Direct Conversion of Bovine Dermal Fibroblasts into Myotubes by Viral Delivery of Transcription Factor bMyoD
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
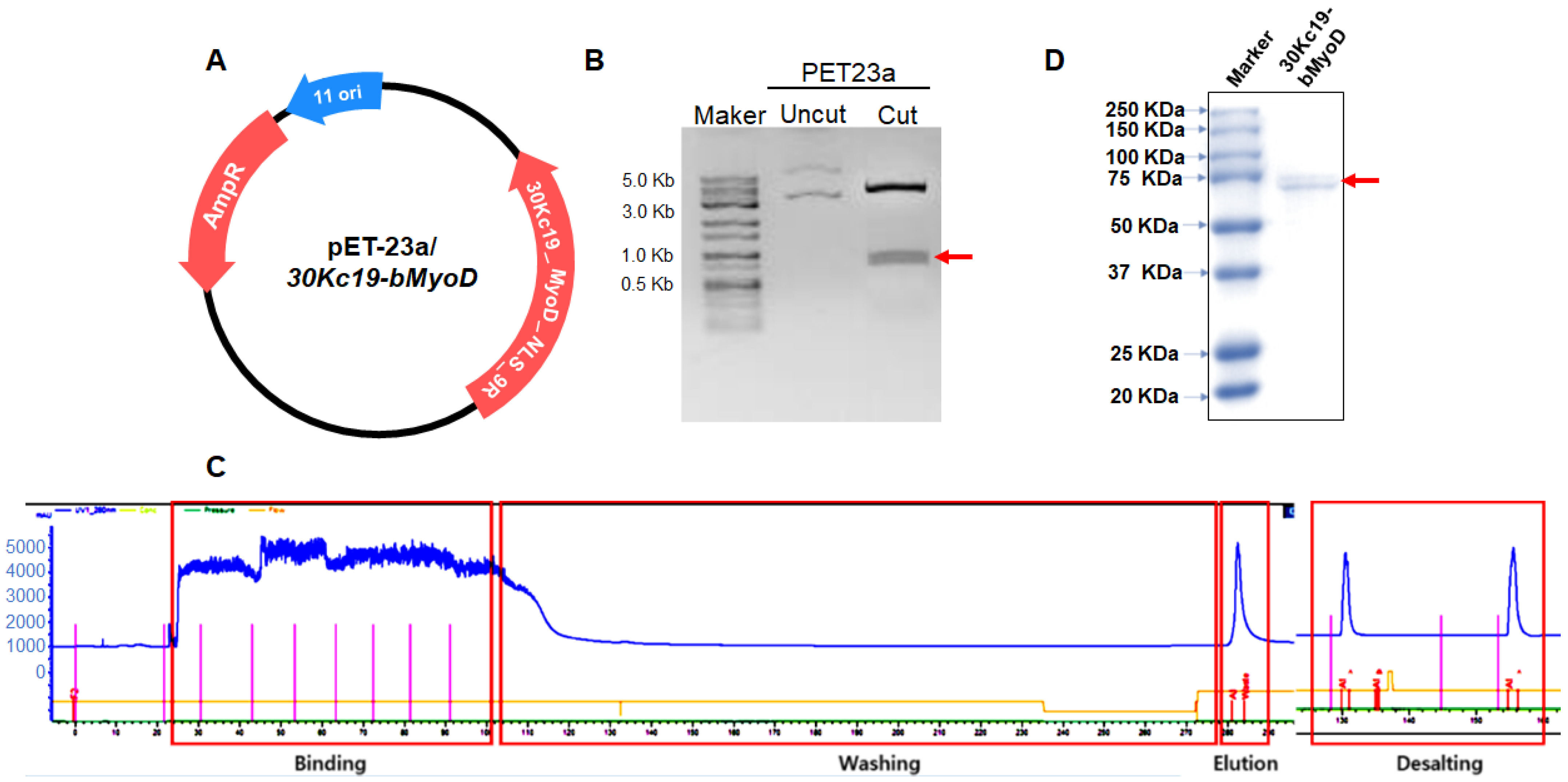
2.2. Plasmid Construction and Production of 30Kc19-bMyoD-NLS-R9 (30MNR)
2.3. SDS-PAGE and Immunoblot Analysis
2.4. Cell-Penetration Assay
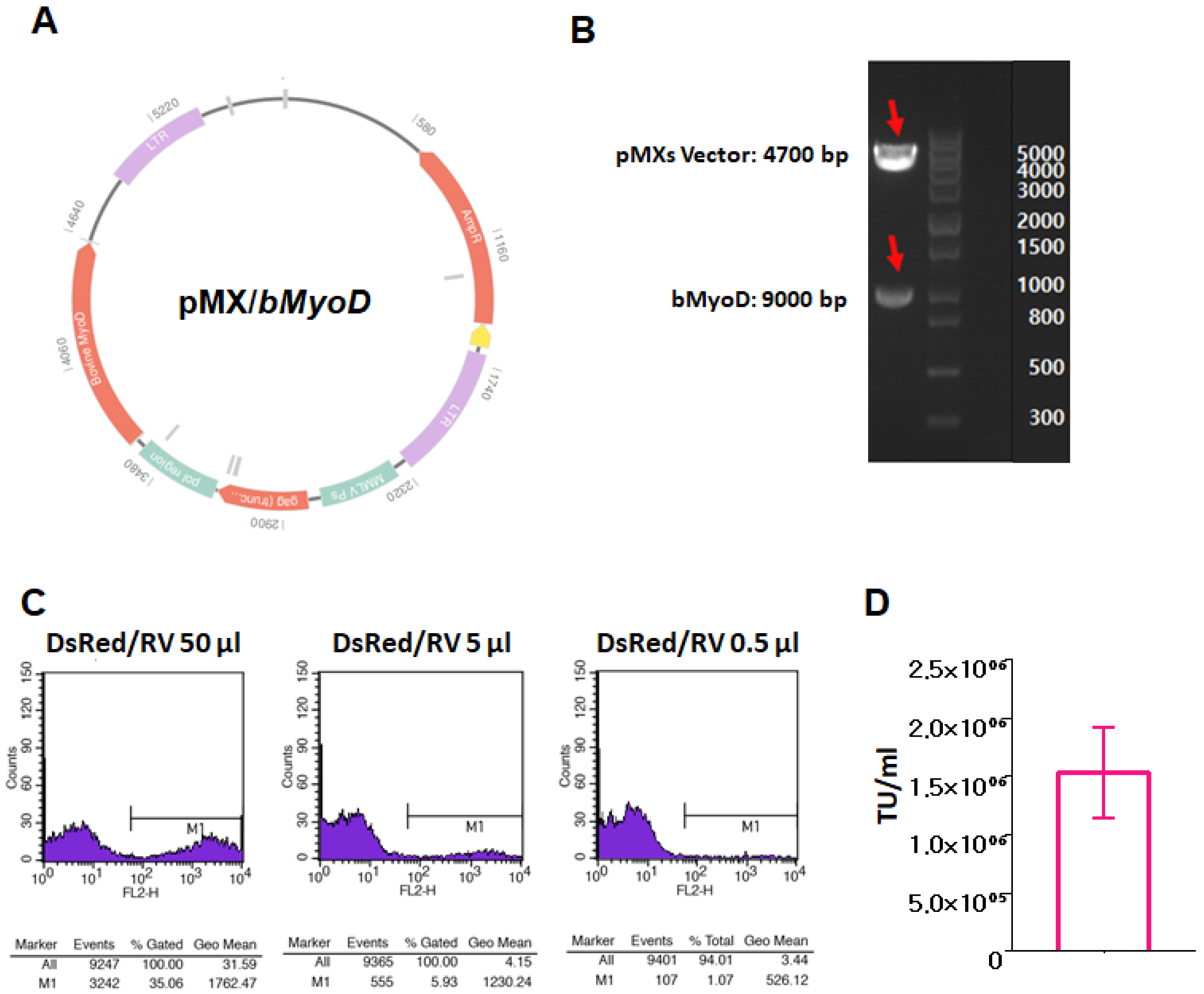
2.5. Retrovirus Production, Titration, and Transduction
2.6. RNA Isolation, cDNA Synthesis, and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.7. Immunocytochemical Analysis
2.8. Statistical Analysis
3. Results
3.1. Soluble Expression and Purification of 30MNR Protein
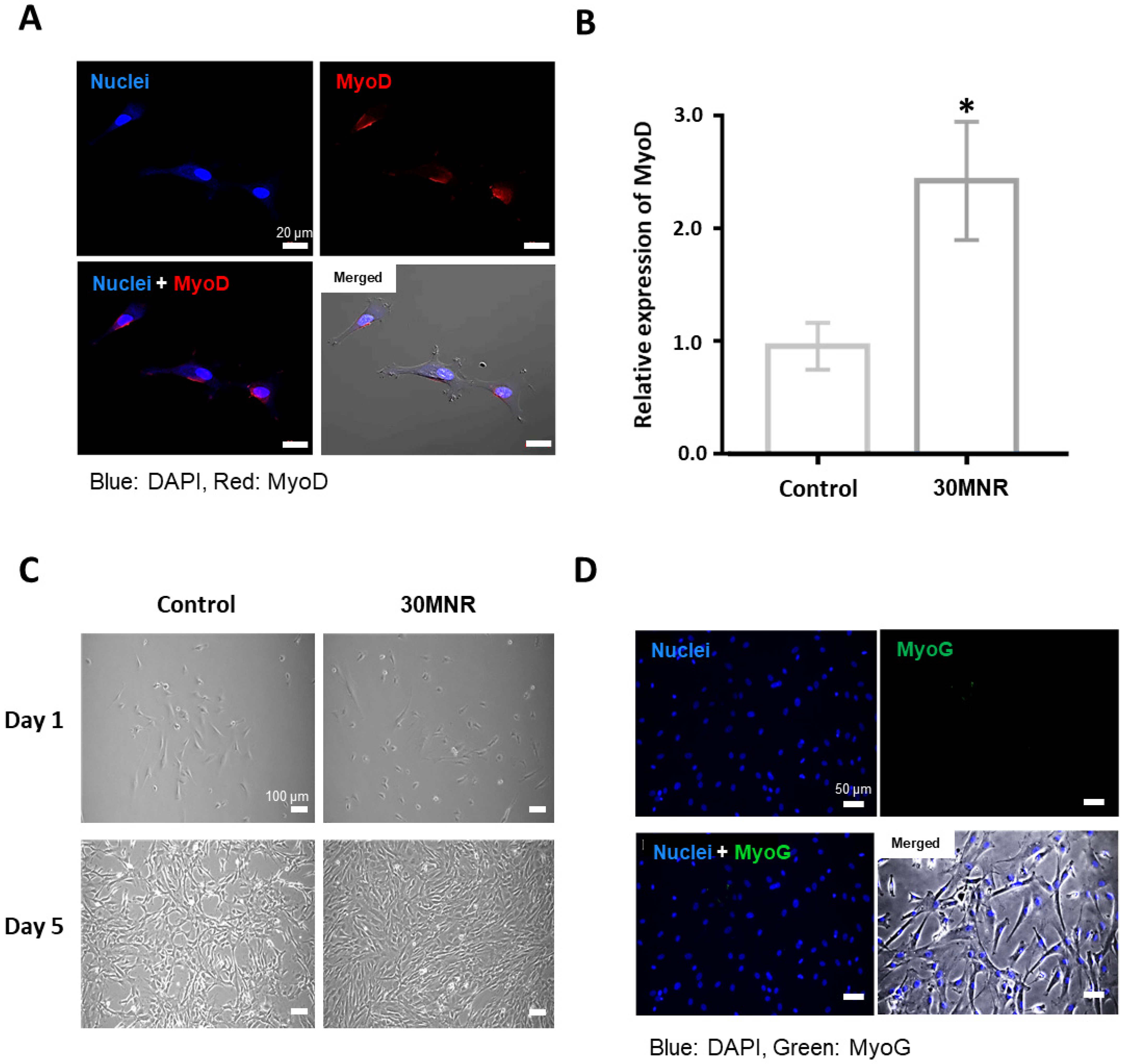
3.2. Intracellular Delivery but Inefficient Direct Reprogramming of 30MNR Protein
3.3. Production of bMyoD Encoding Retrovirus, RV/bMyoD
3.4. Myogenic Gene Expression of Directly Reprogrammed Myogenic cells by RV/bMyoD
3.5. Myogenic Protein Expression of Directly Reprogrammed Myogenic Cells by RV/bMyoD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kent-Braun, J.; Ng, A.; Castro, M.; Weiner, M.; Gelinas, D.; Dudley, G.; Miller, R. Strength, skeletal muscle composition, and enzyme activity in multiple sclerosis. J. Appl. Physiol. 1997, 83, 1998–2004. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.P.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.; Yang, T.; Citri, A.; Sebastiano, V.; Marro, S.; Südhof, T.; et al. Induction of human neuronal cells by defined transcription factors. Nature 2011, 476, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Thier, M.; Wörsdörfer, P.; Lakes, Y.B.; Gorris, R.; Herms, S.; Opitz, T.; Seiferling, D.; Quandel, T.; Hoffmann, P.; Nöthen, M.; et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 2012, 10, 473–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiramatsu, K.; Sasagawa, S.; Outani, H.; Nakagawa, K.; Yoshikawa, H.; Ysumaki, N. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J. Clin. Investig. 2011, 121, 640–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Xiao, D.; Liu, X.; Zhang, M.; Zou, M.; Deng, Q.; Sun, D.; Bian, X.; Cai, Y.; Guo, Y.; Liu, S.; et al. Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 2018, 9, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Peake, J.M.; Neubauer, O.; Della Gatta, P.A.; Nosaka, K. Muscle damage and inflammation during recovery from exercise. J. Appl. Physiol. 2017, 122, 559–570. [Google Scholar] [CrossRef]
