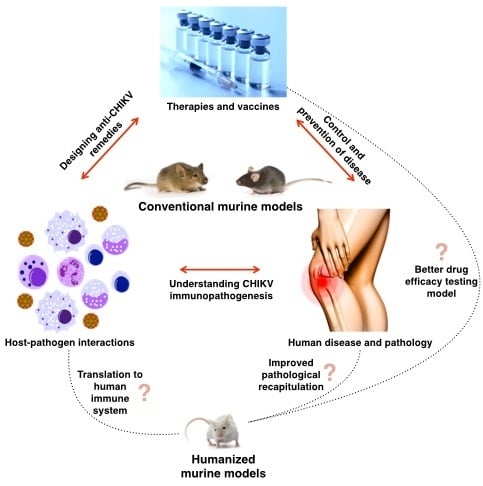
Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis
Abstract
:1. Introduction
1.1. Epidemiology and Global Expansion
1.2. Challenges in Investigating CHIKV Immunopathophysiology
2. Current CHIKV Research on Available Mouse Models
| Type of Mice | Mode of Infection | Outcome Indicators | References |
|---|---|---|---|
| A129, AG129 mice | Intraperitoneal (i.p.) | Weight loss, mortality, viremia, histological damages, antibody titer | Partidos et al. [33] |
| WT C57BL/6 mice | Ventral side of footpad (subcutaneous (s.c.)) | Footpad inflammation, viremia, histological damages | Gardner et al. [34], Morrison et al. [35] |
| Neonatal Swiss albino mice | Intracerebral | Mortality, brain histology | Ross [36], Suckling et al. [37] |
| BALB/c mice | Intranasal infection | Immunohistochemistry | Powers et al. [16] |
| C57BL/6J or NIH Swiss mice | Intranasal infection | Viremia, histological damages | Wang et al. [38] |
| Newborn/neonatal of ICR and CD1 mice | s.c. at the back | Mortality, viral load in organs, histological damage | Ziegler et al. [39] |
| Weaning CD1 mice | s.c. at the rear footpad | In vivo bioluminescence imaging of infectious clone | Ziegler et al. [40] |
| CCR2−/− C57BL/6 mice | s.c. at the rear footpad | Footpad inflammation, viremia, viral load of organs, immunohistochemistry, histological damage | Poo et al. [41] |
| Wild-type, IFNAR−/−, ISG15−/− and UbE1L−/− C57BL/6 mice | s.c. at right flank | Mortality, cytokine analysis, viral load of organs | Werneke et al. [42] |
| IFN-α/βR−/− and IFN-α/βR+/− outbred OF1 or C57BL/6 mice | Intradermal | Mortality, viral load of organs, histological damage | Couderc et al. [43] |
| IFN-α/βR−/−, Cardif−/−, RIG-1−/−, Mda5−/−, Myd88−/−, TLR3−/− outbred OF1 or C57BL/6 mice | Intradermal | Mortality, immunohistochemistry, viral load of organs | Schilte et al. [44] |
| CD4−/−, μMT, IFNγ−/− C57/BL6 mice | Ventral side of footpad (s.c.) | Viremia, footpad inflammation, antibody response | Lum et al. [45] |
| CD4−/−, CD8−/−, IFNγ−/− C57/BL6 mice | Ventral side of footpad (s.c.) | Viremia, footpad inflammation, histological damage, in vivo imaging | Teo et al. [46] |
| Tlr3−/− C57/BL6 mice | Ventral side of footpad (s.c.) | Viremia, footpad inflammation, histological damage, in vivo imaging, immunohistochemistry, cytokine analysis, antibody response | Her et al. [47] |
| IRF3−/−, IRF7−/−, IRF3/7−/− DKO C57BL/6 mice | Intradermal | Mortality, viremia, viral load of organs, IFN expression analysis | Schilte et al. [48] |
| Rsad2−/− C57BL/6 mice | Ventral side of footpad (s.c) | Viremia, footpad inflammation, histological damage, immunohistochemistry, gene expression analysis | Teng et al. [49] |
| DEREG with IL-2 Ab Cx treatment C57BL/6 mice | Ventral side of footpad | Viremia, footpad inflammation, histological damage, lymphocyte profiling | Lee et al. [50] |
| Neonatal CD1 mice | Intradermal ear injection or mosquito inoculation | Cytokines analysis | Thangamani et al. [51] |
| Newborn Swiss albino mice | s.c. at the back | Proteome analysis | Dhanwani et al. [52] |
2.1. Study of CHIKV Infection and Pathology
2.1.1. C57BL/6 Mice
2.1.2. Roles of IFN-α/β and Other Immune Mediators during CHIKV Infection
2.1.3. Roles of Adaptive Immune Responses against CHIKV Infection
2.2. Inadequacy of the Current Mouse Models
3. Humanized Mice as Another Alternative Model
3.1. Current Research Using Humanized Mice
3.2. Current Challenges and Limitations of Humanized Mouse Models
