Identification of Traditional She Medicine Shi-Liang Tea Species and Closely Related Species Using the ITS2 Barcode
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. DNA Extraction, Amplification and Sequencing
3. Results and Discussion
3.1. ITS2 Sequence Characteristics of Shi-Liang Tea Species and Closely Related Species
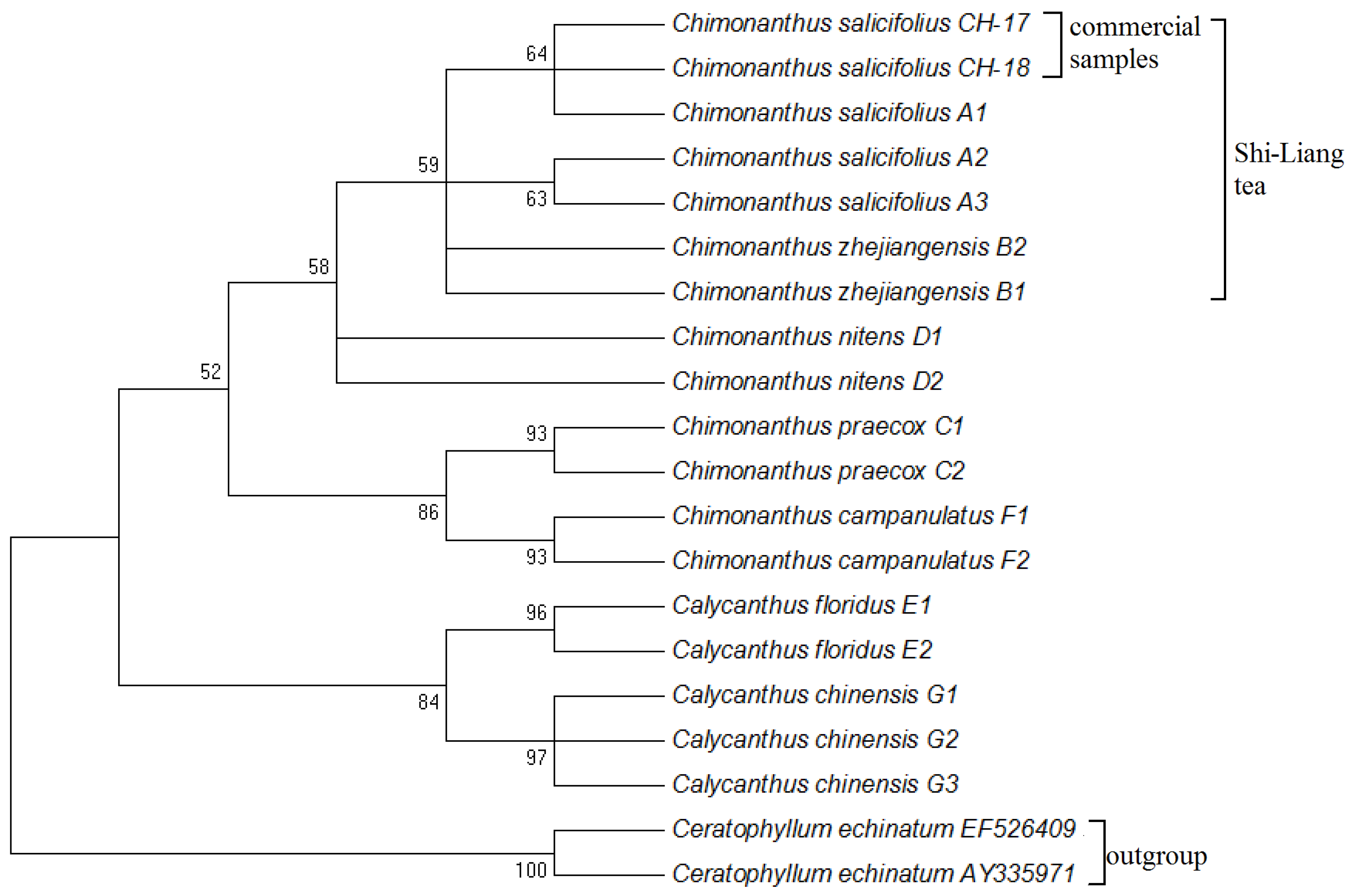
3.2. Clustering Analysis of Shi-Liang Tea Sources and Closely Related Species
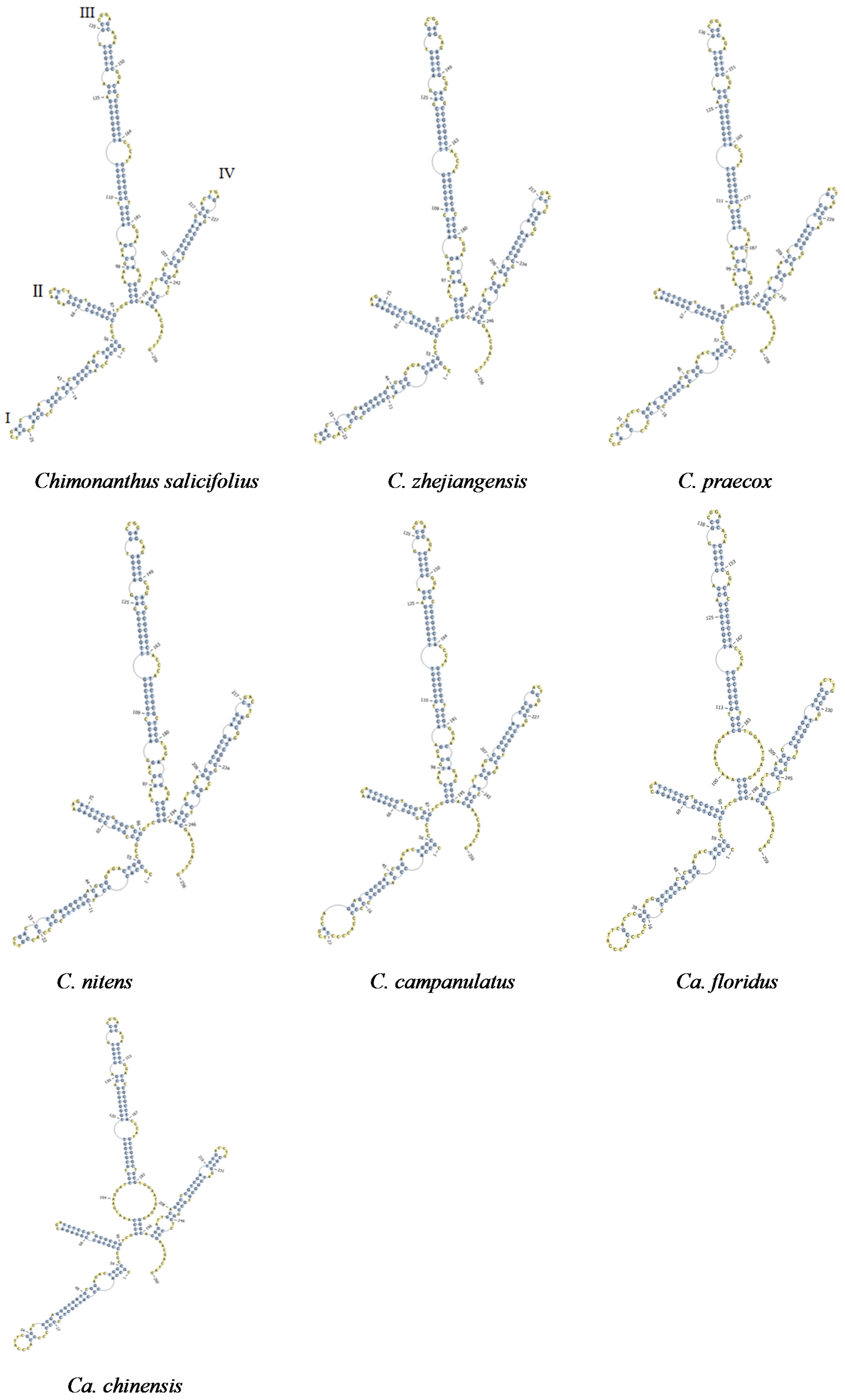
3.3. Comparison of ITS2 Secondary Structures of Shi-Liang Tea Species and Closely Related Species
3.4. Two-Dimensional DNA Barcodes to Monitor Market Circulation and Promote the Modernization of Shi-Liang Tea Species and Closely Related Species
4. Discussion
4.1. Application of ITS2 Sequences in Identification of She Medicine
4.2. DNA Barcoding Identification of Herbal Tea
4.3. Application of the Secondary Structure of ITS2 Sequences in Traditional Chinese Medicine Identification
4.4. Application and Significance of Two-Dimensional DNA Barcodes
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lei, H.; Li, J.; Zheng, S.; Fan, L.; Li, S.; Cheng, W.; Hua, J.; Yu, Y.; Dai, D.; Xie, W. Resources and Application of She’s Nationality Wild Medicinal Plants. Chin. J. Chin. Mater. Med. 2014, 39, 3180–3183. [Google Scholar]
- Lei, H.; Tao, Y.; Yan, L. Recent Progress in Research of She Nationality Medicine. J. Med. Pharm. Chin. Min. 2006, 12, 8–10. [Google Scholar]
- Lei, H.; Li, S. Chinese She Nationality Medicine; China Press of Traditional Chinese Medicine: Beijing, China, 2008. [Google Scholar]
- Wang, L.; Yan, L.; Yang, T.; Zhou, J. HPLC Determination of Rutin Quercein in Traditional She Medicine Diet Cool Tea. Chin. Arch. Tradit. Chin. Med. 2014, 32, 2916–2918. [Google Scholar]
- Zhejiang Province Food and Drug Administration. Zhejiang Province Processing Standard of Traditional Chinese Medicine; Zhejiang Science and Technology Press: Zhejiang, China, 2005; pp. 310–311. [Google Scholar]
- Xiao, K.; Liu, Y. Classification, chemical composition and pharmacological activity research progress of Chimonanthus. Res. Pract. Chin. Med. 2003, 17, 59–61. [Google Scholar]