- Corona, B.T.; Wenke, J.C.; Ward, C.L. Pathophysiology of volumetric muscle loss injury. Cells Tissues Organs 2016, 202, 180–188. [Google Scholar] [CrossRef]
- Ling, X.F.; Peng, X. What is the price to pay for a free fibula flap? A systematic review of donor-site morbidity following free fibula flap surgery. Plast. Reconstr. Surg. 2012, 129, 657–674. [Google Scholar] [CrossRef]
- Asakura, A.; Rudnicki, M.A.; Komaki, M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 2001, 68, 245–253. [Google Scholar] [CrossRef]
- Klein, D.; Benchellal, M.; Kleff, V.; Jakob, H.G.; Ergün, S. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci. Rep. 2013, 3, 2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, L.V.; Alfonso, Z.; Zhang, R.; Leung, J.; Wu, B.; Ignarro, L. Clonogenic multipotent stem cells in human adipose tissue differentiate into functional smooth muscle cells. Proc. Natl. Acad. Sci. USA 2006, 103, 12167–12172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, J.M.; Harley, B.A.; Bailey, R.C. Tunable, photoreactive hydrogel system to probe synergies between mechanical and biomolecular cues on adipose-derived mesenchymal stem cell differentiation. ACS Biomater. Sci. Eng. 2015, 1, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Sim, J.; Kim, H.-J. Neural stem cell differentiation using microfluidic device-generated growth factor gradient. Biomol. Ther. 2018, 26, 380. [Google Scholar] [CrossRef] [PubMed]
- Pires, F.; Ferreira, Q.; Rodrigues, C.A.; Morgado, J.; Ferreira, F.C. Neural stem cell differentiation by electrical stimulation using a cross-linked PEDOT substrate: Expanding the use of biocompatible conjugated conductive polymers for neural tissue engineering. Biochim. Biophys. Acta (BBA) Gen. Subj. 2015, 1850, 1158–1168. [Google Scholar] [CrossRef]
- Sarraf, C.; Otto, W.R.; Eastwood, M. In vitro mesenchymal stem cell differentiation after mechanical stimulation. Cell Prolif. 2011, 44, 99–108. [Google Scholar] [CrossRef]
