Abstract
Bioacoustic research of reptile calls and vocalizations has been limited due to the general consideration that they are voiceless. However, several species of geckos, turtles, and crocodiles are able to produce simple and even complex vocalizations which are species-specific. This work presents a novel approach for the automatic taxonomic identification of reptiles through their bioacoustics by applying pattern recognition techniques. The sound signals are automatically segmented, extracting each call from the background noise. Then, their calls are parametrized using Linear and Mel Frequency Cepstral Coefficients (LFCC and MFCC) to serve as features in the classification stage. In this study, 27 reptile species have been successfully identified using two machine learning algorithms: K-Nearest Neighbors (kNN) and Support Vector Machine (SVM). Experimental results show an average classification accuracy of 97.78% and 98.51%, respectively.
1. Introduction
The taxonomic class Reptilia is formed by turtles, crocodiles, snakes, lizards, and tuataras, of which some are able of vocalize [1], but they do not do it often. As a result, there are few studies in literature about the reptile acoustic communications, and have been considered unimportant until recently. However, some geckos, crocodiles, and turtles are very active in producing vocalizations [2,3], but their social roles are still not fully understood. Crocodiles are probably the most vocal reptiles, with a rich variety of hissing, distress, and threatening calls due to their close relation with birds, they are even capable of vocalizing in the egg before hatching [4]. Moreover, some species such as turtles, crocodiles, and alligators can emit sound both in air and underwater [5].
Reptiles emit vocalizations in a broad range of frequencies—they produce sounds mainly between 0.1–4 kHz—but some turtles, crocodiles, and also lizards are able to generate calls above 20 kHz [5,6]. In addition, as a consequence of their behavior and small size, most reptiles can be very difficult to detect in the field using visual surveys [7], which can lead to an underestimation of species richness.
Bioacoustic technologies are an efficient way to sample populations in extended areas where visibility is limited [8], so they may be able to provide additional data for reptile estimation. Traditional bioacoustic monitoring methods rely on human observers who categorize acoustic patterns according to sound similarities. However, this procedure is slow, and it depends on the observer’s ability to identify species, which leads to bias [9]. Hence, machine learning techniques are being applied in many research areas to design automatic classification intelligent systems, such as mosquito identification based on morphological features [10], carbon fiber fabrics classification to minimize risks in engineering processes [11], automatic recognition of arrhythmias for the diagnosis of heart diseases [12], or this work where bioacustic signals of reptile species are used for taxonomic classification.
In recent decades, several techniques have been proposed to automate the acoustic classification of species through intelligent systems. For instance, Acevedo et al. [13] successfully classified three bird and nine frog species by characterizing their calls with 11 variables: minimum and maximum frequency, call length, maximum power, and frequency of eight highest energy points in the call. Then, the results of three classification algorithms—Decision Tree (DT), Linear Discriminant Analysis (LDA) and Support Vector Machine (SVM)—were compared. In their work, SVM achieved an identification rate of 94.95%, outperforming DT and LDA, but the sample calls were selected manually. Brandes [14] used a Hidden Markov Model (HMM) to recognize the vocalizations of nine bird, ten frog, and eight cricket species. This approach employed the peak frequencies and bandwidth from the spectrogram to characterize the sound samples, getting high classification rates for each animal group individually, though it had difficulties coping with complex broad band calls. Another interesting approach can be found in Le-Qing’s research [15], where 50 different insect sounds were classified with an accuracy of 96.17%. In that work, Mel Frequency Cepstral Coefficients (MFCCs) were employed as features, and a Probabilistic Neural Network (PNN) was applied for classification. However, the sounds were taken from noise-free sections of the recorded files. More recently, Henríquez et al. [16] recognized seven different species of bats by Gaussian Mixture Models (GMM), achieving a low average error of 1.8%, using a combination of linear and non-linear parameters. There is no doubt of the progress made in the field of bioacoustic identification to enable an efficient classification of species. However, a robust machine learning technique to recognize reptile calls has still not been found. Previous studies have been focused on the analysis of spectro-temporal characteristics of reptile calls, but to the best of our knowledge, there has not been any research that has used their acoustic signals for automatic inter-species classification. For this reason, this paper proposes a novel approach of taxonomic identification of reptiles through their acoustic features. To achieve this goal, Linear Frequency Cepstral Coefficients (LFCCs) and MFCCs [17] have been used to parametrize the reptile acoustic signals and they have been fused to obtain a robust characterization of the signal in the frequency domain. In addition, two widely used machine learning algorithms—K-Nearest Neighbors (kNN) [18] and Support Vector Machine (SVM) [19] have been utilized to verify the robustness of the proposed parameters. The approach has been validated in three public collections of reptile sounds selected by experts, which contain 27 different species with several types of calls. Therefore, despite the small corpus, this study may serve as a first reference in the field of automatic acoustic recognition of reptile specimens for researchers.