3.2.1. Boosting Cytokines to Improve Immune System Responses
3.2.2. Human HLA Transgenic Mice and T Cell Education
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lumsden, W.H. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–1953. II. General description and epidemiology. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 33–57. [Google Scholar]
- Robinson, M. An Epidemic of Virus Disease in Southern Province, Tanganyika Territory, in 1952–1953. I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [PubMed]
- Kam, Y.W.; Ong, E.K.S.; Rénia, L.; Tong, J.C.; Ng, L.F.P. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect. 2009, 11, 1186–1196. [Google Scholar] [PubMed]
- Staples, J.E.; Breiman, R.F.; Powers, A.M. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 2009, 49, 942–948. [Google Scholar] [PubMed]
- Dubrulle, M.; Mousson, L.; Moutailler, S.; Vazeille, M.; Failloux, A.-B. Chikungunya virus and Aedes mosquitoes: Saliva is infectious as soon as two days after oral infection. PLoS ONE 2009, 4, e5895. [Google Scholar] [PubMed]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [PubMed]
- Vazeille, M.; Moutailler, S.; Coudrier, D.; Rousseaux, C.; Khun, H.; Huerre, M.; Thiria, J.; Dehecq, J.S.; Fontenille, D.; Schuffenecker, I.; et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS ONE 2007, 2, e1168. [Google Scholar] [PubMed]
- Tsetsarkin, K.A.; Chen, R.; Yun, R.; Rossi, S.L.; Plante, K.S.; Guerbois, M.; Forrester, N.; Perng, G.C.; Sreekumar, E.; Leal, G.; et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat. Commun. 2014, 5, 4084. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, O.; Albert, M.L. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Bodenmann, P.; Genton, B. Chikungunya: An epidemic in real time. Lancet 2006, 368, 258. [Google Scholar] [CrossRef]
- Pialoux, G.; Gaüzère, B.; Jauréguiberry, S.; Strobel, M. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. [Google Scholar] [CrossRef]
- Her, Z.; Malleret, B.; Chan, M.; Ong, E.K.S.; Wong, S.-C.; Kwek, D.J.C.; Tolou, H.; Lin, R.T.P.; Tambyah, P.A.; Rénia, L.; et al. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J. Immunol. 2010, 184, 5903–5913. [Google Scholar] [CrossRef] [PubMed]
- Kou, Z.; Quinn, M.; Chen, H.; Rodrigo, W.W.; Rose, R.C.; Schlesinger, J.J.; Jin, X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 2008, 80, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Simmons, J.D.; Heise, M.T. Complement receptor 3 promotes severe ross river virus-induced disease. J. Virol. 2008, 82, 11263–11272. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.; Brault, A.; Tesh, R.B.; Weaver, S.C. Re-emergence of chikungunya and o’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar] [PubMed]
- Powers, A.M.; Logue, C.H. Changing patterns of chikungunya virus: Re-emergence of a zoonotic arbovirus. J. Gen. Virol. 2007, 88, 2363–2377. [Google Scholar] [CrossRef] [PubMed]
- Sergon, K.; Njuguna, C.; Kalani, R.; Ofula, V.; Onyango, C.; Konongoi, L.S.; Bedno, S.; Burke, H.; Dumilla, A.M.; Konde, J.; et al. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am. Soc. Trop. Med. Hyg. 2008, 78, 333–337. [Google Scholar]
- Khan, K.; Bogoch, I.; Brownstein, J.; Miniota, J.; Nicolucci, A.; Hu, W.; Nsoesie, E.O.; Cetron, M.; Creatore, M.I.; German, M.; et al. Assessing the origin of and potential for international spread of Chikungunya Virus from the Caribbean. PLoS Curr. Outbreaks 2014, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Reiter, P.; Fontenille, D.; Paupy, C. Aedes albopictus as an epidemic vector of chikungunya virus: Another emerging problem? Lancet Infect. Dis. 2006, 6, 463–464. [Google Scholar] [CrossRef]
