Function | Orthologues in C. salexigens (Protein Family) | Orthologues in S. coelicolor |
|---|---|---|
| γ-glutamyl ethanolamine synthetase/ethanolamine γ-glutamylase | PF00120 | SCO1613 |
| γ-glutamyl ethanolamine dehydrogenase/iron-dependent dehydrogenase | PF00465 | SCO1611 |
| γ-glutamyl aldehyde dehydrogenase | PF00171 | SCO1612 |
| γ-glutamyl glycine amidohydrolase /formylglutamate amidohydrolase | PF05013 | SCO1615 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krysenko, S.; Wohlleben, W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Med. Sci. 2022, 10, 40. https://doi.org/10.3390/medsci10030040
Krysenko S, Wohlleben W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Medical Sciences. 2022; 10(3):40. https://doi.org/10.3390/medsci10030040
Chicago/Turabian StyleKrysenko, Sergii, and Wolfgang Wohlleben. 2022. "Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity" Medical Sciences 10, no. 3: 40. https://doi.org/10.3390/medsci10030040