The remainder of this paper is organized as follows. Section 2 presents the proposed technique where the audio signal segmentation and the feature extraction procedure are described. Two classification systems based on kNN and SVM algorithms are described in Section 3, particularized for acoustic recognition. The experimental methodology, the sound dataset, and the results obtained are shown in Section 4, where a comparison of features and classification algorithms is done. Finally, the conclusions and future work are shown in Section 5.
2. Proposed Method
The proposed method is based on the following phases. First, reptile acoustic signals are automatically segmented in syllables and labeled by species. Secondly, for each syllable, the cepstral feature parameters are computed and fused into a unique vector of characteristics per call. Afterwards, these vectors are employed in the classification stage to train and test the two pattern recognition algorithms utilized in this work. Figure 1 illustrates the proposed system technique.
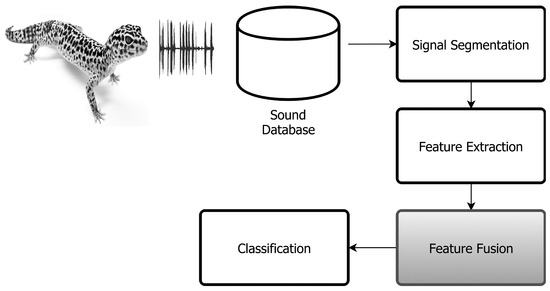
Figure 1.
Automatic acoustic classification system diagram.
2.1. Segmentation
The segmentation stage splits the file recordings into as many syllables as possible to yield useful information for the taxonomy identification. The procedure has been developed based on the Härmä segmentation algorithm [20]. It uses short-time analysis to divide the acoustic signal into a set of frequency and amplitude modulated pulses, where each pulse corresponds to one detected call. For this purpose, the acoustic signal spectrogram was calculated utilizing Short Time Fourier Transform (STFT) with a Hamming window size of 5.8 milliseconds and a 33% overlap, with which they have been heuristically computed. The spectrogram is represented by a matrix , where f is the frequency and t denotes the time. Then, the algorithm explores the matrix using the following strategy:
- Find and such that , placing the nth syllable in . The amplitude of this point is calculated as Equation (1):
- If , the segmentation process is stopped, as the signal amplitude is inferior to the stopping criteria β. For reptile sounds, β has been set to 25 dB.
- From , seek the highest peak of for and , until for both sides. Thus, the starting and ending times of the nth syllable are denoted as, and .
- Save the amplitude trajectories as the nth syllable.
- Delete the nth syllable from the matrix and set .
- Repeat from Step 1 until the end of the spectrogram.
Figure 2 shows four spectrograms belonging to several kinds of reptiles: Crotalus atrox (Diamondback rattlesnake), Gecko, Alligator mississippiensis (American alligator), and Chelonoides nigra (Galapagos giant tortoise). The alligator spectrogram presents a complex and most rich vocalization similar to birds, while the other species present a simpler sound production of hissing and groan calls.
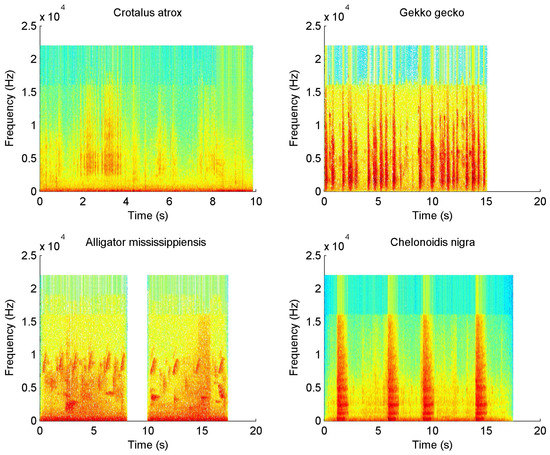
Figure 2.
Example of four reptile call spectrograms.