- Subramony, S.D.; Dargis, B.R.; Castillo, M.; Azeloglu, E.U.; Tracey, M.; Su, A.; Lu, H. The guidance of stem cell differentiation by substrate alignment and mechanical stimulation. Biomaterials 2013, 34, 1942–1953. [Google Scholar] [CrossRef] [Green Version]
- Tomaskovic-Crook, E.; Gu, Q.; Rahim, S.N.A.; Wallace, G.G.; Crook, J.M. Conducting polymer mediated electrical stimulation induces multilineage differentiation with robust neuronal fate determination of human induced pluripotent stem cells. Cells 2020, 9, 658. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.K.; Chen, C.S. Cell adhesion and mechanical stimulation in the regulation of mesenchymal stem cell differentiation. J. Cell. Mol. Med. 2013, 17, 823–832. [Google Scholar] [CrossRef]
- Chen, L.; Liu, J.; Guan, M.; Zhou, T.; Duan, X.; Xiang, Z. Growth factor and its polymer scaffold-based delivery system for cartilage tissue engineering. Int. J. Nanomed. 2020, 15, 6097. [Google Scholar] [CrossRef]
- Gotoh, N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008, 99, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Han, U.; Park, H.H.; Kim, Y.J.; Park, T.H.; Park, J.H.; Hong, J.K. Efficient encapsulation and sustained release of basic fibroblast growth factor in nanofilm: Extension of the feeding cycle of human induced pluripotent stem cell culture. ACS Appl. Mater. Interfaces 2017, 9, 25087–25097. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, D.; Farnia, S.; Kefalas, P.; Maziarz, R.T. Concise review: The high cost of high tech medicine: Planning ahead for market access. Stem Cells Transl. Med. 2017, 6, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ieda, M.; Fu, J.-D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Efe, J.A.; Zhu, S.; Talantova, M.; Yuan, X.; Wang, S.; Lipton, S.; Zhang, K.; Ding, S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA 2011, 108, 7838–7843. [Google Scholar] [CrossRef] [Green Version]
- Szabo, E.; Rampalli, S.; Risueno, R.M.; Schnerch, A.; Mitchell, R.; Fiebig-Comyn, A.; Levadoux-Martin, M.; Bhatia, M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 2010, 468, 521–526. [Google Scholar] [CrossRef]
