Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors
Abstract
:1. Introduction
2. Genetic Abnormalities in Adult Glioblastomas
2.1. Genetic Abnormalities of Pediatric Glioblastomas
2.2. Genetic Abnormalities in Pediatric High-Grade Gliomas
2.3. Gene Expression Studies
2.4. Genetic Abnormalities in WHO Grade II and III Adult Gliomas
2.5. Pediatric Low-Grade Gliomas
2.6. Oligodendrogliomas
2.7. Novel Molecular Classifications of Malignant Gliomas
2.8. Medulloblastoma
2.9. Meningiomas
2.10. Ependymomas
2.11. Genetic Abnormalities in Chordoid Glioma of the Third Ventricle
2.12. Normal Neural Stem Cells
2.13. Brain Tumor Cancer Stem Cells
3. Immunotherapy of Glioblastomas
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K. World Health Organization Histological Classification of Tumors of the Central Nervous System; International Agency for Research on Cancer: Lyon, France, 2016. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Bauger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, D.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 13, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, A.; Lindberg, M.; Faust, G.G.; Leibowitz, M.L.; Clark, R.A.; Layer, R.M.; Quinlan, A.R.; Hall, I.M. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spanned by homology-independent mechanisms. Genome Res. 2013, 23, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.M.; Kancar, R.F.; Michelhaugh, S.K.; Sarkar, F.H.; Mittal, S.; Sloan, A.E.; Barnholtz-Sloan, J.S.; Bahassiel, M. Whole genome sequence analysis links chromotripsis to EGFR, MDM2, MDM4, and CDK4 in glioblastoma. Oncoscience 2015, 2, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Botchen, M. Hitchhking without covalent integration. Cell 2004, 117, 280–281. [Google Scholar] [CrossRef]
- Nikolaev, S.; Santoni, F.; Garieri, M.; Makrythanasis, P.; Falconnet, E.; Guipponi, M.; Vannier, A.; Radovanovic, M.; Bena, T.; Forestier, F.; et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat. Commun. 2014, 5, 5690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, K.M.; Deshpande, V.; Beyter, D.; Koga, T.; Rusert, J.; Lee, H.C.; Li, B.; Arden, K.; Ren, B.; Nathanson, D.A.; et al. Extrachromosomal oncogene amplification drives tumor evolution and genetic heterogeneity. Nature 2017, 543, 122–125. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, A.C.; Kim, H.; Poisson, L.M.; Winn, M.E.; Mueller, C.; Cherba, D.; Koeman, J.; Steh, S.; Protopopov, A.; Felicella, M.; et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat. Genet. 2018, 50, 708–717. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Bredel, M.; Scholtens, D.M.; Yadav, A.K.; Alvarez, A.A.; Renfrow, J.J.; Chandler, J.P.; Yu, I.L.; Carro, M.S.; Dai, F.; Tagge, M.J.; et al. NFKBIA deletion in glioblastomas. N. Engl. J. Med. 2011, 364, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Szerlip, N.; Pedraza, A.; Chatravarty, D.; Azim, M.; McGuire, J.; Fang, Y.; Ozawa, T.; Holland, E.C.; Huse, J.T.; Jhanwar, S.; et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. USA 2012, 109, 3041–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snuderl, M.; Fazlollahi, L.; Le, L.P.; Nitta, M.; Zhelyazkova, B.H.; Davidson, C.J.; Akhavanfard, S.; Cahill, D.P.; Aldape, K.D.; Betensky, R.A.; et al. Mosaic amplification of multiple tyrosine kinase genes in glioblastoma. Cancer Cell 2011, 20, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Little, S.; Popov, S.; Jury, A.; Bax, D.A.; Doey, L.; Al-Sarraj, S.; Jugensmeier, J.M.; Jones, C. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012, 72, 1614–1620. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming fusions of EGFR and TACC genes in human glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.; Verhaak, R.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Nanoguchi, N.; Ohta, T.; Oh, J.E.; Kim, Y.H.; Kleihues, P.; Ohgaki, H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013, 126, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Kilella, P.J.; Reitman, Z.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diplas, B.; He, X.; Brosnan-Cashman, J.; Liu, H.; Chen, L.; Wang, Z.; Moure, C.; Killela, P.; Loriaux, D.; Lipp, E.; et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat. Commun. 2018, 9, 2087. [Google Scholar] [CrossRef] [PubMed]
- Pezmeczi, M.; Molinaro, A.; Wash, K.; Decker, P.; Hansen, H.; Sicotte, H.; Kollmeyer, T.; McCoy, L.; Sarkar, G.; et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: Additional prognostic roles of ATRX and TERT. Acta Neuropahtol. 2017, 133, 1001–1016. [Google Scholar]
- Lee, Y.; Koh, J.; Kim, S.I.; Park, C.K.; Choi, S.H. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol. Commun. 2017, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Johannessen, T.C.; Ohba, S.; Chow, T.T.; Jones, L.; Pandita, A.; Pieper, O. Mutant IDH1 cooperates with ATRX loss to drive the alternative lengthening of telomere phenotype in glioma. Cancer Res. 2018, 78, 2966–2977. [Google Scholar] [CrossRef] [PubMed]
- Cimino, P.J.; Kim, Y.; Wu, H.J.; Alexander, J.; Wirsching, H.G.; Szulzewsky, F.; Pitter, K.; Ozawa, T.; Wang, J.; Vazquez, J.; et al. Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Gene Dev. 2018, 32, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Trifonov, V.; Chan, J.M.; Castano, A.; Lia, M.; Abate, F.; Keir, S.T.; Ji, A.X.; Zoppoli, P.; Niola, F.; et al. The integrated landscape of genomic alterations in glioblastoma. Nat. Genet. 2013, 45, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, C.A.; Giacomini, P.; Vogel, H.; Jensen, K.C.; Florio, T.; Merlo, A.; Pollack, J.R.; Wong, A.J. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene 2013, 32, 2670–2681. [Google Scholar] [CrossRef] [PubMed]
- Emlet, D.R.; Gupta, P.; Holdago-Madruga, M.; Del Vecchio, C.A.; Mitra, S.S.; Han, S.Y.; Li, G.; Jensen, K.C.; Vogel, H.; Xu, L.W.; et al. Targeting glioblastoma cancer stem cell population defined by EGF Receptor Variant III. Cancer Res. 2014, 74, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Stockhausen, M.T.; Kristoffersen, K.; Stobbe, L.; Poulsen, H.S. Differentiation of glioblastoma multiforme stem-like cells leads to downregulation of EGFR and decreased tumorigenic and stem-like cell potential. Cancer Biol. Ther. 2014, 15, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Mechan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Inda, M.M.; Bonavia, R.; Mukasa, A.; Narita, Y.; Sah, D.W.; Vandenberg, S.; Brennan, C.; Johns, T.G.; Bachoo, R.; Hadwiger, P.; et al. Tumor heterogeneity in an active process maintained by mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010, 24, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.W.; Cheng, C.K.; Gustafson, W.C.; Charron, E.; Zipper, P.; Wong, R.A.; Chen, J.; Lau, J.; Knobbe-Thomsen, C.; Weller, M.; et al. EGFR phosphorylates tumor-derived-EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 2013, 24, 438–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Han, L.; Dong, Y.; Tian, J.; Huang, W.; Liu, Z.; Jia, X.; Jiang, T.; Zhang, J.; Li, X.; et al. JAK2/STAT3 targeted therapy suppresses tumor invasion via disruption of the EGFRvIII/JAK2/STAT3 axis and associated focal adhesion in EGFRvIII-expressing glioblastoma. Neuro Oncol. 2014, 16, 1229–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, A.; Dhzuv, H.; Peng, S.; Inge, L.J.; Tuncali, S.; Pineda, M.; Millòard, N.; Mayo, Z.; Eschbacher, J.M.; Laftus, J.C.; et al. EGFRvIII-Stat5 signaling enhances glioblastoma cell migration and survival. Mol. Cancer Res. 2018, 16, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.G.; Volinia, S.; Croce, C.M.; Zanca, C.; Li, M.; Emnett, R.; Gutmann, D.H.; Brennan, C.W.; Furnari, F.B.; Cavenee, W.K. Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. 2014, 74, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Eskilsson, E.; Rosland, G.V.; Solecki, G.; Wang, Q.; Hartewr, P.N.; Graziani, G.; Verhaak, R.; Winkler, F.; Bjerkig, R.; Miletic, H. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol. 2018, 20, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.H.; Heimberger, A.B.; Archer, G.E.; Aldape, K.D.; Friedman, A.H.; Friedman, H.S.; Gilbert, M.R.; Herndon, J.E.; McLendon, R.E.; Mitchell, D.A.; et al. Immunological escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 2010, 28, 4722–4729. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Zheng, S.; Amini, S.S.; Virk, S.M.; Mikkelsen, T.; Brat, D.J.; Grimsby, J.; Sougnez, C.; Muller, F.; Hu, J.; et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015, 25, 316–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavaré, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolution dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014, 343, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.J.; Pisapia, D.J.; Malone, H.R.; Goldstein, H.; Lei, L.; Sonabend, A.; Yun, J.; Samanamud, J.; Sims, J.S.; Banu, M.; et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 12550–12555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, D.S.; et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Eskilsson, E.; Rosland, G.V.; Talasila, K.M.; Knappskog, S.; Keunen, O.; Sottoriva, A.; Foerster, S.; Solecki, G.; Taxt, T.; Jirik, R.; et al. EGFRvIII mutations can emerge as late and heterogeneous events in glioblastoma development and promote angiogenesis through Src activation. Neuro Oncol. 2016, 18, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.M.; Zhang, C.Z.; Maire, C.L.; Jung, J.; Manzo, V.E.; Adalsteinsson, V.A.; Homer, H.; Haidsar, S.; Blumenstiel, B.; Pedamallu, C.S.; et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014, 4, 956–971. [Google Scholar] [CrossRef] [PubMed]
- Van den Bent, M.J.; Gao, Y.; Kerkhof, M.; Kros, J.M.; Gorlia, T.; van Zwieten, K.; Prince, J.; van Duinen, S.; Sillevis Smitt, P.A.; Taphoorn, M.; et al. Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol. 2015, 17, 935–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsberg, J.; Hentschel, B.; Kaulich, K.; Gramatzki, D.; Zacher, A.; Malzkorn, B.; Kamp, M.; Sabel, M.; Simon, M.; Westphal, M.; et al. Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: Prognostic role and comparison between primary and recurrent tumors. Clin. Cancer Res. 2017, 23, 6846–6855. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cazzato, E.; Ladewig, E.; Frattini, V.; Rosenbloom, V.; Zairis, S.; Abate, F.; Liu, Z.; Elliot, O.; Shin, Y.J.; et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016, 48, 768–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimino, P.J.; Bredemeyer, A.; Abel, H.J.; Duncavage, E.J. A wide spectrum of EGFR mutations in glioblastoma is detected by a single clinical oncology targeted next-generation sequencing panel. Exp. Mol. Pathol. 2015, 98, 568–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, Z.A.; Thorne, A.H.; Bakas, S.; Wileyto, E.P.; Biello, M.; Akbari, H.; Rathore, S.; Ha, S.M.; Zhang, L.; Ferguson, C.J.; et al. Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell 2018, 34, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Butowski, N.; Tran, D.D.; Recht, L.D.; Lim, M.; Hirte, H.; Ashby, L.; Mechtler, L.; Goldlust, S.S.; Iwamoto, S.; et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACTIV): A randomized, double-blind, international phase 3 trial. Lancet Oncol. 2017, 18, 1373–1385. [Google Scholar] [CrossRef]
- Bell, E.H.; Pugh, S.; McElroy, J.; Gilbert, M.; Mehtas, M.; Klimowicz, A.; Magliocco, A.; Bredel, M.; Robe, P.; Grasu, A.L.; et al. Molecular-based recursive portioning analysis (RPA) model for glioblastoma in the temozolomide era: A correlative analysis based upon NRG oncology RTOG 0525. JAMA Oncol. 2017, 3, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodeling genes in pediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.W.; Muller, M.M.; Koletsky, M.S.; Cordero, F.; Lin, S.; Banaszynski, L.A.; Garcia, B.A.; Muir, T.W.; Becher, O.J.; Allis, C.D.; et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013, 340, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Natsume, A.; Ito, M.; Katsushima, K.; Ohka, F.; Hatanaka, A.; Shinjo, K.; Sato, S.; Takahashi, S.; Ishikawa, Y.; Takeuchi, I.; et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013, 73, 4559–4570. [Google Scholar] [CrossRef] [PubMed]