- Ting, O.; Mai, X. Analysis of the Chemical Contituents of Essential Oil from Chimonanthus zhejiangensis by GC-MS. J. Chin. Med. Mater. 2010, 33, 385–386. [Google Scholar]
- Zhang, Y.; Hua, J.; Wang, X.; Cheng, W.; Lei, H.; Cheng, K.; Yu, P. Chemical constituents of chloroform fraction from leaves of Chimonanthus salicifolius. China. J. Chin. Mater. Med. 2013, 38, 2661–2664. [Google Scholar]
- Cui, X.; Li, M.; Zhou, B.; Li, J.; Cheng, D.; Zheng, P. Simultaneous Determination of Four Active Compounds in Chimonanthus nitens by HPLC. Chin. J. Exp. Tradit. Med. Formulae 2013, 19, 85–87. [Google Scholar]
- Mao, J.; Wang, W.; Yu, L.; Cheng, K. Study on Methodolgy of Content Determination of Three Flavonoids in Traditional She Medicine Shi Liang Cha. China Pharm. 2014, 17, 1808–1810. [Google Scholar]
- Yang, C.; Cheng, W.; Mei, J.; Yi, Y.; Cheng, J.; Ying, G.; Wang, H. Extraction and purification of total coumarins from Chimonanthus salicifolius Hu with ultrasonic assisted extraction. J. Zhejiang Univ. Technol. 2012, 40, 247–249. [Google Scholar]
- Ma, G.; Yang, G.; Xiong, J.; Cheng, K.; Hu, J. Salicifoxazines A and B, new cytotoxic tetrahydro-1,2-oxazine-containing tryptamine-derived alkaloids from the leaves of Chimonanthus salicifolius. Tetrahedron Lett. 2015, 56, 4071–4075. [Google Scholar] [CrossRef]
- Xu, J.; Cheng, K. Studies on the Alkaloids of the Calycanthaceae and Their Syntheses. Molecules 2015, 20, 6715–6738. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, L.; Chu, X.; Zhou, J. Chemical composition, antioxidant and antimicrobial activity of the extracts of the flowers of the Chinese plant Chimonanthus praecox. Nat. Prod. Res. 2012, 6, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhao, M.; Lan, T.; He, S. Research on China’s Chimonanthus plant taxonomy and Germplasm Resources. J. Beijing For. Univ. 1992, 14, 112–118. [Google Scholar]
- Zhu, Y.; Huang, Q. Studies on the identification of plants of Chimonanthus genus in China. J. Shanghai Med. Univ. 1987, 14, 451–455. [Google Scholar]