2.2. Feature Extraction
For each syllable, two cepstral features were extracted to characterize the acoustic signal. MFCCs and LFCCs have been broadly used in speech recognition with success [21], and they have also been applied in animal bioacoustic classification [22,23,24,25], due to their easy implementation and high performance. Reptiles mainly produce sounds in low frequencies within the human auditory range. For this reason, MFCCs have been adopted to get high resolution in the low frequency region. However, reptiles are capable of producing sound above 20 kHz, so LFCCs have also been computed to obtain the information in high frequency ranges.
They are calculated using STFT (a 25 milliseconds Hamming window with an overlap of 50%) and applying Discrete Fourier Transform (DFT) over each frame of the signal. The resultant magnitude spectrum is wrapped by a bank of 40 triangular band pass filters. For MFCCs, the filters are non-uniformly sparse to perform the mel scale transformation. Finally, the coefficients are retrieved taking the lowest Discrete Cosine Transform (DCT) values from the log-magnitude filter outputs, . They are computed following Equation (2):
where j denotes the index of the cepstral coefficient, B is the number of triangular filters, and N denotes the number of cepstral coefficients to compute.
LFCCs are calculated similarly, but using a linear sparse triangular filter bank instead of the mel-scale filters. Furthermore, the number of coefficients, N, has been established by experimentation in order to reach the highest classification rate in the last stage. As a result, 18 coefficients have been taken for both features.
In this work, the feature vectors have been fused, appending the characteristics horizontally as in Label (3), where each row represents a syllable from the segmentation stage. These features have been combined to hold information of higher as well as lower frequency regions, obtaining a broad spectral representation of the reptile calls. Therefore, these rows contain 36 coefficients, which are used as inputs to the classification stage.
3. Classification System
For the classification stage, the performance of two machine learning algorithms have been compared (kNN and SVM), which have been parametrized to resolve the acoustic signal classification.
3.1. K-Nearest Neighbor
This algorithm determines the classification of new observations based on the closest training samples in the feature space. It matches the class measuring the distances of the k nearest data points to the test data. Then, simple majority of neighbors is used to determine the class prediction. In this study, the number of neighbors has been established to where N is the length of the feature coefficients for a syllable.
3.2. Support Vector Machine
SVM has been used to some extent in bioacoustic species recognition with success [13,26]. It discriminates the data by seeking the optimal hyperplane that separates the training data into two classes. However, the reptile call features are not lineally separable, so a non-linear kernel function has been used to divide the features in a higher dimensional space. For the experiments, a Gaussian or Radial Basis Function (RBF) kernel has been selected, where the parameter γ was optimized using a grid approach. For multiclass classification, the strategy “one-versus-one” [27] has been implemented, which trains a binary SVM classifier for each pair of classes. Therefore, for N different classes, binary classifiers are required to distinguish the samples, where N represents the number of reptile species. The SVM decision function is defined as in (4), where b is a numeric offset threshold and are Lagrange multipliers. The magnitude of α is determined by the C parameter, which imposes a penalty on misclassified samples ().
4. Experimental Procedure
This section describes the datasets and the experimental methodology used in the experiments to evaluate the effectiveness of the proposed method.
To ensure independence between the training and testing sets in each experiment (at least 100 simulations by experiment), the syllables obtained automatically from the segmentation of each sound have been randomly shuffled and split 50/50 into two datasets—one for training and another for testing (k-fold cross-validation with k = 2)—to achieve significant results.
Furthermore, accuracy has been calculated following Equation (5) for each class and averaging the results. F-Measure value [28] has also been calculated as , where P (precision) is the number of correct positive results divided by the number of all positive results, and R (recall) is the number of correct positive results divided by the number of positive results:
The acoustic classification system was implemented in Matlab, where the SVM implementation was based on the libsvm library [29], applying a C-Support Vector Classification (C-SVC) [30]. In addition, a non-dedicated standard laptop with an Intel Core i7-4510 2.0 GHz CPU and 16 GB RAM under Windows 8.1 operating system was used to carry out the experiments.
As indicated in Section 2.2, the number of cepstral features was obtained by experimentation. They were selected applying a wrapper method by varying the number of coefficients, N, from 6 to 25 for each feature until there was no improvement in prediction.
As for the SVM parameters, to find the optimum values of the penalty parameter of the error term (C) and the kernel gamma (γ) parameter, a grid-search was utilized with exponentially growing sequences of the parameters (; ) and employing cross-validation. Finally, a finer grid search was conducted, establishing a gamma value of 0.45 and a penalty term of 30 for all experiments.