- Aihara, K.; Mukasa, A.; Gotoh, K.; Saito, K.; Nagae, G.; Tsuji, S.; Tatsuno, K.; Yamamoto, S.; Takayanagi, S.; Narita, Y.; et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol. 2014, 16, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Strum, D.; Bender, S.; Jones, D.; Lichter, P.; Grill, J.; Becher, O.; Howkins, C.; Majewski, J.; Jones, C.; Costello, J.F.; et al. Pediatric and adult glioblastoma: Multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 2014, 14, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Ballester, T.Y.; Wang, Z.; Shandilya, A.; Miettinen, M.; Burger, P.C.; Eberhart, C.G.; Rodriguez, F.J.; Raabe, E.; Nazarian, J.; Warren, K.; et al. Morphologic characteristics and immunohistochemical profile of diffuse intrinsic pontine gliomas. Am. J. Surg. Pathol. 2013, 37, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and in brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.R.; Mackay, A.; Truffaux, N.; Butterfield, Y.; Morozova, O.; Philippe, C.; Castel, D.; Grasso, C.S.; Vinci, M.; Carvalho, D.; et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat. Genet. 2014, 46, 457–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontebasso, A.M.; Papillon-Cavanagh, S.; Schwartzentruber, J.; Nikbakht, H.; Gerges, N.; Fiset, P.O.; Bechet, D.; Faury, D.; De Jay, N.; Ramkissoon, L.A.; et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat. Genet. 2014, 46, 462–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buczkowicz, P.; Hoeman, C.; Rakopoulos, P.; Pajovic, S.; Letourneau, L.; Dzamba, M.; Morrison, A.; Lewis, P.; Bouffet, E.; Bartels, U.; et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat. Genet. 2014, 46, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Diaz, A.; Paugh, B.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castel, D.; Cathy, P.; Calmon, R.; LeDret, L.; Truffaux, N.; Boddaert, N.; Pages, M.; Taylor, K.R.; Saulnier, P.; Lacroix, L.; et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015, 130, 815–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karremann, M.; Gielen, G.H.; Hoffmann, M.; Wiese, M.; Colditz, N.; Warmuth-Metz, M.; Bison, B.; Claviez, A.; van Vuurden, D.G.; von Bueren, A.O.; et al. Diffuse high-grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro Oncol. 2018, 20, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Meyronet, D.; Esteban-Mader, M.; Bonnet, C.; Joly, M.O.; Uro-Coste, E.; Amiel-Benouaich, A.; Forest, F.; Rousselot-Denis, C.; Burel-Vandenbos, F.; Bourg, V.; et al. Characteristics of H3 K27M-mutant gliomas in adults. Neuro. Oncol. 2017, 19, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Pagès, M.; Beccaria, K.; Boddaert, N.; Saffroy, R.; Besnard, A.; Castel, D.; Fina, F.; Barets, D.; Barret, E.; Lacroix, L.; et al. Co-occurrence of histone H3 K27M and BRAF V600E mutations in pediatric midline grade I ganglioglioma. Brain Pathol. 2018, 28, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Nikbakht, H.; Panditharatna, E.; Mikael, L.G.; Li, R.; Gayden, T.; Osmond, M.; Ho, C.Y.; Kambhampati, M.; Hwang, E.I.; Faury, D.; et al. Spatial and temporal heterogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat. Commun. 2016, 6, 11185. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.M.; DeWire, M.; Ryall, S.; Buczkowicz, P.; Leach, J.; Miles, L.; Ramani, A.; Brudno, M.; Kumar, S.S.; Drissi, R.; et al. Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: Implications for diagnostic biology and targeted therapeutics. Acta Neuropathol. Commun. 2016, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Salloum, R.; McConechy, M.K.; Mikael, L.; Fuller, C.; Drissi, R.; DeWire, M.; Nikbakht, H.; De Jay, N.; Yang, X.; Boue, D.; et al. Characterizing temporal genomic heterogeneity in pediatric high-grade gliomas. Acta Neuropathol. Commun. 2017, 5, 78. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nadhabalan, M.; et al. Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 2017, 32, 520–537. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Pfister, S.M.; Jones, D. Pediatric gliomas: Current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 2017, 35, 2370–2377. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Schrimpf, D.; Ryzhova, M.; Sturm, D.; Chavez, L.; Hovestadt, V.; Sharma, T.; Habel, A.; Burford, A.; Jones, C.; et al. H3-/IDH-wild type pediatric glioblastoma in comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017, 134, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.; Gronych, J.; Warnatz, H.J.; Hutter, B.; Grabner, S.; Ryzhova, M.; Pfaff, E.; Hovestadt, V.; Weinberg, F.; Halbach, S.; et al. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat. Med. 2016, 11, 1314–1320. [Google Scholar]
- Vinci, M.; Burford, A.; Molinari, V.; Kessler, K.; Popov, S.; Clarke, M.; Taylor, K.R.; Pemberton, K.N.; Lord, C.J.; Gutteridge, A.; et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastomas and diffuse intrinsic pontine glioma cells. Nat. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, F.; Weissman, S.; Leblanc, B.; Pandey, D.; Hojfeldt, J.; Comet, I.; Zheng, C.; Johansen, J.V.; Rapin, N.; Porse, B.T.; et al. EZH2 is potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 2017, 23, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Hashizuma, R.; Morgan, M.; Bartom, E.; Harbiuski, C.; Marshall, S.A.; Rendleman, E.J.; Ma, Q.; Takahashi, Y.; Woodfin, A.; et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 2017, 23, 493–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaraja, S.; Vitanza, N.A.; Wou, P.J.; Taylor, K.R.; Liu, F.; Zgang, L.; Li, M.; Meng, W.; Ponnuswami, A.; Sun, W.; et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 2017, 31, 635–652. [Google Scholar] [CrossRef] [PubMed]
- Chheda, Z.; Kohanbash, G.; Okada, K.; Jahan, N.; Sidney, J.; Pecoraro, M.; Yang, X.; Carrera, D.; Downey, K.; Shrivastav, S.; et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J. Exp. Med. 2018, 215, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Grill, J.; Massimino, M.; Bouffet, E.; Azizi, A.A.; McCowage, G.; Canete, A.; Saran, F.; Le Deley, M.C.; Varlet, P.; Morgan, P.S.; et al. Phase II, open-label, randomized, multicenter trial (HERBY) of bevacizumab in pediatric patients with newly diagnosed high-grade glioma. J. Clin. Oncol. 2018, 36, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Molinari, V.; Jones, D.; Izquierdo, E.; Brouwer-Visser, J.; Giangaspero, F.; Haberler, C.; Pietsch, T.; Jacques, T.; et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell 2018, 33, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.G.; Tirosh, I.; Hovestadt, V.; Shaw, M.K.; Escalante, L.; Mathewson, N.D.; Neftel, C.; Frank, N.; Pelton, K.; Hebert, C.M.; et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018, 360, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumors. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.; Momota, H.; Hambardzumyan, D.; Ozawa, T.; Tandon, A.; Pedranza, A.; Holland, E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE 2009, 4, e7752. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Brennan, C.; Wang, L.; Squatrito, M.; Sasayama, T.; Nakada, M.; Huse, J.T.; Pedraza, A.; Utsuki, S.; Yasui, Y.; et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes. Dev. 2010, 24, 2205–2218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducray, F.; Idbaih, A.; De Reyniès, A.; Bieche, I.; Thillet, J.; Mokhtari, K.; Lair, S.; Marie, Y.; Paris, S.; Vidaud, M.; et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol. Cancer 2008, 7, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noushmehr, H.; Weisenberger, D.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG island methylation phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-kB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Mathew, L.K.; Skuli, N.; Mucaj, V.; Lee, S.S.; Zinn, P.O.; Sathyan, P.; Imtiyaz, H.Z.; Zhang, Z.; Davuluri, R.V.; Rao, S.; et al. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Yan, W.; Gao, K.; Wang, Y.; Zhang, J.; You, Y. Prevalence and clinicopathologic characteristics of the molecular subtypes in malignant glioma: A multi-institutional analysis of 941 cases. PLoS ONE 2014, 9, e94871. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Squatrito, M.; Scarpace, L.; de Carvalho, A.; Lyn, S.; Li, P.; Li, Y.; Barthel, F.; et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, W.; Chen, D.; Lv, Y.; Zheng, J.; Lilljebjörn, H.; Ran, L.; Bao, Z.; Soneson, C.; Sjögren, H.O.; et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc. Natl. Acad. Sci. USA 2014, 111, 3538–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Funari, F.B.; Fenton, T.; Bachoo, R.M.; Mukasa, A.; Stommel, J.M.; Stegh, A.; Hahn, W.C.; Ligon, K.L.; Louis, D.N.; Brennan, C.; et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007, 21, 2683–2710. [Google Scholar] [CrossRef] [PubMed]
- Gorovets, D.; Kannan, K.; Shen, R.; Kastenhuber, E.R.; Islamdoust, N.; Campos, C.; Pentsova, E.; Heguy, A.; Jhanwar, S.C.; Mellinghoff, I.K.; et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin. Cancer Res. 2012, 18, 2490–2501. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Kharbanda, S.; Pope, W.B.; Tran, A.; Solis, O.E.; Peale, F.; Forrest, W.F.; Pujara, K.; Carrillo, J.A.; Pandita, A.; et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J. Clin. Oncol. 2011, 29, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Inagaki, A.; Silber, J.; Gorovets, D.; Zhang, J.; Kastenhuber, E.R.; Heguy, A.; Petrini, J.H.; Chan, T.A.; Huse, J.T. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget 2012, 3, 1194–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killela, P.J.; Pirozzi, C.J.; Reitman, Z.; Jones, S.; Rasheed, B.A.; Lipp, E.; Friedman, H.; Friedman, A.H.; He, Y.; McLendon, R.E.; et al. The genetic landscape of neoplastic astrocytoma. Oncotarget 2014, 5, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Serin Harmanci, A.; Erson-Omary, E.Z.; Li, J.; Coşkun, S.; Simon, M.; Krischek, B.; Özduman, K.; Omay, S.B.; Sorensen, E.A.; et al. Integrated genomic characterization if IDH1-mutant glioma malignant progression. Nat. Genet. 2016, 48, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Iafrate, J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, T.A.; Mukherjee, J.; Viswanath, P.; Ohba, S.; Ronen, S.M.; Bjervig, R.; Pieper, R.O. Rapid conversion of mutant IDH1 from driver in a model of human gliomagenesis. Mol. Cancer Res. 2016, 14, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Mazor, T.; Chesnelong, C.; Pankov, A.; Jalbert, L.E.; Hong, C.; Hayes, J.; Smirnov, I.V.; Marshall, R.; Souza, C.F.; Shen, Y.; et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc. Natl. Acad. Sci. USA 2017, 114, 10743–10748. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.F.; Sabedot, T.; Malta, T.; Stetson, L.; Morozova, O.; Sokolov, A.; Laird, P.; Wirnerawicz, M.; Iavarone, A.; Snyder, J.; et al. A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018, 23, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Drier, Y.; Liau, B.B.; Gillespie, S.M.; Venteicher, A.S.; Stemmer-Rachaminov, A.; Suvà, M.; Bernstein, B.E. Insulator dysfunction and oncogene activation in IDH-mutant gliomas. Nature 2016, 529, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Bardella, C.; Al-Dalahman, O.; Krell, D.; Brazauskas, P.; Al-Qahtani, K.; Tomkova, M.; Adam, J.; Serres, S.; Lockstone, H.; Freeman-Mills, L.; et al. Expression of IDH1(R132H) in the murine subventricular zone stem cell niche recapitulates features of early gliomagenesis. Cancer Cell 2016, 30, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Modrek, A.S.; Golub, D.; Khan, T.; Braedy, D.; Prado, J.; Bowman, C.; Deng, J.; Zhang, G.; Rocha, P.P.; Raviram, R.; et al. Low-grade astrocytoma mutations in IDH1, P53, and ATRX cooperate to block differentiation of human neural stem cells via repression of SOX2. Cell Rep. 2017, 21, 1267–1280. [Google Scholar] [CrossRef] [PubMed]
- Venteicher, A.; Tirosh, I.; Hebert, C.; Yizhak, K.; Neftel, C.; Filbin, M.; Hovestadt, V.; Escalante, L.; Shaw, M.K.; Rodman, C.; et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017, 355, eaai8478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, K.; Nakamura, H.; Suzuki, H.; Matsuo, K.; Kataoka, K.; Shimamura, T.; Matomura, H.; Ohka, F.; Shiina, S.; Yamamaoto, T.; et al. Prognostic relevance of genetic alterations in diffuse lower gliomas. Neuro Oncol. 2018, 20, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.; Mondal, G.; Hyun, M.; Ruiz, G.; Lin, H.S.; Van Ziffle, J.; Joseph, N.M.; Onodera, C.; Talevich, E.; Grenert, J.P.; et al. A recurrent kinase domain mutation in PRKCA defines choroid glioma of the third ventricle. Nat. Commun. 2018, 9, 810. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Chang, E.F.; Lamborn, K.R.; Chang, S.M.; Prodos, M.D.; Cha, S.; Tihan, T.; Vanderberg, S.; MacDermott, M.W.; Berger, M.S. Role of extent of resection in long-term outcome of low-grade hemispheric gliomas. J. Clin. Oncol. 2008, 26, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.C.; Show, E.G.; Pugh, S.L.; Chakravatti, A.; Gilbert, M.R.; Berger, G.R.; Coones, S.; Ricci, P.; Bullard, D.; Brown, P.D.; et al. Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N. Engl. J. Med. 2016, 374, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.H.; Zhang, P.; Fisher, B.J.; Mac Donald, D.R.; McElroy, J.P.; Lesser, G.J.; Fleming, J.; Chakrabarty, A.R.; Liu, Z.; Becker, A.P.; et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with temozolomide and radiotherapy. JAMA Oncol. 2018. [Google Scholar] [CrossRef]
- Hawkins, C.; Walker, E.; Mohamed, N.; Zhang, C.; Jacob, K.; Shirinian, M.; Alon, N.; Kahn, D.; Fried, I.; Scheinemann, K.; et al. BRAF-KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin. Cancer Res. 2011, 17, 4790–4798. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Hutter, B.; Jager, N.; Korshunov, A.; Kool, M.; Warnatz, H.J.; Zichner, T.; Lambert, S.R.; Ryzhova, M.; Quang, D.A.; et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 2013, 45, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.; Jones, D.; Giannini, C. Pilocytic astrocytoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.H.; McLellan, M.D.; Hussain, I.; Wallis, J.W.; Fulton, L.L.; Fulton, R.S.; Magrini, V.; Demeter, R.; Wylie, T.; Kandoth, C.; et al. Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res. 2013, 23, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Gronych, J.; Korshunov, A.; Bageritz, J.; Milde, T.; Jugold, M.; Hambardzumyan, D.; Remke, M.; Hartmann, C.; Witt, H.; Jones, D.T.; et al. An activated mutant BRAF kinase domain is sufficient to induce pilocytic astrocytoma in mice. J. Clin. Investig. 2011, 121, 1344–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaul, A.; Chen, Y.H.; Emnett, R.J.; Dahlya, S.; Gutmann, D.H. Pediatric glioma-associated KIAA1549: BRAF expression regulates neuroglial cell growth in a cell specific and mTOR-dependent manner. Genes Dev. 2012, 26, 2561–2566. [Google Scholar] [CrossRef] [PubMed]