- Liu, Z.; Xi, J.; Schröder, S.; Wang, W.; Xie, T.; Wang, Z.; Bao, S.; Fei, J. Chimonanthus nitens var. salicifolius Aqueous Extract Protects against 5-Fluorouracil Induced Gastrointestinal Mucositis in a Mouse Model. Evid. Based Complement. Altern. Med. 2013, 4, 789263. [Google Scholar]
- Li, M.; Cao, H.; But, P.P.; Shaw, P.C. Identification of herbal medicinal materials using DNA barcodes. J. Syst. Evol. 2011, 49, 271–283. [Google Scholar] [CrossRef]
- Yang, P.; Li, X.; Zhou, H.; Hu, H.; Zhang, H.; Sun, W.; Wang, Y.; Yao, H. Molecular Identification of Chinese Materia Medica and Its Adulterants Using ITS2 and psbA-trnH Barcodes: A Case Study on Rhizoma Menispermi. Chin. Med. 2014, 5, 190–198. [Google Scholar] [CrossRef]
- Hou, D.; Song, J.; Yao, H.; Han, J.; Pang, X.; Shi, L.; Wang, X.; Chen, S. Molecular identification of Corni fructus and its adulterants by ITS/ITS2 sequences. Chin. J. Nat. Med. 2013, 11, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 regional a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef] [PubMed]
- Koetschan, C.; Förster, F.; Keller, A.; Schleicher, T.; Ruderisch, B.; Schwarz, R.; Müller, T.; Wolf, M.; Schultz, J. The ITS2 Database III-sequences and structures for phylogeny. Nucleic Acids Res. 2010, 38, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Schleicher, T.; Schultz, J.; Müller, T.; Dandekar, T.; Wolf, M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Xu, X.; Li, H.; Xing, H.; Liang, D.; Jiang, K.; Pang, X.; Song, J.; Chen, S. DNA Barcode Goes Two-Dimensions: DNA QR Code Web Server. PLoS ONE 2012, 7, e35146. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Selig, C.; Wolf, M.; Müller, T.; Dandekar, T.; Schultz, J. Homology modelling RNA structure for molecular systematics. Nucleic Acids Res. 2008, 36, D377–D380. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Muller, T.; Achtziger, M.; Seibel, P.N.; Dandekar, T.; Wolf, M. The internal transcribed spacer 2 database-a web server for (not only) low level phylogenetic analyses. Nucleic Acids Res. 2006, 34, W704–W707. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Standard DNA Barcodes of Chinese Materia Medica in Chinese Pharmacopoeia; Science Press: Beijing, China, 2015. [Google Scholar]