Initially, the features have been analyzed individually to determine their effectiveness. However, different species of reptiles can produce sounds in frequencies anywhere in the spectrum. Therefore, to obtain the correct useful information, the two types of cepstral coefficients have been fused to better represent the acoustic information of the sounds.
4.1. Sound Dataset
The number of reptile sound repositories is quite limited. As a consequence, the dataset has been constructed using three internet sound collections. The main source of audio recordings has come from the Natural Museum of Berlin [31], which contains 120.000 audio recordings of diverse species. A third of them were recorded in controlled conditions, employing animals in captivity, and the rest in natural habitats with background noise. In addition, two on-line collections of reptiles from California have also been used: California Herps [32] and the California Tortoise Club [33]. Hence, the dataset is finally composed of 1,895 samples, which correspond to 27 different reptile species. Table 1 shows the list of species employed in this work, indicating the number of segmented syllables extracted from each species and their family group. All files have been sampled to 44.1 kHz.

Table 1.
Dataset classes.
4.2. Results and Discussion
In order to validate the proposed data fusion, the features have been analyzed individually to compare their performance. At the same time, the acoustic features have been combined with the classification algorithms to seek the best model. The algorithms have been run 100 times to obtain significant results. Additionally, the dataset has been randomly scrambled in each repetition, dividing the data 50/50 for training and testing purposes.
In Table 2, the experimental results show that MFCCs are more suitable for the identification of reptile calls than LFCCs. Mel features present more resolution at the lowest frequencies, emphasizing these spectrum regions where most reptile acoustic energy occurs. In fact, most of the sounds produced by reptiles are in the range 0.1 to 4 kHz. However, some reptiles—mainly lizards—can generate harmonic components at high frequency even into the ultrasound range (>20 kHz). At these frequencies, MFCCs hold insufficient information because the area under the triangular filters used in the mel-filterbank analysis increases at higher frequencies. Therefore, LFCCs are more suitable to model these reptile calls, since no frequency warping is applied. Thus, LFCC surpasses MFCC in some experiments, for instance in class 19 (Kinixys belliana or Hinge-back tortoise), where the mating calls contain high levels of low frequency noise. It can also be observed that Alligator sinensis (class 15) was poorly classified in all experiments, as this class was created by appending several audio recordings because each of them only contains one or two sample calls. Therefore, it presents diverse types of calls, which hinders the classification process. The distress calls emitted by the alligator present a complex harmonic pattern in a wide bandwidth (see Figure 2), occasionally extending over 15 kHz. Hence, the linear cepstral coefficients show a superior performance in both classifiers on class 15.

Table 2.
Classification results. kNN: K-Nearest Neighbors; LFCC: Linear Frequency Cepstral Coefficient; MFCC: Mel Frequency Cepstral Coefficient; SVM: Support Vector Machine.
On the other hand, Crotalus durissus (South American rattlesnake) also achieved low recognition rates, caused by its distinctive rattle noise that overlaps with the hissing call in several syllables. On the other hand, nine species reached a classification accuracy of 100%, regardless of the features used, due to the spectrum distribution of those reptile species being clearly different from others.
Regarding the classifiers, it is observed that SVM performs slightly better than kNN. This result is because the SVM approach is able to separate the classes more efficiently using the Gaussian kernel. However, as a consequence of the small corpus, the difference is not significant. In fact, for MFCC features, kNN outperforms SVM by 0.12%. However, it is expected that the difference will increase when new species are added to the corpus.
The feature fusion technique (MFCC/LFCC) exhibits high classification results in both algorithms, outperforming each of the individual features. In most cases, the resultant accuracy per class is equal to or greater than those achieved by single features. This confirms that this method provides a further characterization of the reptile calls by appending information of high and low frequency regions, which leads to a higher accuracy in classification. Furthermore, 13 classes were identified with a success rate of 100% when this approach was applied.
In the second experiment, the training samples were reduced from 50% to 5% to validate the robustness of the method using the best model found in the first experiment (MFCC/LFCC fusion + SVM). Table 3 shows that the fusion approach is able to deal with the low number of training samples, keeping accuracy and F-Measure values above 90% in almost all cases. This proves that the fusion of both cepstral coefficients is effective for modeling the discriminant information in the reptile calls. Moreover, the reduction in training set size offers savings in time needed to calculate the support vectors. However, when 5% of training is reached, the system clearly declines in effectiveness because most of the reptile species are only characterized by one syllable; thus, SVM has serious difficulties finding discriminant information to identify the classes. Nevertheless, this approach is able to maintain the classification results above 85%, confirming the robustness of the feature fusion method.