- Raabe, E.H.; Lim, K.S.; Kim, J.M.; Meeker, A.; Mao, X.G.; Nikkhah, G.; Maciaczyk, J.; Kahlert, U.; Jain, D.; Bar, E.; et al. BRAF activation induces transformation and then senescence in human neural stem cells: A pilocytic astrocytoma model. Clin. Cancer Res. 2011, 17, 3590–3599. [Google Scholar] [CrossRef] [PubMed]
- Schindler, G.; Capper, D.; Meyer, J.; Janzarik, W.; Omran, H.; Herold-Mende, C.; Schmieder, K.; Wesseling, P.; Mawrin, C.; Hasselblatt, M.; et al. Analysis of BRAF V600E mutation in 1320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extracerebellar pilocytic astrocytoma. Acta Neuropathol. 2011, 121, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Hasselblatt, M.; Riesmeiere, B.; Lechtape, B.; Brentrup, A.; Stummer, W.; Albert, F.K.; Sepehrnia, A.; Ebel, H.; Gerss, J.; Paulus, W. BRAF-KIAA1549 fusion transcripts are less frequent in pilocytic astrocytomas diagnosed in adults. Neuropathol. Appl. Neurobiol. 2011, 37, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.F.; Scheithauer, B.W.; Giannini, C.; Rynearson, A.; Cel, L.; Hoesley, B.; Gilmer-Flynn, H.; Sarkaria, J.N.; Jenkins, S.; Long, J.; et al. PI3K/AKT pathway alterations are associated with clinically aggressive and histologically anaplastic subsets of pilocytic astrocytoma. Acta Neuropathol. 2011, 121, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Stichel, D.; Schrimpf, D.; Sahm, F.; Korshunov, A.; Reuss, D.E.; Koelsche, C.; Huang, K.; Wefers, A.K.; Hovestadt, V.; et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wild-type glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ramkissoon, L.; Horowitz, P.; Craig, J.; Ramkissoon, S.H.; Rich, B.E.; Schumacher, S.E.; McKenna, A.; Lawrence, M.S.; Bergthold, G.; Brastianos, P.K.; et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl. Acad. Sci. USA 2013, 110, 8188–8193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wu, G.; Miller, C.P.; Tatevossian, R.G.; Dalton, J.D.; Tang, B.; Orisme, W.; Punchihewa, C.; Parker, M.; Qaddoumi, I.; et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013, 45, 602–612. [Google Scholar] [PubMed] [Green Version]
- Johnson, A.; Severson, E.; Gay, L.; Vergilio, G.A.; Elvin, J.; Suh, J.; Daniel, S.; Covert, M.; Frampton, G.; Hsu, S.; et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist 2017, 22, 1478–1490. [Google Scholar] [CrossRef] [PubMed]
- Dias-Santagata, D.; Lam, Q.; Vernovsky, K.; Vena, N.; Lennerz, J.K.; Borger, D.R.; Batchelor, T.T.; Ligon, K.L.; Iafrate, A.J.; Ligon, A.H.; et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: Diagnostic and therapeutic implications. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassaletta, A.; Zapotocky, M.; Mistry, M.; Ramaswamg, V.; Honnorat, M.; Krishmarty, R.; Stucklin, A.G.; Zhukova, N.; Arnoldo, A.; Ryall, S.; et al. Therapeutic and prognostic implications of BRAF V600E in pediatric low-grade gliomas. J. Clin. Oncol. 2017, 35, 1934–2941. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.C.; Sturm, D. Gliomas in children. Semin. Neurol. 2018, 38, 121–130. [Google Scholar] [PubMed]
- Vaubel, R.A.; Caron, A.A.; Yamada, S.; Decker, P.A.; Eckel-Passow, J.; Rodriguez, F.J.; Rao, A.; Lachancxe, D.; Parney, I.; Jenkins, R.; et al. Recurrent copy number alterations on low-grade and anaplastic pleomorphic xanthoastrocytoma with and without BRAF V600E mutation. Brain Pathol. 2018, 28, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Pekmeczi, M.; Villanueva-Mayer, J.; Goode, B.; Van Ziffle, J.; Ondera, C.; Grenest, J.P.; Bastian, B.C.; Chamyan, G.; Maher, D.; Zhatib, Z.; et al. The genetic landscape of ganglioma. Acta Neuropathol. Commun. 2018, 6, 47. [Google Scholar]
- Touse, R.; Gupta, T.; Epari, S.; Shastri, J.G.; Gurau, M.; Bano, N.; Jalali, R. Impact of WHO 2016 update of brain tumor classification, molecular markers and clinical outcomes in pleomorphic xanthoastrocytoma. J. Neuroncol. 2018, 136, 343–350. [Google Scholar]
- Rivera, B.; Gayden, T.; Carrot-Zhang, J.; Nadaf, J.; Boshari, T.; Faury, D.; Zeinieh, M.; Blanc, R.; Burk, D.L.; Fahiminiya, S.; et al. Germile and somatic FGFR1 abnormalities in dysembryoplastic neuroepithelial tumors. Acta Neuropathol. 2016, 131, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Qaddoumi, I.; Orisme, W.; Wen, J.; Santiago, T.; Gupta, K.; Dalton, J.D.; Tang, B.; Haupfear, K.; Punchihewa, C.; Easton, J.; et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1 and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016, 131, 833–8435. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.J.; Keeley, A.; Virasami, A.; Harkness, W.; Tisdall, M.; Delgado, E.I.; Gutteridge, A.; Brooks, T.; Kristiansen, M.; Chalker, J.; et al. Comprehensive molecular characterization of epilepsy-associated glioneuronal tumours. Acta Neuropathol. 2018, 135, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Brandopadhayay, P.; Ramkissoon, L.A.; Jain, P.; Bergthold, G.; Wala, J.; Zeid, R.; Schumacher, S.E.; Urbanski, L.; O’Rourke, R.; Gibson, W.J.; et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenesis through a tripartite mechanism. Nat. Genet. 2016, 48, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Bongaarts, A.; Giannikov, K.; Rainten, R.; Aniuk, J.; Mills, J.D.; Jansen, F.E.; Spliet, W.; den Dunnen, W.; Coras, R.; et al. Subependymal giant cell astrocytomas in Tuberous Sclerosis Complex have consistent TSC1/TSC2 biallelic inactivation, and no BRAF mutations. Oncotarget 2017, 8, 95516–95529. [Google Scholar] [CrossRef] [PubMed]
- Zordan, P.; Carminell, M.; Cascino, F.; Tratta, E.; Pollani, P.L.; Galli, R. Tuberous sclerosis complex-associated CNS abnormalities depend on hyperactivation of mTORC1 and AKT. J. Clin. Investig. 2018, 128, 1688–1706. [Google Scholar] [CrossRef] [PubMed]
- Mistry, M.; Zhukova, N.; Merico, D.; Rakopoulos, P.; Krishnatry, R.; Shago, M.; Stavropoulos, J.; Alon, N.; Pole, J.D.; Ray, P.N.; et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J. Clin. Oncol. 2015, 33, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Sausen, M.; Wood, L.D.; Hruban, R.H.; Rodriguez, F.J.; Cahill, D.P.; McLendon, R.; Riggins, G.; et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011, 333, 1453–1455. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Butterfield, Y.; Morozova, O.; Chittaranjan, S.; Blough, M.D.; An, J.; Birol, I.; Chesnelong, C.; Chiu, R.; Chuah, E.; et al. Concurrent CIC mutations and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012, 226, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, H.; Tanaka, S.; Curry, W.T.; Loebel, F.; Zhao, D.; Tateishi, K.; Chen, J.; Klofas, L.K.; Lelic, N.; Kim, J.C.; et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin. Cancer Res. 2014, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, P.; Zhang, C.; Zhang, W.; Liu, Y.; Bao, Z.; Liu, X.; Du, W.; Wang, H.; Jiang, T.; et al. ATRX mRNA expression combined with IDH1/2 mutation status and Ki-67 expression refines the molecular classification of astrocytic tumors: Evidence from the whole transcriptional sequencing of 169 samples. Oncotarget 2014, 5, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Labreche, K.; Simeonova, I.; Kamoun, A.; Gleize, V.; Chubb, D.; Letouzé, E.; Riazalhosseini, Y.; Dobbins, S.E.; Elarouci, N.; Ducray, F.; et al. TCF12 is mutated in anaplastic oligodendroglioma. Nat. Commun. 2015, 6, 7207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubbink, H.J.; Atmodimedjo, P.N.; Kros, J.M.; French, P.J.; Sanson, M.; Idbaih, A.; Wesseling, P.; Enting, R.; Spliet, W.; Tijssen, C.; et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: A report of the prospective randomized EORT brain tumor group 26951 phase III trial. Neuro Oncol. 2016, 18, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Hewer, E.; Prebil, N.; Berezowska, S.; Gutt-Will, M.; Schcht, P.; Dettmer, M.S.; Vassella, E. Diagnostic implications of TERT promoter mutation status in diffuse gliomas in a routine clinical setting. Virchows Arch. 2017, 471, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Kreiger, P.A.; Okada, Y.; Simon, S.; Rorke, L.B.; Louis, D.N.; Golden, J.A. Losses of chromosomes 1p and 19q are rare in pediatric oligodendrogliomas. Acta Neuropathol. 2005, 109, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Nauen, D.; Haley, L.; Lin, M.T.; Perry, A.; Giannini, C.; Burger, P.C.; Rodriguez, F.J. Molecular analysis of pediatric oligodendrogliomas highlights genetic differences with adult counterparts other pediatric gliomas. Brain Pathol. 2016, 26, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Pathak, P.; Purkait, S.; Faruq, M.; Jha, P.; Mallick, S.; Suri, V.; Sharma, M.C.; Suri, A.; Sarkar, C. Oncogenic KIAA1549-BRAF fusion and activation of the MAPK/ERK pathway in pediatric oligodendrogliomas. Cancer Genet. 2015, 208, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, F.J.; Schniederjan, M.J.; Nicolaides, T.; Tihan, T.; Burger, P.C.; Perry, A. High rate of concurrent BRAF-KIAA1549 gene fusion and 1p deletion in disseminated oligodendroglioma-like leptomeningeal neoplasms (DOLN). Acta Neuropathol. 2015, 129, 609–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, M.Y.; Sill, M.; Chiang, J.; Schittenhelm, J.; Ebinger, M.; Schuhman, M.U.; Monoranu, C.M.; Milde, T.; Wittmann, A.; Hartmann, C.; et al. Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol. 2018, 136, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma groups based on 1p/19q, IDH and TERT promoter mutations in tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Mischel, P.S.; Reifenberger, G. Molecular classification of gliomas. Hnad. Clin. Neurol. 2016, 134, 97–120. [Google Scholar]
- Xiu, T.; Piccioni, D.; Juarez, T.; Pingle, S.C.; Hu, J.; Rudnick, J.; Fink, K.; Spetzler, D.B.; Maney, T.; Ghazalpour, A.; et al. Multi-platform molecular profiling of large cohort of glioblastomas reveals potential therapeutic strategies. Oncotarget 2016, 7, 21556–21569. [Google Scholar] [CrossRef] [PubMed]
- Campos, B.; Olsen, L.R.; Poulsen, M.S. A comprehensive profile of recurrent glioblastoma. Oncogene 2016, 35, 5819–5825. [Google Scholar] [CrossRef] [PubMed]
- Andor, N.; Harness, J.V.; Muller, S.; Mewes, H.W.; Petritsch, C. EXPANDS: Expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 2014, 30, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Koster, J.; Bunt, J.; Hasselt, N.E.; Lakeman, A.; van Sluis, P.; Troost, D.; Meeteren, N.S.; Caron, H.N.; Cloos, J.; et al. Integrated genomic identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinopathological features. PLoS ONE 2008, 28, e3088. [Google Scholar]
- Cho, Y.J.; Tsherniak, A.; Tamayo, P.; Santagata, S.; Ligon, A.; Greulich, H.; Berhoukim, R.; Amani, V.; Goumnerova, L.; Eberhart, C.G.; et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011, 29, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.J.; Koster, J.; Schouten-van Meeteren, A.; van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations and clinical data of WNT, SHH, Group3 and Group4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Shih, D.J.; Peacock, K.J.; Garzia, L.; Morrissy, A.S.; Zichner, T.; Stütz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup specific structural variation across 1000 medulloblastoma genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, D.W.; Li, M.; Zhang, X.; Jones, S.; Leary, R.J.; Lin, J.C.; Boca, S.M.; Carter, H.; Samayoa, J.; Bettegowda, C.; et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011, 331, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.; Pomeranz Krummel, D.A.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.; Jager, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, B.; Cho, Y.J.; Pugh, T.J.; Hovestadt, V.; Stütz, A.M.; et al. ICGC Ped Brain: Dissecting the genomic complexity underlying medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.; Parker, M.; Kranenburg, T.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remke, M.; Ramaswamy, V.; Peacock, J.; Shih, D.J.; Koelsche, C.; Northcott, P.A.; Hill, N.; Cavalli, F.M.; Kool, M.; Wang, X.; et al. TERT promoter mutations are highly recurrent in SHH medulloblastoma. Acta Neuropathol. 2013, 126, 917–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Buchhalter, I.; Morissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Grobner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.Y.; Erkek, S.; Tong, Y.; Yin, L.; Federation, A.J.; Zapatka, M.; Haldipur, P.; Kawauchi, D.; Risch, T.; Warnatz, H.J.; et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 2016, 530, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandopadhayay, P.; Bergthold, G.; Nguyen, B.; Schubert, S.; Gholamin, S.; Tang, Y.; Bolin, S.; Schumacher, S.E.; Zeid, R.; Masoud, S.; et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 2014, 20, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Vo, B.H.; Wolf, E.; Kawauchi, D.; Gebhardt, A.; Rehg, J.E.; Finkelstein, D.; Walz, S.; Murphy, B.L.; Youn, Y.H.; Han, Y.G.; et al. The interaction of Myc with Miz1 defines medulloblastoma subgroup identity. Cancer Cell 2016, 29, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Liu, K.W.; Wang, J.; Garancher, A.; Tao, R.; Esparza, L.A.; Maier, D.L.; Udaka, Y.T.; Murad, N.; Morrissy, S.; et al. HDAC and PI3K antagonists cooperate to inhibit the growth of MYC-driven medulloblastoma. Cancer Cell 2016, 29, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Waszak, S.; Northcott, P.; Buchhalter, I.; Robinson, G.W.; Sutter, C.; Groebner, S.; Grund, K.B.; Bugieres, L.; Jones, D.; Pajter, C.; et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: A retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018, 19, 785–798. [Google Scholar] [CrossRef]
