Implementation and Validation of the Roche Light Cycler 480 96-Well Plate Platform as a Real-Time PCR Assay for the Quantitative Detection of Cytomegalovirus (CMV) in Clinical Specimens Using the Luminex MultiCode ASRs System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Experimental Procedures
2.3. PCR Amplification and Detection
2.4. Data Analysis
3. Results
3.1. Analytical Performance
3.1.1. Specimens in Compliance
3.1.2. Reportable Range: The Linearity Study
3.2. Sensitivity
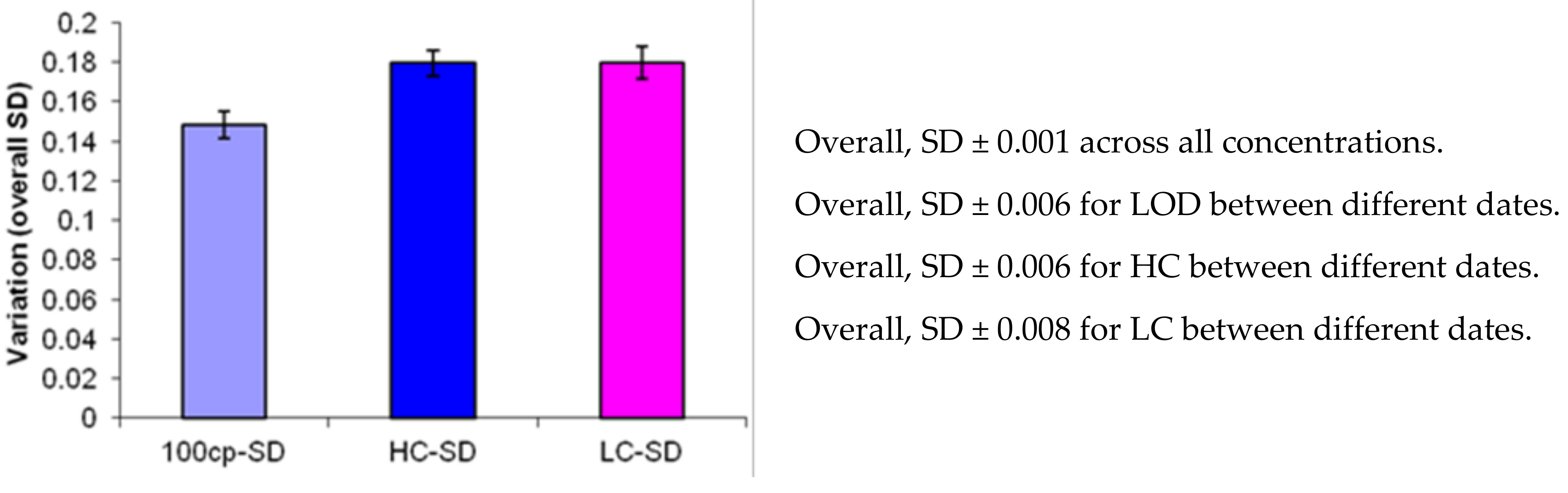
3.3. Precision Determination by Replication Experiments
3.4. Specificity
3.5. Accuracy
3.6. Qualitative Performance Using Clinical Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Boeckh, M. Current antiviral strategies for controlling cytomegalovirus in hematopoietic stem cell transplant recipients: Prevention and therapy. Transpl. Infect. Dis. 1999, 1, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Boeckh, M. Complications, Diagnosis, Management, and Prevention of CMV Infections: Current and Future. Hematology 2011, 2011, 305–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeckh, M.; Murphy, W.J.; Peggs, K.S. Recent advances in cytomegalovirus: An update on pharmacologic and cellular therapies. Biol. Blood Marrow Transplant 2015, 21, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Boeckh, M.; Boivin, G. Quantitation of Cytomegalovirus: Methodologic Aspects and Clinical Applications. Clin. Microbiol. Rev. 1998, 11, 533–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferese, R.; Scorzolini, L.; Campopiano, R.; Albano, V.; Griguoli, A.M.; Giardina, E.; Scala, S.; Ryskalin, L.; D’Alessio, C.; Zampatti, S.; et al. PCR-based approach for qualitative molecular analysis of six neurotropic pathogens. Acta Virol. 2017, 61, 273–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tun, G.S.; Raza, M.; Hale, M.F.; Lobo, A.J. Polymerase chain reaction for detection of mucosal cytomegalovirus infection in patients with acute ulcerative colitis. Ann. Gastroenterol. 2018, 32, 81–87. [Google Scholar] [CrossRef]
- Basso, M.; Andreis, S.; Scaggiante, R.; Franchin, E.; Zago, D.; Biasolo, M.A.; Del Vecchio, C.; Mengoli, C.; Sarmati, L.; Andreoni, M.; et al. Cytomegalovirus, Epstein-Barr virus and human herpesvirus 8 salivary shedding in HIV positive men who have sex with men with controlled and uncontrolled plasma HIV viremia: A 24-month longitudinal study. BMC Infect. Dis. 2018, 18, 683. [Google Scholar] [CrossRef]
- Zhukova, L.I.; Kovalevskaya, O.I.; Gorodin, V.N.; Shakhverdyan, Y.G. The clinical laboratory characteristic of primary acute cytomegalovirus infection in non-immunocompromised adult patients. Klin. Lab. Diagn. 2018, 63, 51–54. [Google Scholar]