- Chang, Y.; Lee, E.; Kim, J.; Kwon, Y.-W.; Kwon, Y.; Lim, J. Efficient in vivo direct conversion of fibroblasts into cardiomyocytes using a nanoparticle-based gene carrier. Biomaterials 2019, 192, 500–509. [Google Scholar] [CrossRef]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [Green Version]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.-W.; Zhang, T.-T.; Liu, W.; Yang, J.; Lin, X.-L.; Jia, J.-S.; Shen, H.-F.; Wang, S.-C.; Li, J.; Zha, W.-T.; et al. Direct conversion of pig fibroblasts to chondrocyte-like cells by c-Myc. Cell Death Discov. 2019, 5, 126. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, A.; Oro, M.; Hiraki, Y.; Shukunami, C. Direct conversion of tenocytes into chondrocytes by Sox9. Exp. Cell Res. 2012, 318, 1492–1507. [Google Scholar] [CrossRef]
- Kim, I.; Ghosh, A.; Bundschuh, N.; Hinte, L.; Von Meyenn, F.; Bar-Nur, O. Integrative molecular roadmap for direct conversion of fibroblasts into myocytes and myogenic progenitor cells. bioRxiv 2021, 2–7. [Google Scholar] [CrossRef]
- Wakao, J.; Kishida, T.; Fumino, S.; Kimura, K.; Yamamoto, K.; Kotani, S.; Mizyshima, L.; Naito, Y.; Yoshikawa, T.; Tajiti, T.; et al. Efficient direct conversion of human fibroblasts into myogenic lineage induced by co-transduction with MYCL and MYOD1. Biochem. Biophys. Res. Commun. 2017, 488, 368–373. [Google Scholar] [CrossRef]
- Dey, C.; Narayan, G.; Krishna Kumar, H.; Borgohain, M.P.; Lenka, N.; Thummer, R. Thummer. Cell-Penetrating Peptides as a Tool to Deliver Biologically Active Recombinant Proteins to Generate Transgene-Free Induced Pluripotent Stem Cells. Stud. Stem Cells Res. Ther. 2016, 3, 006–015. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zeng, F.; Zhang, M.; Huang, F.; Wang, J.; Guo, J.; Liu, C.; Wang, H. Emerging landscape of cell penetrating peptide in reprogramming and gene editing. J. Control. Release 2016, 226, 124–137. [Google Scholar] [CrossRef]
- Seo, B.J.; Hong, Y.J.; Do, J.T. Cellular reprogramming using protein and cell-penetrating peptides. Int. J. Mol. Sci. 2017, 18, 552. [Google Scholar] [CrossRef] [Green Version]
- Takashina, T.; Koyama, T.; Nohara, S.; Hasegawa, M.; Ishiguro, A.; Iijima, K.; Lu, J.; Shimura, M.; Okamura, T.; Sakuma, T.; et al. Identification of a cell-penetrating peptide applicable to a protein-based transcription activator-like effector expression system for cell engineering. Biomaterials 2018, 173, 11–21. [Google Scholar] [CrossRef]