- Mavalankar, D.; Shastri, P.; Bandyopadhyay, T.; Parmar, J.; Ramani, K.V. Increased Mortality Rate Associated with Chikungunya Epidemic, Ahmedabad, India. Emerg. Infect. Dis. 2008, 14, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.C.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; Silvi, G.; et al. Infection with chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; le Roux, K.; Prevost, M.-C.; Fsihi, H.; et al. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007, 3, e89. [Google Scholar] [CrossRef] [PubMed]
- Her, Z.; Kam, Y.W.; Lin, R.T.P.; Ng, L.F.P. Chikungunya: A bending reality. Microbes Infect. 2009, 11, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- AbuBakar, S.; Sam, I.C.; Wong, P.F.; MatRahim, N.; Hooi, P.S.; Roslan, N. Reemergence of endemic Chikungunya, Malaysia. Emerg. Infect. Dis. 2007, 13, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Wanlapakorn, N.; Thongmee, T.; Linsuwanon, P.; Chattakul, P.; Vongpunsawad, S.; Payungporn, S.; Poovorawan, Y. Chikungunya outbreak in Bueng Kan province, Thailand, 2013. Emerg. Infect. Dis. 2014, 20, 1404–1406. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, K.D.; Suarez, L.-A.C.; Labayo, H.K.M.; Liles, V.R.; Salvoza, N.C.; Klinzing, D.C.; Daroy, M.L.G.; Matias, R.R.; Natividad, F.F. Complete genome sequence of chikungunya virus isolated in the Philippines. Genome Announc. 2014, 2, e00336-14. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, U.; Nelson, M.; Su, Y.-C.; Mahalingam, S. Mechanisms of Chikungunya virus disease informed by Ross River virus research. Future Virol. 2008, 3, 509–511. [Google Scholar] [CrossRef]
- Devoy, A.; Bunton-Stasyshyn, R.K.A.; Tybulewicz, V.L.J.; Smith, A.J.H.; Fisher, E.M.C. Genomically humanized mice: Technologies and promises. Nat. Rev. Genet. 2012, 13, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C.W. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.D.; Wainszelbaum, M.J. Human-specific genes may offer a unique window into human cell signaling. Sci. Signal. 2009, 2, e59. [Google Scholar] [CrossRef] [PubMed]
- Emes, R.D. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 2003, 12, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Xu, T. The expanding role of mouse genetics for understanding human biology and disease. Dis. Model. Mech. 2008, 1, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Partidos, C.D.; Weger, J.; Brewoo, J.; Seymour, R.; Borland, E.M.; Ledermann, J.P.; Powers, A.M.; Weaver, S.C.; Stinchcomb, D.T.; Osorio, J.E. Probing the attenuation and protective efficacy of a candidate chikungunya virus vaccine in mice with compromised interferon (IFN) signaling. Vaccine 2011, 29, 3067–3073. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.; Anraku, I.; Le, T.T.; Larcher, T.; Major, L.; Roques, P.; Schroder, W.A.; Higgs, S.; Suhrbier, A. Chikungunya virus arthritis in adult wild-type mice. J. Virol. 2010, 84, 8021–8032. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Oko, L.; Montgomery, S.A.; Whitmore, A.C.; Lotstein, A.R.; Gunn, B.M.; Elmore, S.A.; Heise, M.T. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: Evidence of arthritis, tenosynovitis, myositis, and persistence. Am. J. Pathol. 2011, 178, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.W. The Newala epidemic. III. The virus: Isolation, pathogenic properties and relationship to the epidemic. J. Hyg. (Lond). 1956, 54, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Suckling, A.J.; Jagelman, S.; Webb, H.E. A comparison of brain lysosomal enzyme activities in four experimental togavirus encephalitides. J. Neurol. Sci. 1978, 35, 355–364. [Google Scholar] [CrossRef]