- Coleman, A. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3329. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Li, X.; Yao, H.; Han, J.; Song, J.; Chen, S. A two-dimensional DNA barcode system for circulation regulation of traditional Chinese medicine. Sci. Sin. Vitae 2015, 45, 695–702. [Google Scholar]
- Yao, H.; Song, J.; Ma, X.; Liu, C.; Li, Y.; Xu, H.; Han, J.; Duan, L.; Chen, S. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast psbA-trnH intergenic region. Planta Med. 2009, 75, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Li, X.; Yao, H.; Lin, Y.; Ma, X.; Cheng, R.; Song, J.; Ni, L.; Fan, C.; Chen, S. Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 2015, 5, 8337. [Google Scholar] [CrossRef] [PubMed]
- Sass, C.; Little, D.; Stevenson, D.; Specht, C. DNA barcoding in the cycadales: Testing the potential of proposed barcoding markers for species identification of cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.; Erickson, D. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shi, L.; Li, D.; Sun, Y.; Niu, Y.; Chen, Z.; Luo, H.; Pang, X.; Sun, Z.; Liu, C.; Lv, A.; Deng, Y.; Larson-Rabin, Z.; Wilkinson, M.; Chen, S. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS ONE 2012, 7, e43971. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Song, J.; Gao, Y.; Sun, Q.; Yao, H.; Wu, Q.; Chao, J.; Zhou, J.; Xue, W.; Duan, J. Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PLoS ONE 2013, 8, e67818. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y. The Study of Tea Culture in Contemporary China and the Future Direction of Chinese. Jiangxi Soc. Sci. 2005, 7, 7–18. [Google Scholar]
- Chen, S.; Guo, B.; Zhang, G.; Yan, Z.; Luo, G.; Sun, S.; Wu, H.; Huang, L.; Pang, X.; Chen, J. Advances of studies on new technology and method for identifying traditional Chinese medicinal materials. Chin. J. Chin. Mater. Med. 2012, 37, 1043–1055. [Google Scholar]