- Motohashi, T.; Kunisada, T. Direct conversion of mouse embryonic fibroblasts into neural crest cells. In Skin Stem Cells; Springer: Berlin/Heidelberg, Germany, 2018; pp. 307–321. [Google Scholar]
- Sowa, Y.; Kishida, T.; Tomita, K.; Yamamoto, K.; Numajiri, T.; Mazda, O. Direct conversion of human fibroblasts into Schwann cells that facilitate regeneration of injured peripheral nerve in vivo. Stem Cells Transl. Med. 2017, 6, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Hoshiba, T.; Yokoyama, N. Decellularized extracellular matrices derived from cultured cells at stepwise myogenic stages for the regulation of myotube formation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2020, 1867, 118658. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Choudhary, S.K.; Milner, D.J.; Munir, M.I.; Kuisk, I.R.; Capetanaki, Y. Inhibition of desmin expression blocks myoblast fusion and interferes with the myogenic regulators MyoD and myogenin. J. Cell Biol. 1994, 124, 827–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muscaritoli, M.; Anker, S.; Argiles, J.; Aversa, Z.; Bauer, J.M.; Biolo, G.; Boirie, Y.; Bosaeus, I.; Cederholm, T.; Costelli, P.; et al. Consensus definition of sarcopenia, cachexia and pre-cachexia: Joint document elaborated by Special Interest Groups (SIG)“cachexia-anorexia in chronic wasting diseases” and “nutrition in geriatrics”. Clin. Nutr. 2010, 29, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Fayaz, H. Prospectus of cultured meat—advancing meat alternatives. J. Food Sci. Technol. 2011, 48, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Stephens, N.; Di Silvio, L.; Dunsford, I.; Ellis, M.; Glencross, A.; Sexton, A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol. 2018, 78, 155–166. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, H.H.; Kim, J.A.; Park, J.H.; Ryu, J.; Choi, J.; Lee, J.; Rhee, W.J.; Park, T.H. Enzyme delivery using the 30Kc19 protein and human serum albumin nanoparticles. Biomaterials 2014, 35, 1696–1704. [Google Scholar] [CrossRef]