- Wang, E.; Volkova, E.; Adams, A.P.; Forrester, N.; Xiao, S.Y.; Frolov, I.; Weaver, S.C. Chimeric alphavirus vaccine candidates for chikungunya. Vaccine 2008, 26, 5030–5039. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.A.; Lu, L.; da Rosa, A.P.; Xiao, S.-Y.; Tesh, R.B. An animal model for studying the pathogenesis of chikungunya virus infection. Am. J. Trop. Med. Hyg. 2008, 79, 133–139. [Google Scholar] [PubMed]
- Ziegler, S.A.; Nuckols, J.; McGee, C.E.; Huang, Y.-J.S.; Vanlandingham, D.L.; Tesh, R.B.; Higgs, S. In vivo imaging of chikungunya virus in mice and Aedes mosquitoes using a Renilla luciferase clone. Vector Borne Zoonotic Dis. 2011, 11, 1471–1477. [Google Scholar] [CrossRef] [PubMed]
- Poo, Y.S.; Nakaya, H.; Gardner, J.; Larcher, T.; Schroder, W.A.; Le, T.T.; Major, L.D.; Suhrbier, A. CCR2 Deficiency Promotes Exacerbated Chronic Erosive Neutrophil-Dominated Chikungunya Virus Arthritis. J. Virol. 2014, 88, 6862–6872. [Google Scholar] [CrossRef] [PubMed]
- Werneke, S.W.; Schilte, C.; Rohatgi, A.; Monte, K.J.; Michault, A.; Arenzana-Seisdedos, F.; Vanlandingham, D.L.; Higgs, S.; Fontanet, A.; Albert, M.L.; et al. ISG15 is critical in the control of chikungunya virus infection independent of UbE1l mediated conjugation. PLoS Pathog. 2011, 7, e1002322. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Chrétien, F.; Schilte, C.; Disson, O.; Brigitte, M.; Guivel-Benhassine, F.; Touret, Y.; Barau, G.; Cayet, N.; Schuffenecker, I.; et al. A mouse model for Chikungunya: Young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008, 4, e29. [Google Scholar] [CrossRef] [PubMed]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lum, F.M.; Teo, T.H.; Lee, W.W.L.; Kam, Y.W.; Rénia, L.; Ng, L.F.P. An essential role of antibodies in the control of Chikungunya virus infection. J. Immunol. 2013, 190, 6295–6302. [Google Scholar] [CrossRef] [PubMed]
- Teo, T.H.; Lum, F.M.; Claser, C.; Lulla, V.; Lulla, A.; Merits, A.; Rénia, L.; Ng, L.F.P. A pathogenic role for CD4+ T cells during Chikungunya virus infection in mice. J. Immunol. 2013, 190, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Her, Z.; Teng, T.; Tan, J.J.L.; Teo, T.; Kam, Y.; Lum, F.; Lee, W.W.L.; Gabriel, C.; Melchiotti, R.; Andiappan, A.K.; et al. Loss of TLR 3 aggravates CHIKV replication and pathology due to an altered virus-specific neutralizing antibody response. EMBO Mol. Med. 2015, 7, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Schilte, C.; Buckwalter, M.R.; Laird, M.E.; Diamond, M.S.; Schwartz, O.; Albert, M.L. Cutting edge: Independent roles for IRF-3 and IRF-7 in hematopoietic and nonhematopoietic cells during host response to Chikungunya infection. J. Immunol. 2012, 188, 2967–2971. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.; Foo, S.; Simamarta, D.; Lum, F.M.; Teo, T.H.; Lulla, A.; Yeo, N.K.; Koh, E.G.; Chow, A.; Leo, Y.S. Viperin restricts chikungunya virus replication and pathology. J. Clin. Investig. 2012, 122, 4447–4460. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.W.L.; Teo, T.-H.; Her, Z.; Lum, F.-M.; Kam, Y.-W.; Haase, D.; Rénia, L.; Rötzschke, O.; Ng, L.F.P. Expanding regulatory T cells alleviates chikungunya virus-induced pathology in mice. J. Virol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Higgs, S.; Ziegler, S.; Vanlandingham, D.; Tesh, R.; Wikel, S. Host immune response to mosquito-transmitted chikungunya virus differs from that elicited by needle inoculated virus. PLoS ONE 2010, 5, e12137. [Google Scholar] [CrossRef] [PubMed]