- Wu, X.; Northcott, P.; Dubuc, A.; Dupuy, A.J.; Shih, D.J.; Witt, H.; Croul, S.; Bouffet, E.; Fults, D.W.; Eberhart, C.G.; et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 2012, 482, 529–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grazia, L.; Kijima, N.; Morissy, A.S.; De Antonellisi, P.; Guerriero-Stucklin, A.; Holgado, B.L.; Wu, X.; Wang, X.; Parson, M.; Zayne, K.; et al. A hematogenous route for leptomeningeal metastases. Cell 2018, 172, 1052–1062. [Google Scholar]
- Zapotocky, M.; Mata-mmeba, D.; Sumerauer, D.; Liby, P.; Lasalletta, A.; Zamecnik, J.; Kreskova, L.; Kyncl, M.; Stary, J.; Laughlin, S.; et al. Differential patterns of metastatic dissemination across medulloblastoma subgroups. J. Neurosurg. Pediatr. 2018, 21, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Mumert, M.; Dubuc, A.; Wu, X.; Northcott, P.A.; Chin, S.S.; Pedone, C.A.; Taylor, M.D.; Fults, D.W. Functional genomics identifies drivers of medulloblastoma dissemination. Cancer Res. 2012, 72, 4944–4953. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.M.; Kuijper, S.; Lindsey, J.C.; Petrie, K.; Schwalbe, E.C.; Barker, K.; Boult, J.K.; Williamson, D.; Ahmad, Z.; Hallsworth, A.; et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015, 27, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Garzia, L.; Shih, D.; Zuyderduyn, S.; Huang, X.; Skowron, P.; Remke, M.; Cavalli, F.M.; Ramaswamy, V.; Lindsay, P.E.; et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, D.; Northcott, P.; Remke, M.; Korshunov, A.; Ramaswamy, V.; Kool, M.; Luu, B.; Yao, Y.; Wang, X.; Dubuc, A.M.; et al. Cytogenetic prognostication within medulloblastoma subgroups. J. Clin. Oncol. 2014, 32, 886–896. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, L.; Chen, Y.; Remke, M.; Shih, D.; Lu, F.; Wang, H.; Deng, Y.; Yu, Y.; Xia, Y.; et al. The G-protein alpha subunit Gsα is a tumor suppressor in sonic hedgehog-driven medulloblastoma. Nat. Med. 2014, 20, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Von Bueren, A.; Kortmann, R.D.; von Hoff, K.; Friedrich, C.; Mynarek, M.; Muller, K.; Goscchzik, T.; zur Muhlen, A.; Gerber, N.; Warmuth-Metz, M.; et al. Treatment of children and adolescent with metastatic medulloblastoma and prognostic relevance of clinical and biologic parameters. J. Clin. Oncol. 2016, 34, 4151–4160. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, E.C.; Lindsey, J.C.; Nakjang, S.; Crosier, S.; Smith, A.J.; Hicks, D.; Rafiee, G.; Hill, R.M.; Iliasova, A.; Stone, T.; et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet 2017, 18, 958–971. [Google Scholar] [CrossRef]
- Robinson, G.W.; Rudneva, V.A.; Buchhalter, I.; Bilups, C.A.; Waszak, S.M.; Smith, K.S.; Bowers, D.C.; Bendel, A.; Fisher, P.G.; Partap, S.; et al. Risk-adapted therapy for youg children with medulloblastoma (SJYC07): Therapeutic and molecular outcomes from a multicenter, phase 2 trial. Lancet Oncol. 2018, 19, 768–784. [Google Scholar] [CrossRef]
- Cavalli, F.; Rmeke, M.; Rampasek, L.; Peacock, J.; Shih, D.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morissy, S.; et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 2017, 31, 737–754. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Hinojosa, S.; Lau, L.S.; Stampar, M.; Staal, J.; Zhang, H.; Gordish-Gressman, H.; Northcott, P.A.; Pfister, S.M.; Taylor, M.D.; Brown, K.J.; et al. Proteomic analysis of medulloblastoma reveals functional biology with translational potential. Acta Neuropathol. Commun. 2018, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.M.; Hielscher, T.; Bouffet, E.; Remke, M.; Luu, B.; Gururangan, S.; McLendon, R.E.; Bigner, D.D.; Lipp, E.S.; Perreault, S.; et al. Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup: A retrospective integrated clinical and molecular analysis. Lancet Oncol. 2017, 14, 484–495. [Google Scholar] [CrossRef]
- Kuzan-Fisher, C.M.; Jurashka, K.; Taylor, M.D. Medulloblastoma in the molecular era. J. Korean Neurosurg. Soc. 2018, 61, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Garrido-Garcia, A.; Garcia-Gerique, L.; Lemos, I.; Sunal, M.; de Torres, C.; Kulis, M.; Perez-Jaume, S.; Carcaboso, A.; Luu, B.; et al. A novel method for rapid molecular subgrouping of medulloblastoma. Clin. Cancer Res. 2018, 24, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Merk, D.J.; Ohli, J.; Merk, N.D.; Thatikonda, V.; Morissy, S.; Schoof, M.; Schmid, S.N.; Harrison, L.; Filser, S.; Ahfeld, J.; et al. Opposing effect of CREBBP mutations govern the phenotype of Rubinstein-Taybi syndrome and adult SHH medulloblastoma. Dev. Cell 2018, 44, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.L.; Wojcinski, A.; Rallapalli, H.; Leo, Z.; Sangrajka, R.M.; Stephen, D.; Volkova, E.; Korshunov, A.; Remke, M.; Taylor, M.D.; et al. Lateral cerebellum is preferentially sensitive to high sonic hedgehog signaling and medulloblastoma formation. Proc. Natl. Acad. Sci. USA 2018, 119, 3392. [Google Scholar] [CrossRef] [PubMed]
- Raleigh, D.R.; Choksi, P.K.; Krup, A.L.; Myaer, W.; Santos, N.; Reiter, J.F. Hedgehog signaling drives medulloblastoma growth via CDK6. J. Clin. Investig. 2018, 128, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Jones, D.; Jager, N.; Northcott, P.A.; Pugh, T.J.; Hovestadt, V.; Piro, R.M.; Esparza, L.A.; Markant, S.L.; Remke, M.; et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014, 25, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.E.; Zhang, L.; Peri, S.; Kou, Y.M.; Du, F.; Egleston, B.L.; Ng, J.; Andrews, A.; Attsaturov, I.; Curran, T.; et al. Statins synergize with hedgehog pathway inhibitors for treatment of medulloblastoma. Clin. Cancer Res. 2018, 24, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.W.; Orr, B.A.; Wu, G.; Gururangan, S.; Lin, T.; Qaddoumi, I.; Packer, R.J.; Goldman, S.; Prados, M.D.; Desjardins, A.; et al. Vismodegib exerts target efficacy against recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J. Clin. Oncol. 2015, 33, 2646–2654. [Google Scholar] [CrossRef] [PubMed]
- Morfouace, M.; Shelat, A.; Jacus, M.; Freeman, B.B.; Turner, D.; Robinson, S.; Zindy, F.; Wang, Y.D.; Finkelstein, D.; Ayrault, O.; et al. Pemetrexed and gemcitabine as combination therapy for the treatment of group 3 medulloblastoma. Cancer Cell 2014, 25, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Garancher, A.; Lin, C.Y.; Morabito, M.; Richer, W.; Rocques, N.; Larcher, M.; Bihannic, L.; Smith, K.; Miquel, C.; Leboucher, S.; et al. NRL and CRX define photoreceptor identity-specific dependencies in medulloblastoma. Cancer Cell 2018, 33, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Hovestadt, V.; Jones, D.T.; Picelli, S.; Wang, W.; Kool, M.; Northcott, P.A.; Sultan, M.; Stachurski, K.; Ryzhova, M.; Warnatz, H.J.; et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014, 510, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, S.C.; Doz, F.; Kool, M.; Dufour, C.; Vassal, G.; Milde, T.; et al. Risk stratification of childhood medulloblastoma in the molecular era: The current consensus. Acta Neuropathol. 2016, 131, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Ohgaki, H.; Xu, L.; Giangaspero, F.; Li, C.; Li, P.; Yang, Z.; Wang, B.; Wang, X.; Wang, Z.; et al. Molecular subgroups of adult medulloblastomas: Long-term single-institution study. Neuro Oncol. 2016, 18, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Preusser, M.; Brastianos, P.K.; Mawrin, C. Advances in meningioma genetics: Novel therapeutic opportunities. Nat. Rev. Neurol. 2018, 14, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Erson-Omay, Z.; Serin, A.; Yin, J.; Cotney, J.; Ozduman, K.; Avşar, T.; Li, J.; Murray, P.B.; Henegariu, O.; et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1 and SMO. Science 2013, 339, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Yuzawa, S.; Nishihara, H.; Tanaka, S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016, 33, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Brastianos, P.; Horowitz, P.; Santagata, S.; Jones, R.T.; McKenna, A.; Getz, G.; Ligon, K.L.; Palescandolo, E.; Van Hummelen, P.; Ducar, M.D.; et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 2013, 45, 285–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedalthagafi, M.; Bi, W.L.; Aizer, A.A.; Merrill, P.H.; Brewster, R.; Agarwalla, P.K.; Listewnik, M.L.; Dias-Santagata, D.; Thorner, A.R.; Van Hummelen, P.; et al. Oncogenic PIK3CA mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016, 18, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Schrimpf, D.; Olar, A.; Koelsche, C.; Reuss, D.; Bissel, J.; Kratz, A.; Capper, D.; Schefzyk, S.; Hielscher, T.; et al. TERT promoter mutations and risk of recurrence in meningioma. J. Natl. Cancer Inst. 2016, 108, djv377. [Google Scholar] [CrossRef] [PubMed]
- Goutagny, S.; Nault, J.C.; Mallet, M.; Henin, D.; Rossi, J.Z.; Kalamarides, M. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014, 24, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Harmanci, A.S.; Bai, H.; Youngblood, M.W.; Lee, T.I.; Baranoski, J.F.; Ercan-Snecicek, A.G.; Abraham, B.J.; Weintraub, A.S.; Hnisz, D.; et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat. Genet. 2016, 48, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Harmanci, A.S.; Youngblood, W.W.; Clark, V.E.; Coskun, S.; Henegariu, O.; Duran, D.; Erson-Omay, E.Z.; Kaulen, L.D.; Lee, T.I.; Abraham, B.J.; et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat. Commun. 2017, 14, 14433. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.L.; Greenwald, N.F.; Abedalthagafi, M.; Wala, J.; Gibson, W.J.; Agarwalla, P.K.; Horowitz, P.; Schumcher, S.E.; Esaulava, E.; Mai, Y.; et al. Genomic landscape of high-grade meningiomas. NPJ Genom. Med. 2017, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Biczok, A.; Kraus, T.; Suchorska, B.; Terpolilli, N.A.; Torsteinsdottir, J.; Giese, A.; Tonn, J.C.; Schichor, C. Tert promoter mutation is associated with worse prognosis in WHO grade II and III meningiomas. J. Neuroncol. 2018, 139, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Galani, V.; Lampri, E.; Varouktsi, A.; Alexiou, G.; Mitselou, A. Genetic and epigenetic alterations in meningiomas. Clin. Neurol. Neurosurg. 2017, 158, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.J.; Aminoso, C.; Lopez-Marin, I.; Arjona, D.; Gonzalez-Gomez, P.; Alonso, M.E.; Lomas, J.; de Campos, J.M.; Kusak, M.E.; Vaquero, J.; et al. DNA methylation of multiple promoter-associated CpG islands in meningiomas: Relationship with the allelic status at 1p and 22q. Acta Neuropathol. 2004, 108, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Schrimpf, D.; Stichel, D.; Jones, D.; Hielscher, T.; Schefzyk, S.; Okonechikov, K.; Koelsche, C.; Reuss, D.E.; Capper, D.; et al. DNA methylation-based classification and grading system for meningioma: A multicenter, retrospective analysis. Lancet Oncol. 2017, 18, 682–694. [Google Scholar] [CrossRef]
- Smith, M.J. Germline and somatic mutations in meningiomas. Cancer Genet. 2015, 208, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.J.; O’Sullivan, J.; Bhaskar, S.S.; Hadfield, K.D.; Poke, G.; Caird, J.; Shareif, S.; Eccles, D.; Fitzpatrick, D.; Rawluk, D.; et al. Loss-of-function mutations in SMACE1 cause an inherited disorder of multiple spinal meningiomas. Nat. Genet. 2013, 45, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Poppleton, H.; Fuller, C.; Su, X.; Liu, Y.; Jensen, P.; Magdaleno, S.; Dalton, J.; Calabrese, C.; Board, J.; et al. Radial glial cells are candidate stem cells of ependymoma. Cancer Cell 2005, 8, 232–335. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.A.; Wright, K.D.; Poppleton, H.; Mohankumar, K.M.; Finkelstein, D.; Pounds, S.B.; Rand, V.; Leary, S.E.; White, E.; Eden, C.; et al. Cross-species genomics matches driver mutations and cell compartments to model ependymomas. Nature 2010, 466, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Mack, S.C.; Ryzhova, M.; Bender, S.; Sill, M.; Isserlin, R.; Benner, A.; Hielscher, T.; Milde, T.; Remke, M.; et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymomas. Cancer Cell 2011, 20, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Andreiuolo, F.; Puget, S.; Peyre, M.; Dantas-Barbosa, C.; Boddaert, N.; Philippe, C.; Mauguen, A.; Grill, J.; Varlet, P. Neuronal differentiation distinguishes supratentorial and infratentorial childhood ependymomas. Neuro Oncol. 2010, 12, 1126–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, M.; Mohankumar, K.M.; Punchihewa, C.; Weinlich, R.; Dalton, J.D.; Li, Y.; Lee, R.; Tatevossian, R.G.; Phoenix, T.N.; Thiruvenkatam, R.; et al. C11orf95-RELA fusions drive oncogenic NF-kB signaling in ependymoma. Nature 2014, 506, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Arora, S.; Szulewsky, F.; Juric-Sekhar, G.; Miyajima, Y.; Bolouri, H.; Yasui, Y.; Barber, J.; Kupp, R.; Dalton, J.; et al. A de novo mouse model of C11orf95-RELA fusion-driven ependymoma identifies driver functions in addition to NF-kB. Cell Rep. 2018, 23, 3787–3797. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.C.; Witt, H.; Piro, R.M.; Gu, L.; Zuyderduyn, S.; Stütz, A.M.; Wang, X.; Gallo, M.; Garzia, L.; Zayne, K.; et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014, 506, 445–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Wang, Y.; Wood, L.D.; Rodriguez, F.J.; Hruban, R.H.; Gallia, G.L.; Binder, Z.A.; Riggins, C.J.; et al. Exome sequencing of four rare control nervous system tumor types. Oncotarget 2013, 4, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, K.M.; Currle, D.S.; White, E.; Boulos, N.; Dapper, J.; Eden, C.; Nimmervoll, B.; Thiruvenkatam, R.; Connelly, M.; Kranenburg, T.A.; et al. An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. Nat. Genet. 