- Boeckh, M.; Stevens-Ayers, T.; Travi, G.; Huang, M.-L.; Cheng, G.-S.; Xie, H.; Leisenring, W.; Erard, V.; Seo, S.; Kimball, L.; et al. Cytomegalovirus (CMV) DNA Quantitation in Bronchoalveolar Lavage Fluid From Hematopoietic Stem Cell Transplant Recipients With CMV Pneumonia. J. Infect. Dis. 2017, 215, 1514–1522. [Google Scholar] [CrossRef]
- Buelow, D.; Sun, Y.; Tang, L.; Gu, Z.; Pounds, S.; Hayden, R. Comparative Evaluation of Four Real-Time PCR Methods for the Quantitative Detection of Epstein-Barr Virus from Whole Blood Specimens. J. Mol. Diagn. 2016, 18, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.K.; Chai, C.N.; Capinpin, S.M.; Ang, A.; Ng, S.Y.; Lee, P.L.; Ng, C.W.S.; Yan, G.; Lee, H.K.; Chiu, L.-L.; et al. Evaluation of the Luminex ARIES HSV 1&2 Assay and Comparison with the FTD Neuro 9 and In-house Real-Time PCR Assays for Detecting Herpes Simplex Viruses. Ann. Lab. Med. 2018, 38, 440–445. [Google Scholar] [PubMed]
- Mulligan, E.K.; Germer, J.J.; Arens, M.Q.; D’Amore, K.L.; Di Bisceglie, A.; Ledeboer, N.A.; Moser, M.J.; Newman, A.C.; O’Guin, A.K.; Olivo, P.D.; et al. Detection and Quantification of Hepatitis C Virus (HCV) by MultiCode-RTx Real-Time PCR Targeting the HCV 3′ Untranslated Region. J. Clin. Microbiol. 2009, 47, 2635–2638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caliendo, A.M.; Schuurman, R.; Yen-Lieberman, B.; Spector, S.A.; Andersen, J.; Manjiry, R.; Crumpacker, C.; Lurain, N.S.; Erice, A.; Cmv Working Group of the Complications of Hiv Disease Rac ACTG. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 2001, 39, 1334–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, B.; Larsson, V.C.; Rubin, C.-J.; Sund, F.; Eriksson, B.-M.; Arvidson, J.; Yun, Z.; Bondeson, K.; Blomberg, J. Comparison of a Duplex Quantitative Real-Time PCR Assay and the COBAS Amplicor CMV Monitor Test for Detection of Cytomegalovirus. J. Clin. Microbiol. 2004, 42, 1909–1914. [Google Scholar] [CrossRef] [Green Version]
- Kalpoe, J.S.; Kroes, A.C.; de Jong, M.D.; Schinkel, J.; de Brouwer, C.S.; Beersma, M.F.; Claas, E.C. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J. Clin. Microbiol. 2004, 42, 1498–1504. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Kirov, I.; Klassen, H.K.; Schwartz, P.H. Characterization of stem cells using reverse transcriptase-polymerase chain reaction. In Human Stem Cell Manual: A Laboratory Guide; Academic Press: New York, NY, USA; London, UK, 2007; pp. 127–148. [Google Scholar]
- Klenner, J.; Kohl, C.; Dabrowski, P.W.; Nitsche, A. Comparing Viral Metagenomic Extraction Methods. Curr. Issues Mol. Boil. 2017, 24, 59–70. [Google Scholar] [CrossRef]
- Burd, E.M. Validation of Laboratory-Developed Molecular Assays for Infectious Diseases. Clin. Microbiol. Rev. 2010, 23, 550–576. [Google Scholar] [CrossRef] [Green Version]