- Dhanwani, R.; Khan, M.; Alam, S.I.; Rao, P.V.L.; Parida, M. Differential proteome analysis of Chikungunya virus-infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics 2011, 11, 1936–1951. [Google Scholar] [CrossRef] [PubMed]
- Teo, T.-H.; Lum, F.-M.; Lee, W.W.L.; Ng, L.F.P. Mouse models for Chikungunya virus: Deciphering immune mechanisms responsible for disease and pathology. Immunol. Res. 2012, 53, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.F.P.; Chow, A.; Sun, Y.J.; Kwek, D.J.C.; Lim, P.L.; Dimatatac, F.; Ng, L.C.; Ooi, E.E.; Chao, K.H.; Her, Z.; et al. IL-1β, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS ONE 2009, 4, e4261. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Her, Z.; Ong, E.K.S.; Chen, J.; Dimatatac, F.; Kwek, D.J.C.; Barkham, T.; Yang, H.; Rénia, L.; Leo, Y.-S.; et al. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J. Infect. Dis. 2011, 203, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, J.-J.; Jaffar Bandjee, M.-C.; Krejbich Trotot, P.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef] [PubMed]
- Chirathaworn, C.; Rianthavorn, P.; Wuttirattanakowit, N.; Poovorawan, Y. Serum IL-18 and IL-18BP levels in patients with Chikungunya virus infection. Viral Immunol. 2010, 23, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Chaaitanya, I.K.; Muruganandam, N.; Sundaram, S.G.; Kawalekar, O.; Sugunan, A.P.; Manimunda, S.P.; Ghosal, S.R.; Muthumani, K.; Vijayachari, P. Role of Proinflammatory Cytokines and Chemokines in Chronic Arthropathy in CHIKV Infection. Viral Immunol. 2011, 24, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Labadie, K.; Larcher, T.; Joubert, C.; Mannioui, A.; Delache, B.; Brochard, P.; Guigand, L.; Dubreil, L.; Lebon, P.; Verrier, B.; et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J. Clin. Investig. 2010, 120, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Scholte, F.E.M.; Chiang, C.; Albulescu, I.C.; Nichols, C.; He, Z.; Lin, R.; Snijder, E.J.; van Hemert, M.J.; Hiscott, J. Inhibition of dengue and chikungunya virus infection by RIG-I-mediated type I IFN-independent stimulation of the innate antiviral response. J. Virol. 2014, 88, 4180–4194. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Lecuit, M. Focus on Chikungunya pathophysiology in human and animal models. Microbes Infect. 2009, 11, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Hawman, D.W.; Stoermer, K.A.; Montgomery, S.A.; Pal, P.; Oko, L.; Diamond, M.S.; Morrison, T.E. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J. Virol. 2013, 87, 13878–13888. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.S.; Soong, L.; Girard, Y.A.; Campbell, G.; Mason, P.; Higgs, S. Potentiation of West Nile encephalitis by mosquito feeding. Viral Immunol. 2006, 19, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.F.; Higgs, S.; Beaty, B.J. Mosquito feeding-induced enhancement of Cache Valley Virus (Bunyaviridae) infection in mice. J. Med. Entomol. 