2015, 47, 878–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Panwalkar, P.; Calrk, J.; Ramaswamy, V.; Hawes, D.; Yang, F.; Dunham, C.; Yip, S.; Hukin, J.; Sun, Y.; Schipper, M.J.; et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017, 134, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, J.; Mukherjee, P.; Lu, C.; Jain, S.U.; Chung, C.; Martinez, D.; Sabari, B.; Matgol, A.S.; Panwalkar, P.; Parolia, A.; et al. Lowered H3K27me3 and DNA hypomethylastion define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl. Med. 2016, 8, 366ra161. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Wen, J.; Sill, M.; Lin, T.; Orisme, W.; Tang, B.; Hubner, J.M.; Ramaswamy, V.; Jia, S.; Dalton, J.D.; et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018, 136, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, F.M.G.; Hubner, J.M.; Sharma, T.; Luu, B.; Sill, M.; Zapotochy, M.; Mock, K.; Witt, H.; Lin, T.; Shih, D.J.H.; et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018, 136, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Gojo, J.; Lotsch, D.; Spiegl-Kreinecker, S.; Pajtler, W.W.; Neumayer, K.; Korbel, P.; Araki, A.; Brondstetter, A.; Mokr, T.; Hovestadt, V.; et al. Telomerase activation in posterior fossa group A ependymomas is associated with dismal prognosis and chromosome 1q gain. Neuro Oncol. 2017, 19, 1183–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajtler, K.W.; Mack, S.C.; Ramaswamy, V.; Smith, C.A.; Witt, H.; Smith, A.; Hausford, J.R.; von Hoff, K.; Wright, K.D.; Hwang, E.; et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017, 133, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Dahiya, S.; Li, T.; Fulton, R.S.; Smyth, M.D.; Dunn, G.P.; Rubin, J.B.; Mardis, E.R. Resistance-promoting effects of ependymoma treatment revealed through genomic analysis of multiple recurrences in a single patient. Cold Spring Harb. Mol. Case Stud. 2018, 4, a002444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mack, S.C.; Pajtler, K.W.; Chavez, L.; Okonechnikov, K.; Bertrand, K.C.; Wang, X.; Erkek, S.; Federation, A.; Song, A.; Lee, C.; et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 2018, 553, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Chung, C.K.; Ohn, J.H.; Kim, C.H. The similarities and differences between intracranial and spinal ependymomas: A review from a genetic research perspective. J. Korean Neurosurg. Soc. 2016, 59, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Pomper, M.G.; Passe, T.J.; Burger, P.C.; Scheithauer, B.W.; Brat, D.J. Chordoid glioma: A neoplasm unique to the hypothalamus and anterior third ventricle. Am. J. Neuroradiol. 2001, 22, 464–469. [Google Scholar] [PubMed]
- Bielle, F. Chordoid gliomas of the third ventricle share TTF-1 expression with organum vascolosum of the lamina terminalis. Am. J. Surg. Pathol. 2015, 39, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.; Simeonova, I.; Bielle, F.; Verreault, M.; Bauce, B.; Le Roux, I.; Daniau, M.; Nadaradjane, A.; Gleize, V.; Paris, S.; et al. A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas. Nat. Commun. 2018, 9, 2371. [Google Scholar] [CrossRef] [PubMed]
- Fuentealba, L.C.; Obernier, K.; Alvarez-Buylla, A. Adult neural stem cells bridge their niche. Cell Stem Cell 2012, 10, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Gaga, F.H.; Temple, S. Neural stem cells: Generating and regenerating the brain. Neuron 2013, 80, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.; Gascon, S.; Masserdotti, G.; Deshpande, A.; Simon, C.; Fischer, J.; Dimou, L.; Chichung Lie, D.; Schroeder, T.; et al. Oligodendrogliogenic and neurogenic adult subependymal zone neural stem cells constitute distinct lineages and exhibit differential responsiveness to Wnt signaling. Nat. Cell Biol. 2013, 15, 602–613. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, F.; Michel, J.; Baumgart, E.V. Fast clonal expansion and limited neural stam cell-renewal in the adult subpendymal zone. Nat. Neurosci. 2015, 18, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Bonfanti, L. Adult neurogenesis 50 years later: Limits and opportunities in mammals. Front. Nurosci. 2016, 10, 443. [Google Scholar] [CrossRef] [PubMed]
- Sanai, N.; Alvarez-Buylla, A.; Berger, M.S. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005, 313, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Raff, M. Oligodendrocyte cells reprogrammed to become multipotential CNS stem cells. Science 2009, 289, 1754–1757. [Google Scholar] [CrossRef]
- Boulanger, J.J.; Messier, C. From precursors to myelinating oligodendrocytes: Contribution of intrinsic and extrinsic factors to white matter plasticity in the adult brain. Neuroscience 2014, 269, 343–366. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.P.; Miyawaki, A.; Gage, F.H.; Jan, Y.N.; Jan, L.Y. Local generation of glia as a major astrocyte source in postnatal cortex. Nature 2012, 484, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Laug, D.; Galsgow, S.M.; Deneen, B. A glial blueprint for gliomagenesis. Nat. Rev. Nuerosci. 2018, 19, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Ravanelli, A.M.; Appel, B. Motor neurons and oligodendrocytes arise from distinct lineages by progenitor recruitment. Genes Dev. 2015, 29, 2504–2515. [Google Scholar] [CrossRef] [PubMed]
- Dimou, L.; Gallo, V. NG2-glia and their functions in the central nervous system. Glia 2015, 63, 1429–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiyama, A.; Boshans, L.; Goncalves, C.M.; Wegzryn, J.; Patel, K.D. Lineage, fate, and fate potential of NG2-glia. Brain Res. 2016, 1638, 116–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, P.S.; Perfilieva, E.; Bjort-Eriksson, T.; Alborn, A.M.; Nodborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in human adult hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Goritz, C.; Frisal, J. Neural stem cells. Neurogenesis in the adult. Cell Stem Cell 2012, 10, 637–659. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, R.D.; Curtis, M.A.; Spalding, K.L.; Bucholz, B.A.; Fink, D.; Bjork-Eriksson, T.; Nordborg, C.; Gage, F.H.; Druid, H.; Eirksson, P.S.; et al. Neocortical neurogenesis in humans is restricted to development. Proc. Natl. Acad. Sci. USA 2006, 103, 12564–12568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spalding, K.L.; Bergmann, O.; Alkass, K.; Bernard, S.; Salehpar, M.; Hottner, H.B.; Bostrom, E.; Westerlund, I.; Vial, C.; Bucholr, B.A.; et al. Dynamics of hippocampal neurogenesis in human adults. Cell 2013, 153, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Boldrini, M.; Fulmore, C.A.; Tartt, A.N.; Simeon, L.R.; Pavlova, I.; Poposka, I.; Poposka, V.; Rosoklija, G.B.; Stankov, A.; Arango, V.; et al. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 2018, 22, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, S.F.; Paredes, M.F.; Cabriau-Silla, A.; Sandoval, K.; Qu, D.; Kelly, K.W.; James, D.; Mayer, S.; Chang, J.; Augusta, K.I.; et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 2018, 355, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, B.A.; Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eye wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, T.N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human cortical glial tumors contain neural stem like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, H.D.; Nakano, I.; Lazareff, J.A.; Masterman-Smith, M.; Geschwind, D.H.; Bronner-Fraser, M.; Kornblum, H.I. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 15178–15183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Cattaneo, E. Neural stem cell systems: Physiological players or in vitro entities? Nat. Rev. Neurosci. 2010, 11, 167–187. [Google Scholar]
- Wurdak, H.; Zhu, S.; Romero, A.; Lorger, M.; Watson, J.; Chiang, C.Y.; Zhang, J.; Natu, V.S.; Lairson, L.L.; Walker, J.R. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell 2010, 6, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumor initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Joo, K.M.; Kim, J.; Jin, J.; Kim, M.; Seol, H.J.; Muradov, J.; Yang, H.; Choi, Y.L.; Park, W.Y.; Kong, D.S.; et al. Patient-specific orthoptic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastoma in situ. Cell Rep. 2013, 3, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Beier, D.; Hau, P.; Proescholdt, M.; Lohemeier, A.; Wischhusen, J.; Oefner, P.J.; Aiogner, L.; Brawanski, A.; Bogdahn, U.; Beier, C.P. CD133+ and CD133− glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007, 67, 4010–4015. [Google Scholar] [CrossRef] [PubMed]
- Joo, K.M.; Kim, S.Y.; Jin, X.; Song, S.Y.; Kong, D.S.; Lee, J.I.; Jeon, J.W.; Kim, M.H.; Kang, B.G.; Jung, Y. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastoma. Lab. Investig. 2008, 88, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sakariassen, P.O.; Tsinkalovsky, O.; Imervoll, H.; Boe, S.O.; Svendsen, A.; Prestegarden, L.; Rosland, G.; Thorsen, F.; Stuhr, L. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int. J. Cancer 2008, 122, 761–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Nishimura, M.C.; Bumbaca, S.M.; Kharbanda, S.; Forrest, W.F.; Kasman, I.M.; Greve, J.M.; Soriano, R.H.; Gilmour, L.L.; Rivers, C.S. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell 2010, 17, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Clement, V.; Dutoit, V.; Marino, D.; Dietrich, P.Y.; Radanovic, I. Limits of CD133 as a marker of glioma-renewing cells. Int. J. Cancer 2009, 125, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Clement, V.; Marino, D.; Cudalbu, C.; Hamou, M.F.; Mlynarik, V.; De Tribolet, N.; Dietrich, P.Y.; Gruetter, R.; Hegi, M.E.; Radanovic, I. Marker-independent identification of glioma-initiating cells. Nat. Metods 2010, 7, 224–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brescia, P.; Ortensi, B.; Fornosari, L.; Levi, D.; Broggi, G.; Pelicci, G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells 2013, 31, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jiang, Y.; Zou, F.; Liu, Y.; Wang, S.; Xu, N.; Xu, W.; Cui, C.; Xing, Y.; Liu, Y.; et al. Activation of PI3K/AKT pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 6829–6834. [Google Scholar] [CrossRef] [PubMed]
- Gaedicke, S.; Braun, F.; Prasad, S.; Machein, M.; Firat, E.; Hettich, M.; Gudihal, R.; Zhu, X.; Klingner, K.; Schüler, J.; et al. Noninvasive positron emission tomography and fluorescence imaging of CD133+ tumor stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E692–E701. [Google Scholar] [CrossRef] [PubMed]
- Dahlroth, R.H.; Hermansen, S.K.; Hansen, S.; Kristensen, B.W. What is the clinical value of cancer stem cells markers in gliomas? Int. J. Clin. Exp. Pathol. 2013, 6, 334–348. [Google Scholar]
- Lubanska, D.; Market-Velker, B.; De Carvalho, A.; Mikkelsen, T.; Fidalgo da Silva, E.; Porter, L.A. The cyclin-like protein Spy1 regulates growth and division characteristics of The CD133+ population in human glioma. Cancer Cell 2014, 25, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Woolard, K.; Nam, D.H.; Lee, J.; Fine, H.A. SSEA-1 is an enrichement marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 2009, 4, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Vora, P.; Venugopal, C.; McFarlane, N.; Singh, S.K. Culture and isolation of brain tumor initiating cells. Curr. Protoc. Stem Cell Biol. 2015, 34, 1–13. [Google Scholar]
- Lathia, J.D.; Gallagher, J.; Heddleston, J.M.; Wang, J.; Eyler, C.E.; Macwords, J.; Wu, Q.; Vasanji, A.; McLendon, R.E.; Hjemeland, A.B. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell 2010, 6, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Acquaviva, J.; Ramachandran, P.; Boskovitrz, A.; Woolfenden, S.; Pfanni, R.; Bronson, R.; Chen, J.; Weeissieder, R.; Housman, D.; et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 2712–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, M.; Nambu, E.; Furuyama, N.; Yoshida, Y.; Takino, T.; Hayashi, Y.; Sato, H.; Sai, Y.; Tsuji, T.; Miyamoto, K.; et al. Integrin α3 is overexpressed in glioma stem-like cells and promotes invasion. Br. J. Cancer 2013, 108, 2516–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podergajs, N.; Motaln, H.; Rajcevic, U.; Verbovšek, U.; Koršič, M.; Obad, N.; Espedal, H.; Vittori, M.; Herold-Mende, C.; Miletic, H.; et al. Transmembrane protein CD9 is glioblastoma biomarker for maintenance of glioblastoma stem cells. Oncotarget 2016, 7, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Li, M.; Sinyuk, M.; Alvarado, A.G.; Flavahan, W.A.; Stoltz, K.; Rosager, A.M.; Hale, J.; Hitomi, M.; Gallagher, J.; et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule A as a cancer stem cell maintenance factor. Cell Rep. 2014, 6, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, A.G.; Turaga, S.M.; Sathyan, P.; Mulkearns-Hubert, E.E.; Otvos, B.; Silver, D.J.; Hale, J.S.; Flavahan, W.A.; Zinn, P.O.; Sinyuk, M.; et al. Coordination of self-renewal in glioblastoma by integration of adhesion and microRNA signaling. Nat. Oncol. 2016, 18, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Jung, J.E.; Beck, S.; Kim, H. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 2013, 433, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Anand, R.; Mandalaparty, C.; Suryadevara, R.; Sriranga, P.U.; Michelhaugh, S.K.; Cazacu, S.; Finniss, S.; Thakur, A.; Lum, L.G.; et al. Preferential expression of functional IL-17R in glioma stem cells: Potential role in self-renewal. Oncotarget 2016, 7, 6121–6135. [Google Scholar] [CrossRef] [PubMed]
- Drachsler, M.; Kleber, S.; Mateos, A.; Volk, K.; Mohr, N.; Chen, S.; Cirovic, B.; Tüttenberg, J.; Gieffers, C.; Sykora, J.; et al. CD95 maintains stem cell-like and non-classical EMT programs in primary human glioblastoma cells. Cell Death Dis. 2016, 7, e2209. [Google Scholar] [CrossRef] [PubMed]