- Stoeckle, M.Y.; Gamble, C.C.; Kirpekar, R.; Young, G.; Ahmed, S.; Little, D.P. Commercial Teas Highlight Plant DNA Barcode Identification Successes and Obstacles. Sci. Rep. 2011, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wong, K.; Chan, W.; Li, J.; But, P.P.; Cao, H. Establishment of DNA barcodes for the identification of the botanical sources of the Chinese ‘cooling’ beverage. Food Control. 2012, 25, 758–766. [Google Scholar] [CrossRef]
- Lei, H.; Li, S.; Li, J. Study of Traditional She’s Medicine Characteristics and Its Application. Chin. Arch. Tradit. Chin. Med. 2013, 31, 1911–1913. [Google Scholar]
- Zhang, Y. Research on Chinese Tea Culture of Ming Dynasty from Historical Perspective of Health Promotion; China Academy of Chinese Medical Sciences: Beijing, China, 2012. [Google Scholar]
- Huang, X. Studies on the Booming of Tea Culture and Its Effects in Modern Time; Zhejiang University: Hangzhou, China, 2003. [Google Scholar]
- Zhao, S.; Chen, X.; Song, J.; Pang, X.; Chen, S. Internal transcribed spacer 2 barcode: A good tool for identifying Acanthopanacis cortex. Front. Plant Sci. 2015, 6, 840. [Google Scholar] [CrossRef] [PubMed]

| Species Name | Amount of Samples | Sample No. or GenBank Accession Number | Sample Sources |
|---|---|---|---|
| Chimonanthus salicifolius | 1 | CH-3 | Liandu, Zhejiang |
| 6 | CH-4, CH-18 *, CH-23~24, CH-26, RC_CH-4 | Kaihua, Zhejiang | |
| 4 | CH-16, CH-17 *, CH-25, RC_CH-16 | Songyang, Zhejiang | |
| 1 | CH-22 | Chun’an, Zhejiang | |
| 2 | CH-5, RC_CH-5 | Wuyuan, Jiangxi | |
| 1 | CH-21 | Dexing, Jiangxi | |
| 2 | CH-6, RC_CH-6 | Xi’an, Shaanxi | |
| 1 | CH-19 | Xiuning, Anhui | |
| 5 | JQ781681-85 | GenBank | |
| C. zhejiangensis | 2 | CH-7, CH-33 | Liandu, Zhejiang |
| 1 | CH-27 | Songyang, Zhejiang | |
| 1 | CH-28 | Qingtian, Zhejiang | |
| 1 | CH-29 | Yunhe, Zhejiang | |
| 1 | CH-30 | Jingning, Zhejiang | |
| 1 | CH-31 | Qingyuan, Zhejiang | |
| 1 | CH-32 | Longquan, Zhejiang | |
| 1 | CH-34 | Taishun, Zhejiang | |
| 1 | CH-9 | Shouning, Fujian | |
| 5 | JQ781668-72 | GenBank | |
| C. praecox | 1 | CH-1 | Liandu, Zhejiang |