- Emmadi, R.; Boonyaratanakornkit, J.B.; Selvarangan, R.; Shyamala, V.; Zimmer, B.L.; Williams, L.; Bryant, B.; Schutzbank, T.; Schoonmaker, M.M.; Amos Wilson, J.A.; et al. Molecular methods and platforms for infectious diseases testing a review of FDA-approved and cleared assays. J. Mol. Diagn. 2011, 13, 583–604. [Google Scholar] [CrossRef] [Green Version]
- Kuang, J.; Yan, X.; Genders, A.; Granata, C.; Bishop, D. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef]
- Wolff, D.J.; Heaney, D.L.; Neuwald, P.D.; Stellrecht, K.A.; Press, R.D. Multi-Site PCR-based CMV viral load assessment-assays demonstrate linearity and precision but lack numeric standardization: A report of the association for molecular pathology. J. Mol. Diagn. 2009, 11, 87–92. [Google Scholar] [CrossRef] [Green Version]
- Lewandowski, K.; Bell, A.; Miles, R.; Carne, S.; Wooldridge, D.; Manso, C.; Hennessy, N.; Bailey, D.; Pullan, S.T.; Gharbia, S.; et al. The Effect of Nucleic Acid Extraction Platforms and Sample Storage on the Integrity of Viral RNA for Use in Whole Genome Sequencing. J. Mol. Diagn. 2017, 19, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelmann, I.; Alidjinou, E.K.; Lazrek, M.; Ogiez, J.; Pouillaude, J.-M.; Chazard, E.; Dewilde, A.; Hober, D. Comparison of two commercial quantitative PCR assays and correlation with the first WHO International Standard for human CMV. Diagn. Microbiol. Infect. Dis. 2018, 91, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Kearns, A.M.; Guiver, M.; James, V.; King, J. Development and evaluation of a real-time quantitative PCR for the detection of human cytomegalovirus. J. Virol. Methods 2001, 95, 121–131. [Google Scholar] [CrossRef]
- Jebbink, J.; Bai, X.; Rogers, B.B.; Dawson, D.B.; Scheuermann, R.H.; Domiati-Saad, R. Development of real-time PCR assays for the quantitative detection of Epstein-Barr virus and cytomegalovirus, comparison of TaqMan probes, and molecular beacons. J. Mol. Diagn. 2003, 5, 15–20. [Google Scholar] [CrossRef]
- Watzinger, F.; Suda, M.; Preuner, S.; Baumgartinger, R.; Ebner, K.; Bašková, L.; Niesters, H.G.; Lawitschka, A.; Lion, T. Real-Time Quantitative PCR Assays for Detection and Monitoring of Pathogenic Human Viruses in Immunosuppressed Pediatric Patients. J. Clin. Microbiol. 2004, 42, 5189–5198. [Google Scholar] [CrossRef] [Green Version]

| Compositions | 1 Target per Reaction (μL) | |
|---|---|---|
| 1 | Nuclease Free Water, 2 mL (PN 1468) | 11 |
| 2 | 5× ISOlution, DNA Use, 500 μL (PN 1790) Lot 1130404 | 5 |
| 3 | CMV Primers (PN 3906) Lot 1204003 | 1 |
| 4 | 25× Control Primers 3 (PN 3804), Lot 1204002 | 1 |
| 5 | DNA Universal Reference, 100 μL (PN 1844), Lot 1114317 | 1 |
| 6 | UNG (Uracil-DNA-N-Glycosylase) (No lot #) Roche_12790500 Sept 2012 | 0.5 |
| 7 | TITANIUM™ Taq DNA polymerase (50x), S1792 Lot 1003099, (Clontech Cat. No. 639208 or 639272) | 0.5 |
| Total volume | 20 |