1998, 35, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Akkina, R. New generation humanized mice for virus research: Comparative aspects and future prospects. Virology 2013, 435, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Drake, A.; Chen, Q.; Chen, J. Engineering humanized mice for improved hematopoietic reconstitution. Cell. Mol. Immunol. 2012, 9, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007, 7, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Hiramatsu, H.; Kobayashi, K.; Suzue, K.; Kawahata, M.; Hioki, K.; Ueyama, Y.; Koyanagi, Y.; Sugamura, K.; Tsuji, K.; et al. NOD/SCID/γ mouse: An excellent recipient mouse model for engraftment of human cells. Blood 2002, 100, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Takahashi, T.; Katano, I.; Ito, M. Current advances in humanized mouse models. Cell. Mol. Immunol. 2012, 9, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Denton, P.W.; Garcia, J.V. Humanized mouse models of HIV infection. AIDS Rev. 2011, 13, 135–148. [Google Scholar] [PubMed]
- Nischang, M.; Gers-Huber, G.; Audigé, A.; Akkina, R.; Speck, R.F. Modeling HIV infection and therapies in humanized mice. Swiss Med. Wkly. 2012, 142, w13618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berges, B.K.; Rowan, M.R. The utility of the new generation of humanized mice to study HIV-1 infection: Transmission, prevention, pathogenesis, and treatment. Retrovirology 2011, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Islas-Ohlmayer, M.; Padgett-Thomas, A.; Domiati-Saad, R.; Melkus, M.W.; Cravens, P.D.; Martin Mdel, P.; Netto, G.; Garcia, J.V. Experimental infection of NOD/SCID mice reconstituted with human CD34+ cells with Epstein-Barr virus. J. Virol. 2004, 78, 13891–13900. [Google Scholar] [CrossRef] [PubMed]
- Yajima, M.; Imadome, K.-I.; Nakagawa, A.; Watanabe, S.; Terashima, K.; Nakamura, H.; Ito, M.; Shimizu, N.; Yamamoto, N.; Fujiwara, S. T cell-mediated control of Epstein-Barr virus infection in humanized mice. J. Infect. Dis. 2009, 200, 1611–1615. [Google Scholar] [CrossRef] [PubMed]
- Rämer, P.C.; Chijioke, O.; Meixlsperger, S.; Leung, C.S.; Münz, C. Mice with human immune system components as in vivo models for infections with human pathogens. Immunol. Cell Biol. 2011, 89, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Ploss, A.; Evans, M.J.; Gaysinskaya, V.A.; Panis, M.; You, H.; de Jong, Y.P.; Rice, C.M. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 2009, 457, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Flint, M.; von Hahn, T.; Zhang, J.; Farquhar, M.; Jones, C.T.; Balfe, P.; Rice, C.M.; McKeating, J.A. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 2006, 80, 11331–11342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Randall, G.; Higginbottom, A.; Monk, P.; Rice, C.M.; McKeating, J.A. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004, 78, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Dorner, M.; Horwitz, J.A.; Donovan, B.M.; Labitt, R.N.; Budell, W.C.; Friling, T.; Vogt, A.; Catanese, M.T.; Satoh, T.; Kawai, T.; et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature 2013, 501, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Washburn, M.L.; Bility, M.T.; Zhang, L.; Kovalev, G.I.; Buntzman, A.; Frelinger, J.A.; Barry, W.; Ploss, A.; Rice, C.M.; Su, L. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 2011, 140, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Brehm, M.A.; Bavari, S.; Greiner, D.L. Humanized mice as a preclinical tool for infectious disease and biomedical research. Ann. N. Y. Acad. Sci. 2011, 1245, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Mrózek, E.; Anderson, P.; Caligiuri, M.A. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood 1996, 87, 2632–2640. [Google Scholar] [PubMed]