| 9 | JQ781691-95, AY786095-98 | GenBank | |
| C. nitens | 1 | CH-11 | Yangshou, Guangxi |
| 5 | JQ781664-67, AY786094 | GenBank | |
| C. campanulatus | 2 | CH-13, RC_CH-13 | Liandu, Zhejiang |
| 5 | JQ781686-90 | GenBank | |
| Calycanthus chinensis | 3 | CH-15, CH-36, RC_CH-15 | Liandu, Zhejiang |
| 2 | KF547941, AY524084 | GenBank | |
| Ca. floridus | 1 | CH-14 | Liandu, Zhejiang |
| 4 | AY524078, AY786084-86 | GenBank |
| Latin Name | Sequence Length (bp) | Average GC Contents (%) |
|---|---|---|
| Chimonanthus salicifolius | 256–258 | 71.2 |
| C. zhejiangensis | 256 | 71.1 |
| C. praecox | 258 | 71.7 |
| C. nitens | 256 | 71.0 |
| C. campanulatus | 256–257 | 72.7 |
| Calycanthus chinensis | 257–260 | 71.5 |
| Ca. floridus | 258–259 | 72.0 |
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 0.000–0.008 | 0.004–0.008 | 0.036 | 0.008–0.016 | 0.040 | 0.073–0.078 | 0.061–0.066 |
| 2 | 0.000–0.004 | 0.032–0.036 | 0.004–0.016 | 0.036–0.040 | 0.070–0.078 | 0.057–0.066 | |
| 3 | 0.000 | 0.028–0.036 | 0.016 | 0.069–0.074 | 0.052–0.057 | ||
| 4 | 0.000–0.012 | 0.032–0.040 | 0.074–0.088 | 0.061–0.075 | |||
| 5 | 0.000 | 0.074–0.078 | 0.057–0.061 | ||||
| 6 | 0.000–0.004 | 0.032–0.040 | |||||
| 7 | 0.000–0.004 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, S.; Lv, Q.; Zhou, H.; Fang, J.; Cheng, W.; Jiang, C.; Cheng, K.; Yao, H. Identification of Traditional She Medicine Shi-Liang Tea Species and Closely Related Species Using the ITS2 Barcode. Appl. Sci. 2017, 7, 195. https://doi.org/10.3390/app7030195
Ma S, Lv Q, Zhou H, Fang J, Cheng W, Jiang C, Cheng K, Yao H. Identification of Traditional She Medicine Shi-Liang Tea Species and Closely Related Species Using the ITS2 Barcode. Applied Sciences. 2017; 7(3):195. https://doi.org/10.3390/app7030195
Chicago/Turabian StyleMa, Shuangjiao, Qundan Lv, Hong Zhou, Jie Fang, Wenliang Cheng, Chengxi Jiang, Kejun Cheng, and Hui Yao. 2017. "Identification of Traditional She Medicine Shi-Liang Tea Species and Closely Related Species Using the ITS2 Barcode" Applied Sciences 7, no. 3: 195. https://doi.org/10.3390/app7030195