- Lee, J.; Park, H.H.; Park, J.H. Efficient production of cell-permeable oct4 protein using 30Kc19 protein originating from silkworm. Biotechnol. Bioprocess Eng. 2019, 24, 964–971. [Google Scholar] [CrossRef]
- Park, H.H.; Sohn, Y.; Yeo, J.W.; Park, J.H.; Lee, H.J.; Ryu, J.; Rhee, W.J.; Park, T.H. Identification and characterization of a novel cell-penetrating peptide of 30Kc19 protein derived from Bombyx mori. Process Biochem. 2014, 49, 1516–1526. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, J.H.; Park, H.H.; Rhee, W.J.; Choi, S.S.; Park, T.H. A protein delivery system using 30Kc19 cell-penetrating protein originating from silkworm. Biomaterials 2012, 33, 9127–9134. [Google Scholar] [CrossRef]
- Ryu, J.; Park, H.H.; Park, J.H.; Lee, H.J.; Rhee, W.J.; Park, T.H. Soluble expression and stability enhancement of transcription factors using 30Kc19 cell-penetrating protein. Appl. Microbiol. Biotechnol. 2016, 100, 3523–3532. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Badu-Mensah, A.; Thomas, M.C.; McAleer, C.W.; Hickman, J.J. Characterization of functional human skeletal myotubes and neuromuscular junction derived—from the same induced pluripotent stem cell source. Bioengineering 2020, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Pircher, T.; Wackerhage, H.; Akova, E.; Böcker, W.; Aszodi, A.; Saller, M. Fusion of Normoxic-and Hypoxic-Preconditioned Myoblasts Leads to Increased Hypertrophy. Cells 2022, 11, 1059. [Google Scholar] [CrossRef] [PubMed]
- Yuzawa, R.; Koike, H.; Manabe, I.; Oishi, Y. VDR regulates simulated microgravity-induced atrophy in C2C12 myotubes. Sci. Rep. 2022, 12, 1377. [Google Scholar] [CrossRef]
- Li, M.T.A.; Willett, N.J.; Uhrig, B.A.; Guldberg, R.E.; Warren, G.L. Functional analysis of limb recovery following autograft treatment of volumetric muscle loss in the quadriceps femoris. J. Biomech. 2014, 47, 2013–2021. [Google Scholar] [CrossRef] [Green Version]
- Norris, R.; Glasby, M.; Gattuso, J.; Bowden, R. Peripheral nerve repair in humans using muscle autografts. A new technique. J. Bone Jt. Surg. Br. Vol. 1988, 70, 530–533. [Google Scholar] [CrossRef] [Green Version]