- Mazzoleni, S.; Politi, L.; Pala, L.; Caminelli, M.; Franzin, A.; Sergi, L.S.; Falini, A.; De Palma, M.; Bulfone, A.; Poliani, P.; et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010, 70, 7500–7513. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chadalavadam, K.; Wilshire, J.; Kowalik, U.; Howinga, K.; Geber, A.; Fligelman, B.; Leverska, M.; Brennan, C.; Tabar, V. Glioblastoma stem-like cells give rise to tumor endothelium. Nature 2010, 468, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Nathanson, D.A.; Gini, B.; Mottahedeh, J.; Visnyei, K.; Koga, T.; Gomez, G.; Eskin, A.; Hwang, K.; Wang, J.; Masui, K.; et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014, 343, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Chu, J.; Chan, W.K.; Zhang, J.; Wang, Y.; Cohen, J.B.; Victor, A.; Meisen, W.H.; Kim, S.H.; Grandi, P.; et al. CAR-engineered NK cells targeting wild-type EGFR and EGFRvIII enhance killing of glioblastoma and patient-derived stem cells. Sci. Rep. 2015, 5, 11483. [Google Scholar] [CrossRef] [PubMed]
- Day, B.; Stringer, B.; Al-Ejeh, F.; Ting, M.J.; Wilson, J.; Ensbey, K.S.; Jamieson, P.R.; Bruce, Z.C.; Lim, Y.C.; Offenhäuser, C.; et al. Eph A3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell 2013, 23, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Binda, E.; Visioli, A.; Giani, F.; Lamorte, G.; Copetti, M.; Pitter, K.L.; Huse, J.T.; Cajola, L.; Zanetti, N.; DiMeco, F.; et al. The Eph A2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell 2012, 22, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Gale, N.W.; Guo, H.; Qian, J.; Petty, A.; Kaspar, J.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.; Hambardzumyan, D.; et al. Eph A2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene 2015, 34, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitani, L.; Pallini, R.; Biffoni, M.; Todaro, M.; Cenci, T.; Maira, G.; Parati, E.A.; Stassi, G.; Larocca, L.M.; De Maria, R. Tumor vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 2010, 468, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.M.; Kim, S.H.; Jin, X.; Park, J.B.; Kim, S.H.; Joshi, K.; Nakano, I.; Kim, H. Crosstalk between glioma-initiating cells and endothelial cells drives tumor progression. Cancer Res. 2014, 74, 4482–4492. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Jorg, G.J.; Cavalli, F.; Richards, L.; Nguyen, L.; Vanner, R.; Guilhemon, P.; Lee, L.; Kushida, M.; Pellacani, D.; et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 2017, 549, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visvader, J.E. Cells of origin in cancer. Nature 2011, 469, 314–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Guignard, F.; Zhao, D.; Liu, L.; Bums, D.K.; Mason, R.P.; Messing, A.; Parada, L.F. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 2005, 8, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AlcantaraLlaguno, S.; Chen, J.; Kwon, C.H.; Jackson, E.L.; Li, Y.; Durns, D.K.; Alvarez-Buylla, A.; Parada, L.F. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 2009, 15, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.; Zheng, H.; Tamasek, G.J.; Zhang, P.; McKeever, P.E.; Lee, E.Y.; Zhu, Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell 2009, 15, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Bachoo, R.M.; Maher, E.A.; Ligon, K.L.; Sharpless, N.E.; Chan, S.S.; You, M.J.; Tang, Y.; DeFrances, J.; Stover, E.; Weissleder, R. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 2002, 1, 269–277. [Google Scholar] [CrossRef]
- Lindberg, N.; Kastemar, M.; Olofsson, T.; Smits, A.; Uhrbom, L. Oligodendrocyte progenitor cells as origin for experimental glioma. Oncogene 2009, 28, 2266–2275. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sage, J.C.; Miller, M.R.; Verhaak, R.; Hoppenmeyer, S.; Vogel, H.; Foreman, O.; Bonson, R.T.; Nishiyama, A.; Luo, L.; et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 2011, 146, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, Q.; Kutlu, B.; Difilippantonio, S.; Bash, R.; Gilbert, D.; Yin, C.; O’Sullivan, T.N.; Yang, C.; Kozlov, S.; et al. Evolutionary etiology of high-grade astrocytomas. Proc. Natl. Acad. Sci. USA 2013, 110, 17933–17938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonabend, A.; Bansal, M.; Guarnieri, P.; Lei, L.; Amendolara, B.; Soderquist, C.; Leung, R.; Yun, J.; Kennedy, B.; Sisti, J.; et al. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 2014, 74, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, P.C.; Hiller, D.J.; Chen, A.J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma/stem progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, F.; Liu, C. Rivisit the candidacy of brain cell types as the cell(s) of origin for human high-grade glioma. Front. Mol. Neurosci. 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J.E.; Kahng, J.Y.; Kim, S.H.; Park, J.S.; Yoon, S.Y.; Um, J.Y.; Kim, W.K.; Lee, J.K.; Park, J.; et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018, 560, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Qin, E.Y.; Cooper, D.; Abbott, K.L.; Lennon, J.; Nagaraja, S.; MacKay, A.; Jones, C.; Vogel, H.; Jackson, P.K.; Menje, M. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell 2017, 170, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Poppleton, H.; Kocak, M.; Hogg, T.L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A perivascular niche for brain tumor cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Folkius, C.; Shaked, Y.; Man, S.; Tang, T.; Lee, C.R.; Zhu, Z.; Hoffman, R.M.; Kerbel, R.S. Glioma tumor stem-like promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res 2009, 69, 7243–7251. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; McKey, R.M.; Parada, L.F. Malignant glioma: Lessons from genomics, mouse models and stem cells. Cell 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Huang, Z.; Zhou, W.; Wu, Q.; Donnola, S.; Liu, J.K.; Fang, X.; Sloan, A.E.; Mao, Y.; Lathia, J.D.; et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 2013, 153, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Infanger, D.W.; Cho, Y.; Lopez, B.S.; Mohanan, S.; Liu, S.C.; Gursel, D.; Boockvar, J.A.; Fischbach, C. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res. 2013, 73, 7079–7089. [Google Scholar] [CrossRef] [PubMed]
- Pietras, A.; Katz, A.; Ekstrom, E.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ke, S.O.Q.; Huang, Z.; Flavahan, W.; Fang, X.; Paul, J.; Wu, L.; Sloan, A.E.; McLendon, R.E.; Li, X.; et al. Periostin secreted by glioblastoma stem cells recruits M2 tumor-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Whang, X.; Proger, B.C.; Wu, Q.; Kim, L.; Gimple, R.C.; Shi, Y.; Yang, K.; Marton, A.R.; Zhou, W.; Zhu, Z.; et al. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell 2018, 22, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth following chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Anido, J.; Saez-Borderias, A.; Gonzalez-Juncà, A.; Rodón, L.; Folch, G.; Carmona, M.A.; Prieto-Sánchez, R.M.; Barba, I.; Martínez-Sáez, E.; Prudkin, L.; et al. TGF-β receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma. Cancer Cell 2010, 18, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, G.; Cesaroni, M.; Senesi, M.; de Vries, N.; Hulsman, D.; Bruggeman, S.W.; Lancini, C.; van Lohuizen, M. In vivo RNAi screen for BMI1 targets identifies identifies TGF-β/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 2013, 23, 660–676. [Google Scholar] [CrossRef] [PubMed]
- Soroceanu, L.; Murase, R.; Limbad, C.; Singer, E.; Allison, J.; Adrados, I.; Kawamura, R.; Pakdel, A.; Fukuyo, Y.; Nguyen, D.; et al. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2012, 73, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.E.; Granot, Z.; Coker, C.; Iavarone, A.; Hambardzumyan, D.; Holland, E.C.; Nam, H.S.; Benezra, R. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell 2012, 21, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jeon, H.M.; Jin, X.; Kim, E.J.; Yin, J.; Jeon, H.Y.; Sohn, Y.W.; Oh, S.Y.; Kim, J.K.; Kim, S.H.; et al. The ID1-CULLIN3 axis regulates intracelluklar SHH and WNT signaling in glioblastoma stem cells. Cell Rep. 2016, 16, 1629–1641. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Kim, L.; Dixit, D.; Jeon, H.Y.; Kim, E.J.; Kim, J.K.; Lee, S.Y.; Yin, J.; Rich, J.N.; et al. Inhibition of ID1-BMPR2 intrinsic signaling sensitizes stem cells to differentiation therapy. Cancer Res. 2017, 24, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.S.; Johung, T.B.; Caretti, V.; Noll, A.; Tang, Y.; Nagaraja, S.; Gibson, E.M.; Mount, C.W.; Polepalli, J.; Mitra, S.S.; et al. Neuronal activity promotes glioma growth through neuroglin-3 secretion. Cell 2015, 161, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.S.; Tam, L.T.; Woo, P.J.; Lennon, J.; Nagaraja, S.; Gillespie, S.M.; Ni, J.; Duveau, D.Y.; Morris, P.J.; Zhao, J.J.; et al. Targeting neuronal activity-regulated nuroligin-3 dependency in high-grade glioma. Nature 2017, 547, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Qin, X.P.; Zhuang, Y.; Zhang, Y.; Liao, H.B.; Tang, J.C.; Pam, M.X.; Zang, F.F.; Lei, Y.; Wang, S.; et al. Glioblastoma recurrence correlates with NLGN3 levels. Cancer Med. 2018, 7, 2848–2859. [Google Scholar] [CrossRef] [PubMed]
- Alavarado, A.G.; Thiagarajan, P.S.; Mulkearns-Hubert, E.E.; Silver, D.J.; Hale, J.S.; Alban, T.J.; Turaga, S.M.; Jarrar, A.; Reizes, O.; Congworth, M.S.; et al. Glioblastoma cancer stem cells evade innate immune suppression of self-renewal through reduced TLR4 expression. Cell Stem Cell 2017, 20, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Liau, B.B.; Sievers, C.; Donohue, L.K.; Gillespie, S.M.; Flavahan, W.A.; Miller, T.E.; Venteicher, A.; Hebert, C.H.; Carey, C.D.; Rodig, S.J.; et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 2017, 20, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.K.; Wang, Y.; Belz, T.; Bock, D.; Takacs, A.; Radlwimmer, B.; Barbus, S.; Reifenberger, G.; Lichter, P.; Schütz, G. The nuclear receptor tailless induces long-term neural stem cells expansion and brain initiation. Genes Dev. 2010, 24, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Khan, M.A.; Weiler, M.; Blaes, J.; Jestaedt, L.; Geibert, M.; Zou, P.; Gronych, J.; Bernhardt, O.; Korshunov, A.; et al. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell 2014, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA 2013, 110, 8644–8649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliday, J.; Helmy, K.; Pattwell, S.; Pitter, K.L.; LaPlant, Q.; Ozawa, T.; Holland, E.C. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc. Natl. Acad. Sci. USA 2014, 111, 5248–5253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; Della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kathagen, A.; Schulte, A.; Balcke, G.; Phillips, H.S.; Martens, T.; Matschke, J.; Günther, H.S.; Soriano, R.; Modrusan, Z.; Sandmann, T.; et al. Hypoxia and oxygenation induce a metabolic switch between pentose phosphate pathway and glycolysis in glioma stem-like cells. Acta Neuropathol. 2013, 126, 763–780. [Google Scholar] [CrossRef] [PubMed]
- Ran, C.; Liu, H.; Hitoshi, Y.; Israel, M.A. Proliferation-independent control of tumor glycolysis by PDGFR-mediated AKT activation. Cancer Res. 2013, 76, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, M.; Suva, M.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clément-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.; Kim, L.; Wang, X.; Wu, Q.; Sanvoranat, T.; Hubert, C.; Prager, B.; Wlaface, L.; Jin, X.; Mack, S.; et al. Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight 2017, 2, e90019. [Google Scholar] [CrossRef] [PubMed]
- Eyler, C.E.; Wu, Q.; Yan, K.; MacSwords, J.M.; Chandler-Militello, D.; Misuraca, K.L.; Lathia, J.D.; Forrester, M.; Lee, J.; Starnier, J.; et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell 2011, 146, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Chhipa, R.R.; Fan, Q.; Anderson, J.; Muraleedharan, R.; Huang, Y.; Ciraolo, G.; Chen, X.; Waclaw, R.; Cow, L.M.; Kchuchua, Z.; et al. AMPKinase promotes glioblastoma bioenergetics and tumor growth. Nat. Cell Biol. 2018, 20, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Mair, R.; Wright, A.; Ros, S.; Ho, D.E.; Booth, T.C.; Kreis, F.; Rao, J.; Watts, C.; Brindle, K.M. Metabolic imaging detects low levels of glycolytic activity that vary with levels of c-Myc expression in patient-derived xenograft models of glioblastoma. Cancer Res. 2018, 78, 5408–5418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, D.A.; Horbinski, C.M.; Hashizume, R.; James, C.D. Therapeutic hypothesis testing with rat brain tumor models. Neurotherapeutics 2017, 14, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Soeda, A.; Hara, A.; Kunisada, T.; Yoshimura, S.; Iwama, T.; Park, D.M. The evidence of glioblastoma heterogeneity. Sci. Rep. 2015, 5, 7979. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Carla da Hora, C.; Kantar, R.S.; Nakano, I.; Wakimoto, H.; Batchelor, T.T.; Chiocca, E.A.; Badr, C.E.; Tannous, B.A. Dissecting inherent intratumor heterogeneity in patient-derived glioblastoma culture models. Neuro Oncol. 2017, 19, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Ha, G.; Tseng, Y.Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, J.S.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2107, 49, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma model using human cerebral organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, C.G. In search of medulloblastoma: Neural stem cells and embryonal brain tumors. Neurosurg. Clin. N. Am. 2007, 18, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.J.; Ellison, D.W. The origins of medulloblastoma subtypes. Ann. Rev. Pathol. 2008, 3, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Ellis, T.; Markant, S.; Reard, T.A.; Kessler, J.; Bourboulas, M.; Schuller, U.; Machold, R.; Fishell, G.; Rowitch, D.; et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008, 14, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Schuller, U.; Heine, V.; Mao, J.; Kho, A.; Dillon, A.; Han, Y.G.; Huillard, E.; Sun, T.; Ligon, A.; Qian, Y.; et al. Acquisition of granule neuron precursor identitity is a critical determinant of progenitor cell competence to form SHH-induced medulloblastoma. Cancer Cell 2008, 14, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Shimada, I.S.; Hwang, S.H.; Somatilaka, B.N.; Wang, X.; Showron, P.; Kim, J.; Kim, M.; Shelton, J.M.; Rajaram, V.; Xuan, Z.; et al. Basal suppression of the sonic hedgehog pathway by the G-protein-coupled receptor Gpr161 restricts medulloblastoma pathogenesis. Cell Rep. 2018, 22, 1169–1184. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Aiken, C.; McClelland, R.; Morrison, L.C.; Tatari, N.; Remke, M. Characterization of novel biomarkers in selecting for subtype specific medulloblastoma phenotypes. Oncotarget 2015, 6, 38881–38900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Morrison Coudiere, L.; Tatari, N.; Stromecki, M.; Fresznova, A.; Porter, C.J.; Del Bigio, M.; Howkins, C.; Chan, J.A.; Ryken, T.C.; et al. CD271+ cells are diagnostic and prognostic and ehibit elevated MAPK activity in SHH medulloblastoma. Cancer Res. 2018, 78, 4745–4759. [Google Scholar] [CrossRef] [PubMed]