Table 3.
Classifier performance by training set size.
5. Conclusions and Future Work
Automated methods to detect and identify species are particularly useful for biodiversity studies and conservation purposes. In this paper, a novel automatic method for the bioacoustic recognition of reptile species by a fusion of frequency cepstral features has been presented. Reptile acoustic characteristics have been analyzed to seek the more discriminant features to parametrize their acoustic signals. It has been concluded that MFCCs are able to represent the reptile call efficiently because their acoustic signals are emitted predominantly in low frequencies. However, some species can also produce sounds in high frequencies; hence, LFCCs have also been utilized to hold information regarding that part of the spectra. The experimental results have demonstrated that the fusion of both features allows a broad characterization of the signal, increasing the classification rate. It has been validated in over 27 different reptile species, achieving an average accuracy of 98.52% ± 3.26. In addition, the proposed solution has been tested under low training sample conditions, proving the strength of the technique.
Traditional reptile surveys rely on visual searching, which is costly and time-consuming. Therefore, this approach can lead to the development of new remote monitoring systems for reptile research. In addition, the authors are not aware of other studies that have assessed the use of reptile acoustic signals for their inter-species recognition. However, despite the promising results of this first research, it is necessary increase the corpus and extend the solution to the entire animal group. Furthermore, it would be useful to enhance the approach by recognizing individuals within the same species. Finally, this approach could be applied to classify animals with similar sound production mechanisms (such as frogs or birds) by adjusting the system parameters.
Author Contributions
Juan J. Noda conceived and performed the experiments. Carlos M. Travieso, David Sánchez-Rodríguez and Juan J. Noda contributed to the study design, analyzed the data, and wrote the manuscript. All authors have reviewed and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gans, C.; Maderson, P.F. Sound producing mechanisms in recent reptiles: Review and comment. Am. Zool. 1973, 13, 1195–1203. [Google Scholar] [CrossRef]
- Bauer, A.M. Geckos: The Animal Answer Guide; JHU Press: Baltimore, MD, USA, 2013. [Google Scholar]
- Galeotti, P.; Sacchi, R.; Fasola, M.; Ballasina, D. Do mounting vocalisations in tortoises have a communication function? A comparative analysis. Herpetol. J. 2005, 15, 61–71. [Google Scholar]
- Vergne, A.; Pritz, M.; Mathevon, N. Acoustic communication in crocodilians: From behaviour to brain. Biol. Rev. 2009, 84, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Giles, J.C.; Davis, J.A.; McCauley, R.D.; Kuchling, G. Voice of the turtle: The underwater acoustic repertoire of the long-necked freshwater turtle, Chelodina oblonga. J. Acoust. Soc. Am. 2009, 126, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Labra, A.; Silva, G.; Norambuena, F.; Velásquez, N.; Penna, M. Acoustic features of the weeping lizard’s distress call. Copeia 2013, 2013, 206–212. [Google Scholar] [CrossRef]
- Bell, T.P. A novel technique for monitoring highly cryptic lizard species in forests. Herpetol. Conserv. Biol. 2009, 4, 415–425. [Google Scholar]
- Blumstein, D.T.; Mennill, D.J.; Clemins, P.; Girod, L.; Yao, K.; Patricelli, G.; Deppe, J.L.; Krakauer, A.H.; Clark, C.; Cortopassi, K.A.; et al. Acoustic monitoring in terrestrial environments using microphone arrays: Applications, technological considerations and prospectus. J. Appl. Ecol. 2011, 48, 758–767. [Google Scholar] [CrossRef]
- Fitzpatrick, M.C.; Preisser, E.L.; Ellison, A.M.; Elkinton, J.S. Observer bias and the detection of low-density populations. Ecol. Appl. 2009, 19, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Fuchida, M.; Pathmakumar, T.; Mohan, R.E.; Tan, N.; Nakamura, A. Vision-Based Perception and Classification of Mosquitoes Using Support Vector Machine. Appl. Sci. 2017, 7, 51. [Google Scholar] [CrossRef]
- Zhao, M.; Li, Z.; He, W. Classifying Four Carbon Fiber Fabrics via Machine Learning: A Comparative Study Using ANNs and SVM. Appl. Sci. 2016, 6, 209. [Google Scholar] [CrossRef]
- Li, H.; Yuan, D.; Wang, Y.; Cui, D.; Cao, L. Arrhythmia Classification Based on Multi-Domain Feature Extraction for an ECG Recognition System. Sensors 2016, 16, 1744. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, M.A.; Corrada-Bravo, C.J.; Corrada-Bravo, H.; Villanueva-Rivera, L.J.; Aide, T.M. Automated classification of bird and amphibian calls using machine learning: A comparison of methods. Ecol. Inform. 2009, 4, 206–214. [Google Scholar] [CrossRef]
- Brandes, T.S. Feature vector selection and use with hidden Markov models to identify frequency-modulated bioacoustic signals amidst noise. IEEE Trans. Audio Speech 2008, 16, 1173–1180. [Google Scholar] [CrossRef]
- Zhu, L.-Q. Insect sound recognition based on mfcc and pnn. In Proceedings of the 2011 International Conference on Multimedia and Signal Processing (CMSP), Guilin, China, 14–15 May 2011; Volume 2, pp. 42–46.