- Rosenzwajg, M.; Canque, B.; Gluckman, J.C. Human dendritic cell differentiation pathway from CD34+ hematopoietic precursor cells. Blood 1996, 87, 535–544. [Google Scholar] [PubMed]
- Huntington, N.D.; Legrand, N.; Alves, N.L.; Jaron, B.; Weijer, K.; Plet, A.; Corcuff, E.; Mortier, E.; Jacques, Y.; Spits, H.; et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009, 206, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Yokota, T.; Kastelein, R.; Zurawski, S.M.; Arai, N.; Takebe, Y. Species-specificity of T cell stimulating activities of IL 2 and BSF-1 (IL 4): Comparison of normal and recombinant, mouse and human IL 2 and BSF-1 (IL 4). J. Immunol. 1987, 138, 1813–1816. [Google Scholar] [PubMed]
- Billerbeck, E.; Barry, W.T.; Mu, K.; Dorner, M.; Rice, C.M.; Ploss, A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rγnull humanized mice. Blood 2011, 117, 3076–3086. [Google Scholar] [CrossRef] [PubMed]
- Rongvaux, A.; Willinger, T.; Takizawa, H.; Rathinam, C.; Auerbach, W.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Eynon, E.E.; Stevens, S.; et al. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 2378–2383. [Google Scholar] [CrossRef] [PubMed]
- Boucherma, R.; Kridane-Miledi, H.; Bouziat, R.; Rasmussen, M.; Gatard, T.; Langa-Vives, F.; Lemercier, B.; Lim, A.; Bérard, M.; Benmohamed, L.; et al. HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02, and HLA-C*07:01 monochain transgenic/H-2 class I null mice: novel versatile preclinical models of human T cell responses. J. Immunol. 2013, 191, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Mangalam, A.K.; Rajagopalan, G.; Taneja, V.; David, C.S. HLA Class II Transgenic Mice Mimic Human Inflammatory Diseases. Adv. Immunol. 2008, 97, 65–147. [Google Scholar] [PubMed]
- Billerbeck, E.; Horwitz, J.A.; Labitt, R.N.; Donovan, B.M.; Vega, K.; Budell, W.C.; Koo, G.C.; Rice, C.M.; Ploss, A. Characterization of human antiviral adaptive immune responses during hepatotropic virus infection in HLA-transgenic human immune system mice. J. Immunol. 2013, 191, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Pearson, T.; Friberg, H.; Shultz, L.D.; Greiner, D.L.; Rothman, A.L.; Mathew, A. Dengue virus infection and virus-specific HLA-A2 restricted immune responses in humanized NOD-scid IL2rγnull mice. PLoS ONE 2009, 4, e7251. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Saito, Y.; Najima, Y.; Tanaka, S.; Ochi, T.; Tomizawa, M.; Doi, T.; Sone, A.; Suzuki, N.; Fujiwara, H.; et al. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma(null) humanized mice. Proc. Natl. Acad. Sci. USA 2010, 107, 13022–13027. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, Y.-H.; Lum, F.-M.; Ng, L.F.P. Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis. Med. Sci. 2015, 3, 64-77. https://doi.org/10.3390/medsci3030064
Chan Y-H, Lum F-M, Ng LFP. Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis. Medical Sciences. 2015; 3(3):64-77. https://doi.org/10.3390/medsci3030064
Chicago/Turabian StyleChan, Yi-Hao, Fok-Moon Lum, and Lisa Fong Poh Ng. 2015. "Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis" Medical Sciences 3, no. 3: 64-77. https://doi.org/10.3390/medsci3030064
APA StyleChan, Y.-H., Lum, F.-M., & Ng, L. F. P. (2015). Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis. Medical Sciences, 3(3), 64-77. https://doi.org/10.3390/medsci3030064