- Sutter, R.; Shakhova, O.; Bhagat, H.; Behesti, H.; Sutter, C.; Peukar, S.; Santuccione, A.; Bernays, P.; Heppuer, F.L.; Schuller, U.; et al. Cerebellar stem cells act as medulloblastoma-initiating cells in a mouse model and a neural stem cell signature characterizes a subset of human medulloblastomas. Oncogene 2010, 29, 1845–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, P.; Tang, Y.; Robinson, G.; Thompson, M.; Guttman, D.; Ellison, D.; Wechsler-Reya, R.; Gilbertson, R.J. Subtypes of medulloblastoma have distinct developmental origins. Nature 2010, 468, 1095–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromecki, M.; Tatari, N.; Coudière Morrison, L.; Kaur, R.; Zagozewski, J.; Palidwor, G.; Ramaswamy, V.; Skowron, P.; Wolff, M.; Milde, T.; et al. Characterization of a novel OTX2-driven stem cell program in group 3 and group 4 medulloblastoma. Mol. Oncol. 2018, 12, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Palm, T.; Figarella-Brauger, D.; Chapon, F.; Lacroix, C.; Gray, F.; Scaravilli, F.; Ellison, D.; Salmon, I.; Vikkula, M.; Goldfana, C. Expression profiling of ependymomas unravels localization and tumor grade-specific tumorigenesis. Cancer 2009, 115, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Baxter, P.A.; Voicu, H.; Gurusidappa, S.; Zhao, Y.; Adesina, A.; Man, T.K.; Shu, Q.; Zhang, Y.; Zhao, X.; et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 2010, 12, 580–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geha, S.; Pallaud, J.; Junier, M.P.; Devaux, B.; Leonard, N.; Chassoux, F.; Chneiweiss, H.; Dumas-Duport, C.; Varlet, P. NG2+/Oligo2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 2010, 20, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Persson, A.; Petrisch, C.; Swartling, F.; Itsara, M.; Sim, F.; Auvergne, R.; Goldenberg, D.; Vandenberg, S.; Nguyen, K.; Yakovenko, S.; et al. Non-stem cell origin for oligodendroglioma. Cell Stem Cell 2010, 18, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Venteicher, A.S.; Hebert, C.; Escalante, L.E.; Patel, A.P.; Yizhak, K.; Fisher, J.M.; Rodman, C.; Mount, C.; Filbin, M.G.; et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016, 539, 309–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalaamrides, M.; Stemmer-Rachamimov, M.; Chereyre, T.; Taranchon, E.; Han, Z.Y.; Martinelli, C.; Lusis, E.A.; Hegedry, B.; Gutmann, D.H.; Giovannini, M. Nf2 gene inactivation in arachnoidal cells is rate limiting for meningioma development in the mouse. Genes Dev. 2002, 16, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Kalamarides, M.; Niwa-Nawakita, M.; Leblois, H.; Abramowski, V.; Perricaudet, M.; Janin, A.; Thomas, G.; Gutmann, D.H.; Giovannini, M. Identification of a progenitor cell of origin capable of generating diverse meningioma histological subtypes. Oncogene 2011, 30, 2333–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyre, M.; Salaud, C.; Clermont-Tarachon, E.; Niwa-Kawakita, M.; Gontagny, S.; Mawrin, C.; Giovannini, M.; Kalamarides, M. PDGF activation in PGDS-positive arachnoid cells induces meningioma formation in mice promoting tumor progression in combination with Nrf2 and Cdkn2ab loss. Oncotarget 2015, 6, 32713–32722. [Google Scholar] [CrossRef] [PubMed]
- Hueng, D.Y.; Sytwu, S.M.; Huang, S.M.; Chang, C.; Ma, H.I. Isolation and characterization of tumor atem-like cells from human meningioma. J. Neuroncol. 2011, 104, 9. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, X.; Zhu, H.; Hua, L.; Li, J.; Xie, Q.; Chen, X.; Zhang, T.; Gong, Y. KLF4 is a tumor suppressor in anaplastic meningioma stem-like cells and human meningiomas. J. Mol. Cell. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.H.; Fang, D.; Kitange, G.; He, L.; Hu, Z.; Parney, I.F.; et al. A novel enhancer regulates MGMT expression and promotes tomozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef] [PubMed]
- Woroniecka, K.I.; Rhodin, K.E.; Chongsathidkiet, P.; Keith, K.A.; Fecci, P.E. T-cell dysfunction in glioblastoma: Applying a new framework. Clin. Cancer Res. 2018, 24, 3792–3802. [Google Scholar] [CrossRef] [PubMed]
- Woroniecka, K.I.; Chongsathidkiet, P.; Rhodin, K.; Kemeny, H.; Dechant, C.; Farber, H.; Elsamodicy, A.; Cui, X.; Kayama, S.; Jackson, C.; et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin. Cancer Res. 2018, 24, 4175–4186. [Google Scholar] [CrossRef] [PubMed]
- Mohme, M.; Schliffke, S.; Maire, C.; Runger, A.; Glan, L.; Mende, K.C.; Matschke, J.; Gehbaueer, C.; Akyuz, N.; Zapf, S.; et al. Immunophenotyping of newly diagnosed and recurrent glioblastoma defines distinct immune exhaustion profiles in peripheral and tumor-infiltrating lymphocytes. Clin. Cancer Res. 2018, 24, 4187–4200. [Google Scholar] [CrossRef] [PubMed]
- Chongsathidkiet, P.; Jackson, C.; Koyama, S.; Loebel, F.; Farber, S.H.; Woroniecka, K.; Elsamadicy, A.A.; Dechant, C.A.; Kemeny, H.R.; Sanchez-Perez, L.; et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 2018, 24, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.J.; Desai, A.S.; Linette, G.P.; June, C.H.; O’Rourke, D.M. CAR T cell therapy for glioblastoma: Recent clinical advances and future challenges. Neuro Oncol. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Brawley, V.; Hedge, M.; Bielamovicz, K.; Kalra, M.; Landi, D.; Robertson, C.; Gray, T.L.; Diof, O.; Wakefield, A.; et al. HER-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: A phase 1 dose-escalation trial. JAMA Oncol. 2017, 3, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Hedge, M.; Mukherjee, M.; Gnow, Z.; Pigna, M.; Navai, S.A.; Wakefield, A.; Fousek, K.; Bielamowicz, K.; Cho, W.; Brawley, V.S.; et al. Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J. Clin. Investig. 2016, 126, 3036–3052. [Google Scholar]
- Bielamowicz, K.; Fousek, K.; Byrd, T.T.; Samaha, H.; Mukheryww, M.; Aware, N.; Wu, M.F.; Orang, J.S.; Sumarein, P.; Man, T.K.; et al. Trivalent CAR T cells overcome interpatient antigenic variability in glioblastomas. Neuro Oncol. 2018, 20, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Badie, B.; Barish, M.E.; Weng, L.; Ostberg, J.R.; Chang, W.C.; Naranjo, A.; Starr, R.; Wagner, J.; Wright, C.; et al. Bioactivity and safety of IL13Rα2-redirected chimeric antigen receptor CD8+ T cells in patients with recurrent glioblastoma. Clin. Cnacer Res. 2015, 21, 4062–4072. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Alizadeh, D.; Starr, B.; Weng, L.; Wagner, J.R.; Naranjo, A.; Ostberg, J.R.; Blanchard, M.S.; Kilpatrick, J.; Simpson, J.; et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N. Engl. J. Med. 2016, 375, 2561–2569. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Aguilar, B.; Starr, B.; Yang, X.; Chang, W.C.; Weng, L.; Chang, B.; Sarkissian, A.; Brito, A.; Sanchez, J.F.; et al. Optimization of IL13Rα2-targeted dimeric antigen receptor T cells for improved anti-tumor efficacy against glioblastoma. Mol. Ther. 2018, 26, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Pituch, K.C.; Miska, J.; Krenciute, G.; Pauek, W.K.; Li, G.; Rodriguez-Cruz, T.; Han, Y.; Leoniak, M.S.; Gottschalk, S.; Balyanikova, I.V.; et al. Adoptive transfer of IL13Rα2-specific chimeric antigen receptor T cells creates a proinflammatory environment in glioblastoma. Mol. Ther. 2018, 26, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Aguilar, B.; Starr, R.; Aliradeh, D.; Brito, A.; Sarkisvian, A.; Ostberg, J.R.; Forman, S.J.; Brown, C.E. Glioblastoma-targeted CD4+ CAR T cells mediate superior antitumor activity. JCI Insigth 2018, 3, e99048. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, D.; Dietrich, P.Y.; Stupp, R.; Linette, G.P.; Posey, A.D.; June, C.H. CAR T-cell therapies in glioblastoma: A first look. Clin. Cancer Res. 2018, 24, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Pellegatta, S.; Savoldo, B.; Di Ianni, N.; Corbetta, C.; Chen, Y.; Patanè, M.; Sun, C.; Pollo, B.; Ferrone, S.; Di Meco, F.; et al. Constitutive and TNF-α-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: Implications for CAR-T cell therapy. Sci. Trnsl. Med. 2018, 10, eaoo2731. [Google Scholar] [CrossRef] [PubMed]
- Mount, C.W.; Majzner, R.G.; Sundaresh, S.; Arnold, E.P.; Kadapakkam, M.; Haile, S.; Labarrich, L.; Hullemn, E.; Woo, P.J.; Rietberg, S.P.; et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3K27M+ diffuse midline gliomas. Nat. Med. 2108, 24, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Weiss, T.; Weller, M.; Guckenberger, M.; Sentman, C.L.; Roth, P. NKG2D-based CAR T cells and radiotherapy exerts synergistic efficacy in glioblastoma. Cancer Res. 2018, 78, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Suryadevana, C.M.; Desai, R.; Abel, M.L.; Riccione, K.A.; Batich, K.A.; Shen, S.H.; Changsathidkiet, P.C.; Elsamidicy, A.A.; Snyder, D.J.; Hemdon, J.E.; et al. Temozolomide lymphodepletion enhances CAR abundance and correlates with antitumor efficacy against established glioblastoma. Oncoimmunology 2018, 7, e1434464. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European association for neuro-oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Wang, Z.F.; Cao, C.J.; Weng, H.; Xu, C.S.; Li, K.; Lan, J.; Zeng, X.T.; Li, Z.Q. The clinical significance of O6-Methyguanine-DNA methyltransferase promoter methylation status in adult patients with glioblastoma: A meta-analysis. Front. Neurol. 2018, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Cimino, P.J.; McFerrin, L.; Wirsching, H.G.; Arora, S.; Bolouri, H.; Rabadan, R.; Weller, M.; Holland, E.C. Copy number profiling across glioblastoma populations has implications for clinical trial design. Neuro Oncol. 2018, 20, 1368–1373. [Google Scholar] [CrossRef] [PubMed]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in gliuoiblastoma: A meta-analysis of clinical trials. J. Cell. Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, R.J.; Verbaan, D.; Lamba, S.; Zanon, C.; Jeuken, J.W.; Boots-Sprenger, S.H.; Wesseling, P.; Hulsebos, T.J.; Troost, D.; van Tilborg, A.A.; et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma beterr than either IDH1 or MGMT alone. Neuro Oncol. 2014, 16, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Lie, A.; Li, T.; Chowdhury, R.; Liu, F.; Ozer, B.; Wei, B.; Green, R.M.; Ellingson, B.M.; Wang, H.J.; et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017, 394, 394–404. [Google Scholar]
- Kessler, T.; Sahm, F.; Sadik, A.; Stichel, D.; Hertenstein, A.; Reifenberger, G.; Zacher, A.; Sabel, M.; Tabatabai, G.; Steinbach, J.; et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro Oncol. 2018, 20, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Pusch, S.; Krausert, S.; Fischer, V.; Balss, J.J.J.; Ott, M.; Schrimpf, D.; Capper, D.; Sahm, F.; Eisel, J.; Beck, A.C.; et al. Pan-IDH1 mutant inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017, 133, 629–644. [Google Scholar] [CrossRef] [PubMed]
- Philip, B.; Yu, D.X.; Silvis, M.R.; Shin, C.H.; Robinson, J.P.; Robinson, G.L.; Welker, A.E.; Angel, S.N.; Tripp, S.R.; Sonnen, J.A.; et al. Mutant IDH1 promotes glioma formation in vivo. Cell Rep. 2018, 23, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Kopinja, J.; Sevilla, R.S.; Levitan, D.; Dai, D.; Vanko, A.; Spooner, E.; Ware, C.; Forget, R.; Hu, K.; Kral, A.; et al. A brain penetrant mutant IDH1 inhibitor provides in vivo survival benefit. Sci. Rep. 2107, 7, 13853. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Levell, J.R.; Liu, G.; Caferro, T.; Sutton, J.; Shafer, C.M.; Coastales, A.; Manning, J.R.; Zhao, Q.; Sendzik, M.; et al. Discovery and evaluation of clinical candidate IDH305, a brain penetrant mutant IDH1 inhibitor. ACS Med. Chem. Lett. 2017, 8, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Manning, J.R.; Sutton, J.; Coastales, A.; Sendzik, M.; Shafer, C.M.; Levell, J.R.; Liu, G.; Caferro, T.; Cho, J.S.; et al. Optimization of 3-Pyridin-4-yl-oxazolidin-2-ones as orally bioavailable and brain penetrant mutant IDH1 inhibitors. Chem. Lett. 2018, 9, 746–751. [Google Scholar]