- Bansal, V.; De, D.; An, J.; Kang, T.M.; Jeong, H.-J.; Kang, J.-S.; Lim, K.K. Chemical induced conversion of mouse fibroblasts and human adipose-derived stem cells into skeletal muscle-like cells. Biomaterials 2019, 193, 30–46. [Google Scholar] [CrossRef]
- Lee, K.; Rafi, M.; Wang, X.; Aran, K.; Feng, X.; Sterzo, C.; Tang, R.; Lingampalli, N.; Kim, H.J.; Murthy, N. In vivo delivery of transcription factors with multifunctional oligonucleotides. Nat. Mater. 2015, 14, 701–706. [Google Scholar] [CrossRef] [Green Version]
- Rilo-Alvarez, H.; Ledo, A.M.; Vidal, A.; Garcia-Fuentes, M. Delivery of transcription factors as modulators of cell differentiation. Drug Deliv. Transl. Res. 2021, 11, 426–444. [Google Scholar] [CrossRef]
- Berkes, C.A.; Tapscott, S.J. Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2005; pp. 585–595. [Google Scholar]
- Hollenberg, S.M.; Cheng, P.F.; Weintraub, H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. USA 1993, 90, 8028–8032. [Google Scholar] [CrossRef] [Green Version]
- Rudnicki, M.A.; Jaenisch, R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays 1995, 17, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Heidt, A.B.; Rojas, A.; Harris, I.S.; Black, B.L. Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol. Cell. Biol. 2007, 27, 5910–5920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shklover, J.; Etzioni, S.; Weisman-Shomer, P.; Yafe, A.; Bengal, E.; Fry, M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007, 35, 7087–7095. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, B.; Lee, S.H.; Hong, S.; Kwon, M.; Joo, J.; Lim, K.S.; Park, H.H. Direct Conversion of Bovine Dermal Fibroblasts into Myotubes by Viral Delivery of Transcription Factor bMyoD. Appl. Sci. 2022, 12, 4688. https://doi.org/10.3390/app12094688
Son B, Lee SH, Hong S, Kwon M, Joo J, Lim KS, Park HH. Direct Conversion of Bovine Dermal Fibroblasts into Myotubes by Viral Delivery of Transcription Factor bMyoD. Applied Sciences. 2022; 12(9):4688. https://doi.org/10.3390/app12094688
Chicago/Turabian StyleSon, Boram, Seong Ho Lee, Seyoung Hong, Miji Kwon, Jinmyoung Joo, Kwang Suk Lim, and Hee Ho Park. 2022. "Direct Conversion of Bovine Dermal Fibroblasts into Myotubes by Viral Delivery of Transcription Factor bMyoD" Applied Sciences 12, no. 9: 4688. https://doi.org/10.3390/app12094688