- Henríquez, A.; Alonso, J.B.; Travieso, C.M.; Rodríguez-Herrera, B.; Bolaños, F.; Alpízar, P.; López-de Ipina, K.; Henríquez, P. An automatic acoustic bat identification system based on the audible spectrum. Expert Syst. Appl. 2014, 41, 5451–5465. [Google Scholar] [CrossRef]
- Zhou, X.; Garcia-Romero, D.; Duraiswami, R.; Espy-Wilson, C.; Shamma, S. Linear versus mel frequency cepstral coefficients for speaker recognition. In Proceedings of the 2011 IEEE Workshop on Automatic Speech Recognition and Understanding (ASRU), Waikoloa, HI, USA, 11–15 December 2011; pp. 559–564.
- Cover, T.M.; Hart, P.E. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Härmä, A. Automatic identification of bird species based on sinusoidal modeling of syllables. In Proceedings of the 2003 IEEE International Conference on Acoustics, Speech, and Signal Processing, Hong Kong, China, 6–10 April 2003; Volume 5, p. V-545.
- Tiwari, V. MFCC and its applications in speaker recognition. Int. J. Emerg. Technol. 2010, 1, 19–22. [Google Scholar]
- Yuan, C.L.T.; Ramli, D.A. Frog sound identification system for frog species recognition. In Context-Aware Systems and Applications; Springer: Berlin, Germany, 2013; pp. 41–50. [Google Scholar]
- Jaafar, H.; Ramli, D.A.; Rosdi, B.A.; Shahrudin, S. Frog identification system based on local means k-nearest neighbors with fuzzy distance weighting. In The 8th International Conference on Robotic, Vision, Signal Processing & Power Applications; Springer: Berlin, Germany, 2014; pp. 153–159. [Google Scholar]
- Xie, J.; Towsey, M.; Truskinger, A.; Eichinski, P.; Zhang, J.; Roe, P. Acoustic classification of Australian anurans using syllable features. In Proceedings of the 2015 IEEE Tenth International Conference on Intelligent Sensors, Sensor Networks and Information Processing (ISSNIP), Singapore, 7–9 April 2015; pp. 1–6.
- Ganchev, T.; Potamitis, I.; Fakotakis, N. Acoustic monitoring of singing insects. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP 2007, Honolulu, HI, USA, 16–20 April 2007; Volume 4, p. IV-721.
- Fagerlund, S. Bird species recognition using support vector machines. EURASIP J. Appl. Signal Proc. 2007, 2007, 64. [Google Scholar] [CrossRef]
- Hsu, C.W.; Lin, C.J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar] [PubMed]
- Powers, D.M. Evaluation: From Precision, Recall and F-measure to ROC, Informedness, Markedness and Correlation; Bioinfo Publications: Pune, India, 2011. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011, 2, 27. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152.
- The Animal Sound Archive. Berlin Natural Museum. Available online: http://www.tierstimmenarchiv.de (accessed on 26 July 2015).
- California Reptiles and Amphibians. Available online: http://www.californiaherps.com (accessed on 26 July 2015).
- California Turtle and Tortoise Club. Available online: http://www.tortoise.org (accessed on 23 July 2015).
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).