- Waitkins, M.S.; Diplas, B.H.; Yan, H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell 2018, 34, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Mellinghoff, I.K.; Tonat, M.; Maher, E.; DeLa Fuente, M.; Cloughesy, T.F.; Holdhoff, M.; Cote, G.M.; Burris, H.; Jnku, F.; Huang, R.; et al. AG-120, a first-in-class mutant IDH1 inhibitor in patients with recurrent or progressive IDH1 mutant glioma: Updated results from the phase 1 non-enhnacing glioma population. Nuro Oncol. 2017, 6, vi10–vi11. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Penas-Prado, M.; Peters, K.B.; Cloughesy, T.F.; Burris, H.A.; Maher, E.A.; Janku, F.; Cote, M.; DeLa Fuente, M.I.; Clarke, J.; et al. Phase 1 study of AG-881, an inhibitor of mutant IDH1/IDH2, in patients with advanced IDH-mutant solid tumors, including gliomas. J. Clin. Oncol. 2018, 36, 2002. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Arrillaga-Romany, I.C.; Ly, K.I.; Boguer, W.; Ratai, E.M.; Reitz, K.; Iafrate, A.J.; Dietrich, J.; Gerstner, E.R.; Chi, A.S.; et al. Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxy-glutarate. Nat. Commun. 2018, 9, 1474. [Google Scholar] [CrossRef] [PubMed]
- Sulkowski, P.L.; Corso, C.D.; Robinson, N.D.; Scanlon, S.E.; Purshouse, K.R.; Bai, H.; Liu, Y.; Sundaram, R.K.; Hegan, D.C.; Fons, N.R.; et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 2017, 9, eaal2463. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Kwintkiewicz, J.; Liu, Y.; Tech, K.; Frady, L.N.; Su, Y.T.; Bautista, W.; Moon, S.I.; MacDonald, J.; Ewend, M.G.; et al. Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mnediated DNA repair. Cancer Res. 2017, 77, 1709–1718. [Google Scholar] [CrossRef] [PubMed]
- Leseur, P.; Chevalier, F.; El-Habr, E.; Junier, M.P.; Chneiweiss, H.; Castera, L.; Muller, E.; Stefan, D.; Saintigny, Y. Radiosensitization effect of talazoparib, a Parp inhibitor, on glioblastoma stem cells exposed to low and high linear energy transfer radiation. Sci. Rep. 2018, 8, 3664. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, A.I.; Short, S.; Watts, C.; Herbert, C.; Morris, A.; Stobo, J. Phase I clinical trials evaluating oliparib in combination with radiotherapy (RT) and/or tremozolomide (TMZ) in glioblastoma patients: Results of OPARATIC and PARADIGM phase I and early results of PARADIGM-2. J. Clin. Oncol. 2018, 36, 2018. [Google Scholar] [CrossRef]
- Forget, A.; Martignetti, L.; Puget, S.; Calzone, L.; Brabetz, S.; Picard, D.; Montagut, A.; Liva, S.; Sta, A.; Dingli, F.; et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell 2018, 34, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Hargrave, D.R.; Moreno, L.; Broniscer, A.; Bouffet, E.; Aerts, I.; Andre, N.; Violet Shen, A.P.; Bertoz, A.I.; Cohen, K.J.; Dunkel, I.J.; et al. Dabrafenib in pediatric patients with BRAF V600-positive high-grade glioma. J. Clin. Oncol. 2018, 36, 10505. [Google Scholar] [CrossRef]
- Archer, T.C.; Ehrenberger, T.; Mundt, F.; Gold, M.P.; Krug, K.; Mah, C.K.; Mahoney, E.L.; Daniel, C.J.; LeNail, A.; Ramamoorthy, D.; et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell 2018, 34, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Zomerman, W.W.; Plasschaert, S.; Conroy, S.; Scherpen, F.J.; Meeuwsen-de Boer, T.; Lourens, H.J.; Llobet, S.G.; Smit, M.J.; Menkem, L.S.; Seitz, A.; et al. Identification of two protein-signaling states delineating transcriptionally heterogeneous human medulloblastoma. Cell Rep. 2018, 22, 3206–3216. [Google Scholar] [CrossRef] [PubMed]
- Abadal, K.B.; Walsh, M.A.; Yachnis, A.T.; Tran, D.D.; Ghiaseddin, A.P. Elen months progression-free survival on vemurafenib monotherapy in a patient with recurrent and metastatic BRAF V600E-mutated glioblastoma WHO grade 4. JCO Precis. Oncol. 2017, 11, 1–5. [Google Scholar]
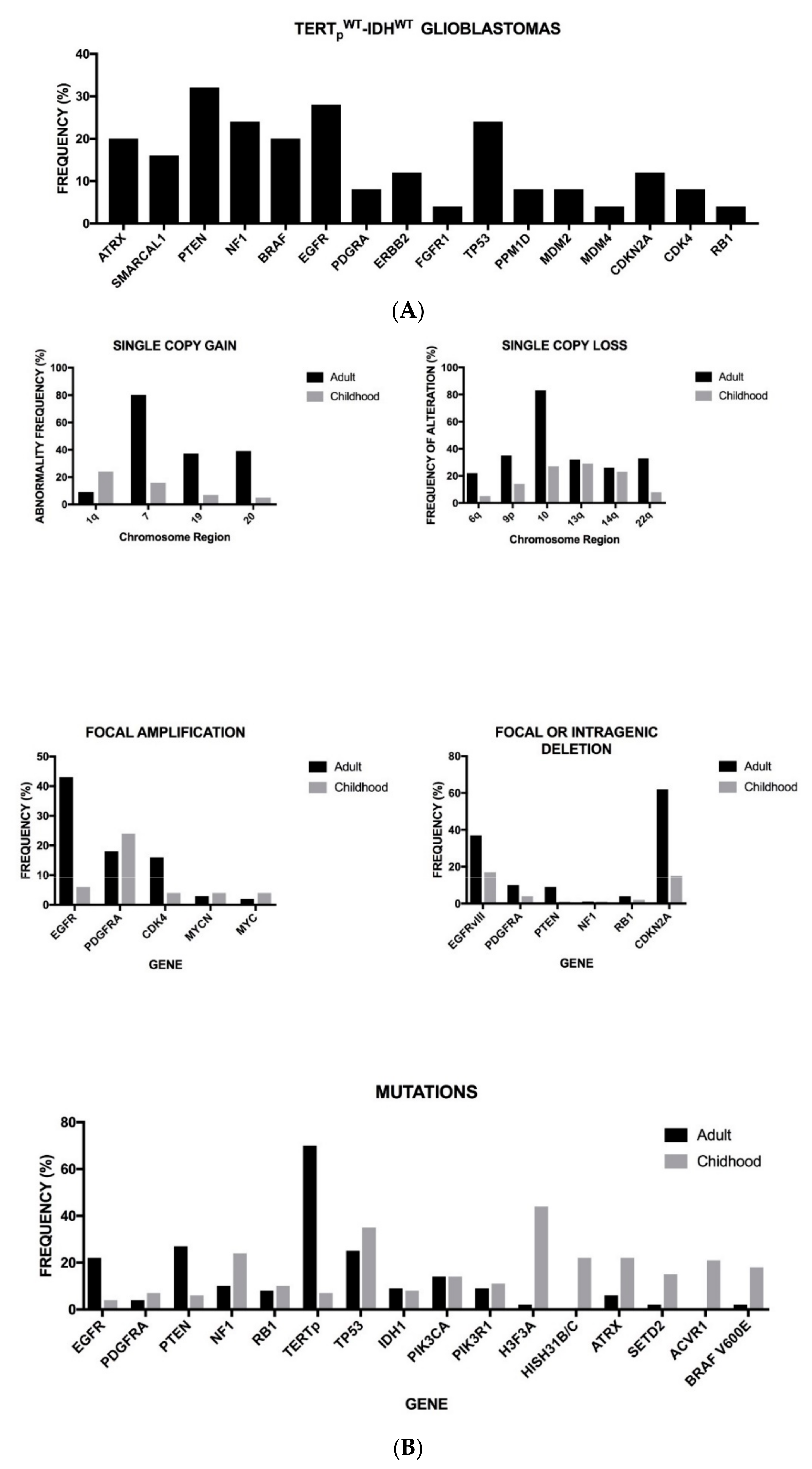

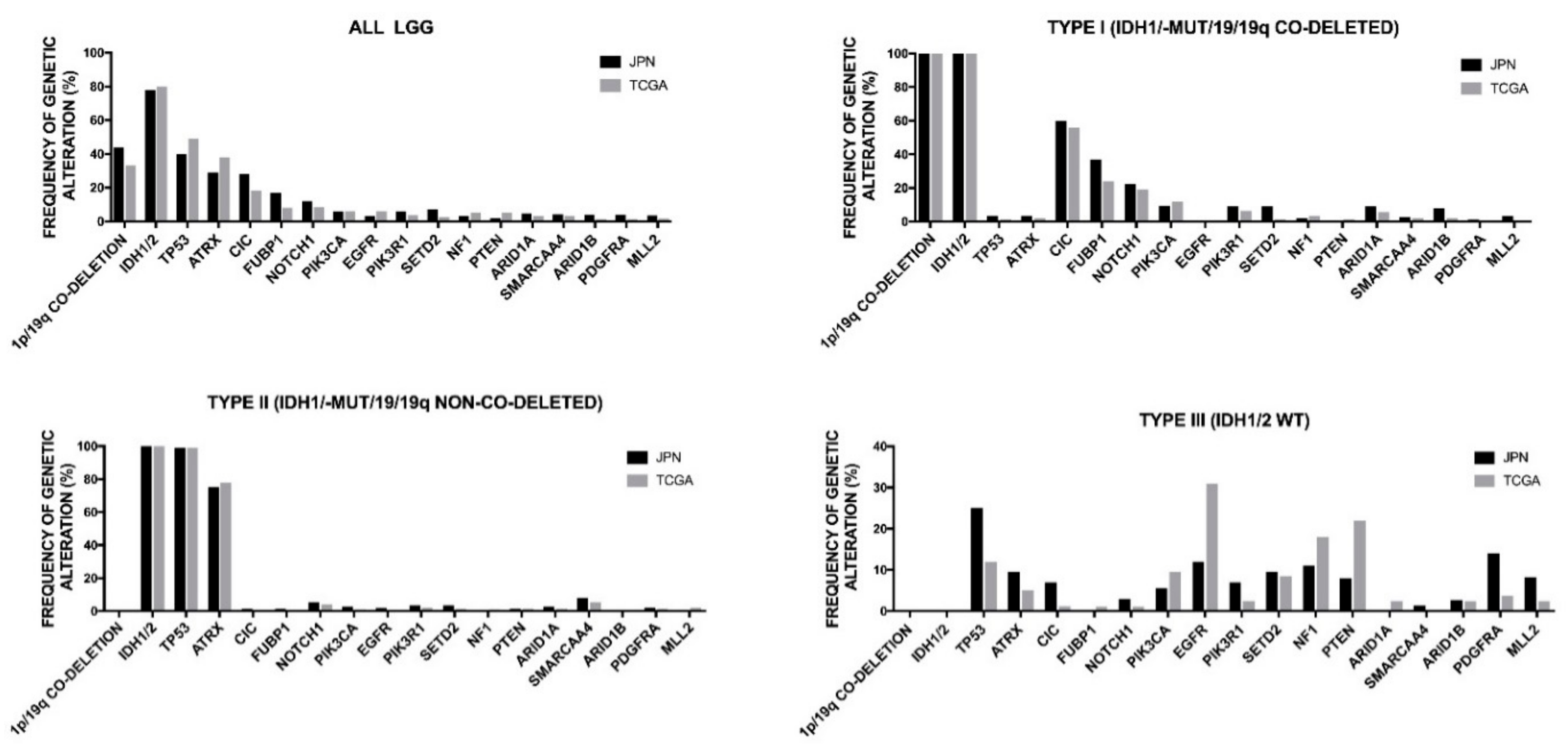





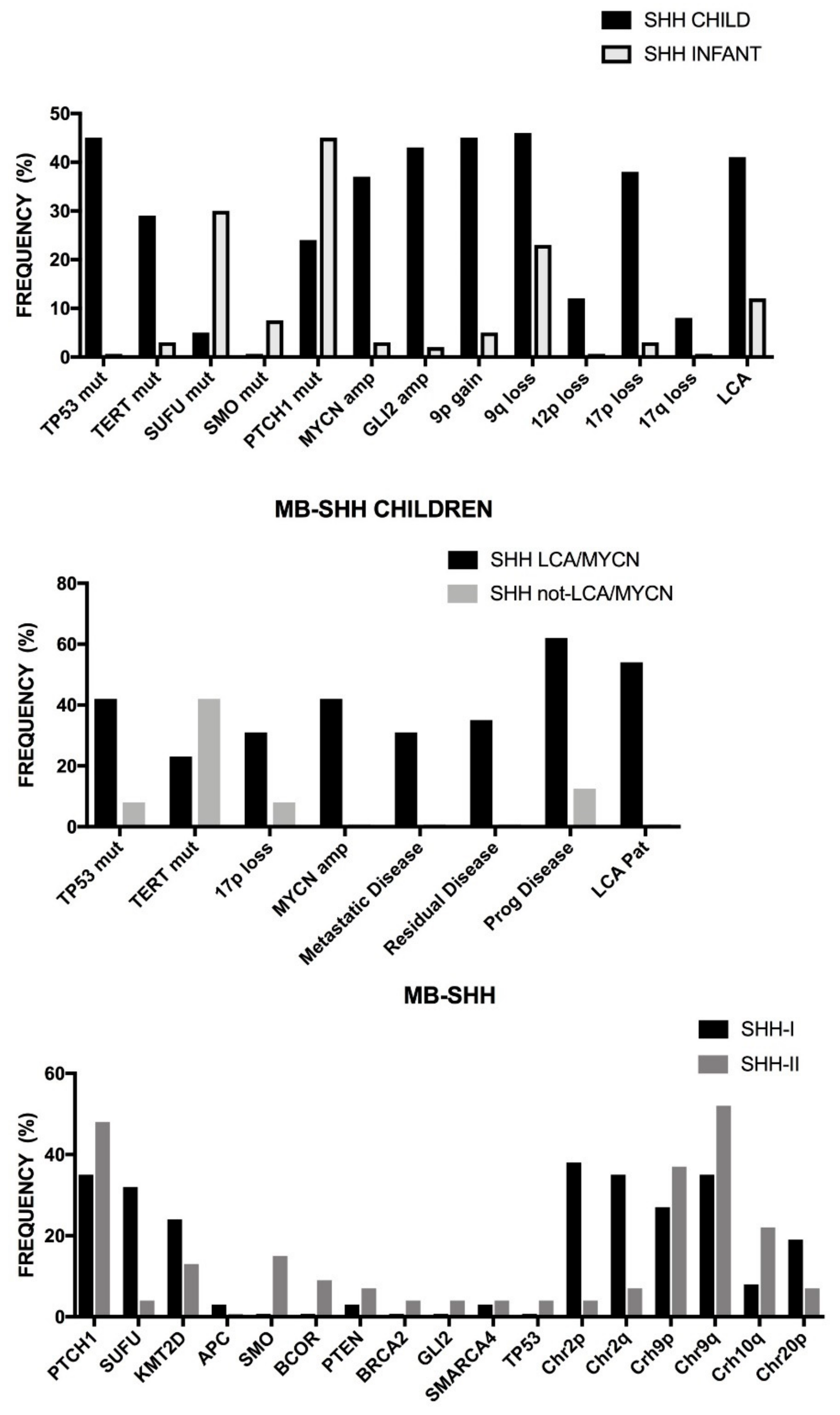

| Tumor Type | WHO Grade |
|---|---|
| Diffuse Astrocytic and oligodendroglial tumors | |
| Diffuse astrocytoma, IDH mutant | II |
| Anaplastic astrocytoma, IDH mutant | III |
| Glioblastoma, IDH wild-type | IV |
| Glioblastoma, IDH-mutant | IV |
| Diffuse midline glioma K27M mutant | IV |
| Oligodendroglioma | II |
| Anaplastic oligodendroglioma | III |
| Oligoastrocytoma | II–III |
| Other Astrocytic tumors | |
| Pilocytic astrocytoma | I |
| Subependymal giant cell astrocytoma | I |
| Pleomorphic xanthoastrocytoma | II |
| Anaplastic pleomorphic xanthoastrocytoma | III |
| Other Gliomas | |
| Chordoid glioma of third ventricle | II |
| Angiocentric glioma | I |
| Ependymal tumors | |
| Ependymoma | II |
| Ependymoma, RELA fusion-positive | II–III |
| Subpendymoma | I |
| Myxopapillary ependymoma | I |
| Anaplastic ependymoma | III |
| Neuronal and mixed neuronal-glial tumors | |
| Gangliocytoma | I |
| Gangliolioma | I |
| Anaplastic ganglioglioma | III |
| Dysembryoplastic neuroepithelial tumor | I |
| Embryonal tumors | |
| Medulloblastoma (all subtypes) | IV |
| Medulloepithelioma | IV |
| Embryonal tumor with multilayered rosettes, C19M altered | IV |
| CNS embryonal tumor | IV |
| Meningiomas | |
| Meningioma | I |
| Atypical meningioma | II |
| Aplastic meningioma | III |
| Tumors of cranial and paraspinal nerves | |
| Neurofibroma | I |
| Perineurinoma | I |
| Schwannoma | I |
| Malignant peripheral nerve sheath tumor | II,II,III |
| Tumor Type | Main Genetic and Chromosomal Abnormalities | Epigenetic Feature |
|---|---|---|
| Diffuse and anaplastic astrocytoma, IDH-mutant | IDH1 or IDH2, ATRX, MYC, CCND2, TP53 mutations. Trisomy 7 or 7q gain; LOH 17p | G-CIMPhigh |
| Oligodendroglioma, IDH-mutant,1p/19q-codeleted | IDH1 or IDH2, TERT, NOTCH1, CIC, FUBP1 mutations; CDKN2A deletion; 1p/19q codeletion | G-CIMPhigh |
| Anaplastic oligodendroglioma, IDH-mutant, 1p/19q codeleted | IDH1 or IDH2, TERT, FUBP1, CIC, TCF12 mutations; CDKN2A deletion; 1p/19q codeletion. | G-CIMPhigh |
| Diffuse astrocytoma, IDH-WT | PTEN, EGFR, NF1, MDM4, TERT mutations; CDKN2A deletion. Trisomy 7; 9p del; 10 monosomy. | G-CIMPlow |
| Glioblastoma, IDH-WT | TP53, PTEN, TERT, PIK3CA, PIK3R1, NF1, H3F34 mutations; CDKN2A, PTEN deletion; EGFR, PDGFRA, CDK4, CDK6, MDM2, MDM4, MET amplification: EGFRvIII rearrangement. Trisomy 7, 7q gain, 10 monosomy, 9p deletion. | G.CIMPlow MGMT-promoter methylation |
| Glioblastoma, IDH-mutant | IDH1 or IDH2. TP53, ATRX mutations. Trisomy 7 or 7q gain: LOH 17p; 10q deletion. | G-CIMPhigh |
| Diffuse midline glioma, H27M-mutant, pediatric, young adult | H3.3 K27M or H3.1 K27M, TP53, PPM1D, FGFR1, ACVR1, PI3K, CCND2 mutations; PTEN, CDKN2A deletion; MYC, PDGFRA, MYCN, CDK4, CDK6, ID2, MET amplification | Loss of histone H3-lysine trimethylation |
| Tumor Subtype | Main Genetic and Chromosomal Abnormalities | Prognosis |
|---|---|---|
| IDHWT-TERTSV | EGFR, PTEN, TP53, and PPM1D mutations; Loss of CDKN2A/B; PDGFRA amplification. Loss of chromosome 4; gain of chromosome 7 and 19. | Poor |
| IDHWT-TERTpWT-ALT | ATRX, SMARCAL1, NF1, and BRAF mutations Loss of chromosome 4; gain of chromosome 7 and 19. | Poor |
| IDHWT-TERTpmut | TERT, EGFR, EGFRvIII, NF1, PTEN, RB1, PIK3CA, And PIK3R1 mutations; amplification of EGFR; Deletion of PTEN; loss of CDKN2A/B (homozygous) Loss of chromosome 4; gain of chromosome 7 and 19. | Poor |
| IDHmut-TERTpWT | IDH1 or IDH2, TP53 and ATRX mutations. Duplication of 7q, duplication of 8q24, loss of CDKN2/B (hemizygous), deletion of 19q. | Better |
| IDHmut-TERTpmut | TERTp, IDH1, or IDH2, TP53 and ATRX mutations. Gain of chromosome 7, duplication of 8q24, loss of CDKN2A/B (hemizygous), deletion of PTEN. | Better |
| Gene (Chromosome) | Histone Subtype | Mutation | Frequency | Tumor | Molecular, Biologic and Clinical Features |
|---|---|---|---|---|---|
| H3F3A (1) | H3.3 | K27M | Up to 93% | DIPG and midline tumors | These mutations are restricted to midline gliomas (pons, thalamus, cerebellum, and spine). All K27M gliomas have a poor prognosis. Geneexpression profile of these tumors is similar to the expression profiles of tumors with a proneural/oligodendroglial phenotype. TP53 mutations and PDGFRA and TOP3A amplifications are frequent. |
| HIST1H3B (17) | H3.1 | K27M | Up to 31% | DIPG | These mutations are highly specific for tumors localized in the pons (DIPG) and the gliomas displaying these mutations show better prognosis than those H3.3K27M-positive. Gene expression profile in these tumors is similar to the expression profiles in adult tumors with a mesenchymal/astrocytic phenotype. ACVR1 and BCOR gene mutations are frequent. Genomic alterations of chr1q and chr2 are frequent. |
| HIST1H3C (6) | H3.1 | K27M | <3% | DIPG | Properties similar to HIST1H3B-mutated tumors. |
| HIST2H3C (6) | H3.2 | K27M | <2% | DIPG | Properties similar to HIST1H3B-mutated tumors. |
| H3F3A (1) | H3.3 | K27I | <1% | DIPG and midline tumors | Properties similar to H3.3K27M-mutated tumors. |
| H3F3A (1) | H3.3 | G34R | 12–14% | HGG | H3.3G34R/V tumors are almost entirely restricted to the cerebral hemispheres (16% in this location), particularly parietal and temporal lobes, are found predominantly in adolescent and young adults (median 15 years) and have a longer overall survival compared with other H3 mutant tumors. These tumors have a higher mutational burden than K27M gliomas. At the molecular level, frequent co-occurring events are TP53/ATRX alterations, PI3K and MAPK pathway mutations, MGMT promoter methylation. Genomic alterations of chr14q24 and chr17p13.1 are frequent. |
| H3F3A (1) | H3.3 | G34L | <2% | HGG | Properties similar to G34R-mutated tumors. |
| Risk Groups | Medulloblastoma Subgroups |
|---|---|
| Low Risk | MBWNT non-metastatic MBGroup 4 non-metastatic, whole chromosome 11 loss or Whole chromosome 17 gain |
| Standard Risk | MBSHH non-metastatic, TP53WT and no MYCN amplification MBGroup 3 non-metastatic and no MYC amplification MBGroup 4 non-metastatic and no chromosome 11 loss |
| High Risk | MBSHH metastatic and TP53WT MBSHH non-metastatic and MYCN amplification MBGroup 4 metastatic |
| Very High Risk | MBSHH TP53 mutation MBGroup3 metastatic and MYC amplification |
| Mutation | Cytogenetics | Histology | Prognosis | |
|---|---|---|---|---|
| MC ben-1 | NF2 | 22q deletion | Fibroblastic Transitional Atypical | Good |
| MC ben-2 | TRAF7, AKT1, KLF4, SMO | Balanced | Secretory Transitional Meningothelial | Good |
| MC ben-3 | 22q deletion 5 gain | Angiomatous Transitional Atypical | Good | |
| MC int-A | NF2 | 22q deletion 1p deletion | Fibroblastic Transitional Atypical | Intermediate |
| MC int-B | NF2, TERT | 22q deletion 1p, CDKN2A deletion | Atypical Anaplastic | Intermediate |
| MC mal | NF2, TERT | 22q deletion 1p, 10, CDKN2A deletion | Anaplastic | Poor |
| Molecular Group | CNS Location | Age of Occurrence | Genetic Abnormalities | Histopathology | Prognosis |
|---|---|---|---|---|---|
| ST-SE | Supratentorial | Adults | Balanced | Subependymoma WHO I | Good |
| ST-EPN-YAP1 | Supratentorial | Children | Focal aberrations Chr 11 YAP1-fusion | Anaplastic ependymoma WHO II/III | Good |
| ST-EPN-RELA | Supratentorial | Children Young adults | Aberrations Chr 11 Chromotripsis RELA-fusion TERTp hypermethylated | L1CAM-positive Anaplastic ependymoma WHO II/III | Poor |
| PF-SE | Posterior Fossa | Adults | Balanced | Subependymoma WHO I | Good |
| PF-EPN-A | Posterior Fossa | Children | Balanced Few CNAs TERTp hypermethylated | Anaplastic ependymoma WHO II/III | Poor |
| PF-EPN-B | Posterior Fossa | Adolescent Adults | Chromosome instability Many CNAs | Anaplastic ependymoma WHO II/III | Good |
| SP-SE | Spine | Adults | 6q deletion | Subependymoma WHO I | Good |
| SP-MPE | Spine | Mainly adults | Chromosome instability | Mixopapillary ependymoma WHO I | Good |
| SP-EPN | Spine | Mainly adults | Chromosome instability NF2 mutation 22q loss | Anaplastic ependymoma WHO II/III | Good |
| Target | Number of Patients | Number of Infusions | Efficacy | Toxicity |
|---|---|---|---|---|
| IL13RA | 4 Recurrent or progressive IL13 R-positive glioblastoma | Post-resection intracavitary infusions ×12 (3 patients). Direct intratumor infusion (1 patient). 2–106 cells | Median OS 11 months. In one patient, regression of intracranial and spinal tumors lasting 7.5 months. Decreased IL13Rα expression after therapy. Increase in necrotic tumor volume by MRI. | Headache Neurologic Lymphopenia |
| HER2 | 17 Recurrent or progressive HER2+-glioblastoma. | Single (10 patients) or multiple peripheral infusions (7 patients) 1 × 106 m2 to 1 × 108 cells/m2 No cell expansion in vivo. CAR cells detected up to 1 year after infusion. | Median OS 11 months. Three patients with stable disease at a follow-up of 24–29 months | Headache Weakness Cerebral edema Hydrocephalus Lymphopenia Neutropenia |
| EGFRvIII | 10 First, second and third line of treatment. | Single peripheral infusion of cells. 1.75 × 108 – 5 × 108 cells Only transient engraftment in all patients. | Median OS 8 months (251 days). One patient alive at >30 months after CAR T infusion. Positive evidence of brain trafficking. | Cerebral edema Seizures Headache Intracranial hemorrhage |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Testa, U.; Castelli, G.; Pelosi, E. Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors. Med. Sci. 2018, 6, 85. https://doi.org/10.3390/medsci6040085
Testa U, Castelli G, Pelosi E. Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors. Medical Sciences. 2018; 6(4):85. https://doi.org/10.3390/medsci6040085
Chicago/Turabian StyleTesta, Ugo, Germana Castelli, and Elvira Pelosi. 2018. "Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors" Medical Sciences 6, no. 4: 85. https://doi.org/10.3390/medsci6040085