| Compositions | Volume (μL) |
|---|---|
| Master mix (see Table 1) | 20 |
| Target DNA | 5 |
| Total volume | 25 |
| Temperature | Control | |||||
|---|---|---|---|---|---|---|
| Program | Cycles | Target (°C) | Acquisition Mode | Hold (hh:mm:ss) | Ramp Rate (°C/s) | Analysis Mode |
| UNG | 1 | 40 | None | 0:10:00 | 4.4 | None |
| Denaturation | 1 | 95 | None | 0:10:00 | 4.4 | None |
| PCR | 45 | 95 | None | 0:00:05 | 4.4 | Quantification |
| 58 | None | 0:00:10 | 2.2 | |||
| 72 | Single | 0:00:20 | 1 | |||
| Melt | 1 | 95 | None | 0:00:10 | 4.4 | Melting Curves |
| 60 | None | 0:00:30 | 2.2 | |||
| 95 | Continuous | |||||
| Melt | 1 | 40 | None | 0:00:30 | 2.2 | Yes |
| Sample ID | Ct | Tm | Roche-LC480 Platform (Copy/mL) | Log Copy Number | |
|---|---|---|---|---|---|
| 6/13/2012 | 2012vsID1-01_VZV (5 μL) | N/A | N/A | N/A | |
| Stnd3 | 2012vsID1-04_EBV (5 μL) | N/A | N/A | N/A | |
| 2012vsID1-05_HSVI (5 μL) | N/A | N/A | N/A | ||
| CAP2011-VLS01_BK (5 μL) | N/A | N/A | N/A | ||
| NC | N/A | N/A | N/A | ||
| ID1-01+CMV5E5 (1:1–2.5 μL:2.5 μL) | 22 | 81 | 2.00E+05 | 5.30103 | |
| ID1-04+CMV5E5 (1:1–2.5 μL:2.5 μL) | 23 | 80.9 | 1.70E+05 | 5.230449 | |
| ID1-05+CMV5E5(1:1–2.5 μL:2.5 μL) | 22 | 80.5 | 2.40E+05 | 5.380211 | |
| CAP2011-VLS01+CMV5E5(1:1–2.5 μL:2.5 μL) | 22 | 81.1 | 3.10E+05 | 5.491362 | |
| mean | 230000 | 5.350763 | |||
| SD | 60553.01 | 0.111929 | |||
| NTC | N/A | N/A | N/A | ||
| CMV5.00E+05(5 μL) | 21 | 81.1 | 6.90E+05 | 5.838849 | |
| CMV5.00E+05(5 μL) | 21 | 81.1 | 5.10E+05 | 5.70757 | |
| CMV5.00E+05(5 μL) | 22 | 81.1 | 2.60E+05 | 5.414973 | |
| CMV5.00E+05(5 μL) | 22 | 81.2 | 3.50E+05 | 5.544068 | |
| mean | 452500 | 5.626365 | |||
| SD | 189098.7 | 0.185475 | |||
| NC | N/A | N/A | N/A | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.C.; Sparks, K.J.; Sender, L.S. Implementation and Validation of the Roche Light Cycler 480 96-Well Plate Platform as a Real-Time PCR Assay for the Quantitative Detection of Cytomegalovirus (CMV) in Clinical Specimens Using the Luminex MultiCode ASRs System. Med. Sci. 2020, 8, 14. https://doi.org/10.3390/medsci8010014
Li SC, Sparks KJ, Sender LS. Implementation and Validation of the Roche Light Cycler 480 96-Well Plate Platform as a Real-Time PCR Assay for the Quantitative Detection of Cytomegalovirus (CMV) in Clinical Specimens Using the Luminex MultiCode ASRs System. Medical Sciences. 2020; 8(1):14. https://doi.org/10.3390/medsci8010014
Chicago/Turabian StyleLi, Shengwen Calvin, Kara J. Sparks, and Leonard S. Sender. 2020. "Implementation and Validation of the Roche Light Cycler 480 96-Well Plate Platform as a Real-Time PCR Assay for the Quantitative Detection of Cytomegalovirus (CMV) in Clinical Specimens Using the Luminex MultiCode ASRs System" Medical Sciences 8, no. 1: 14. https://doi.org/10.3390/medsci8010014






