Atomistic Insight into the Role of Threonine 127 in the Functional Mechanism of Channelrhodopsin-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site-Directed Mutagenesis, Cloning, Expression and Purification of CrChR2
2.2. Molecular Spectroscopy
2.3. Electrophysiology
2.4. QM/MM Calculations
3. Results
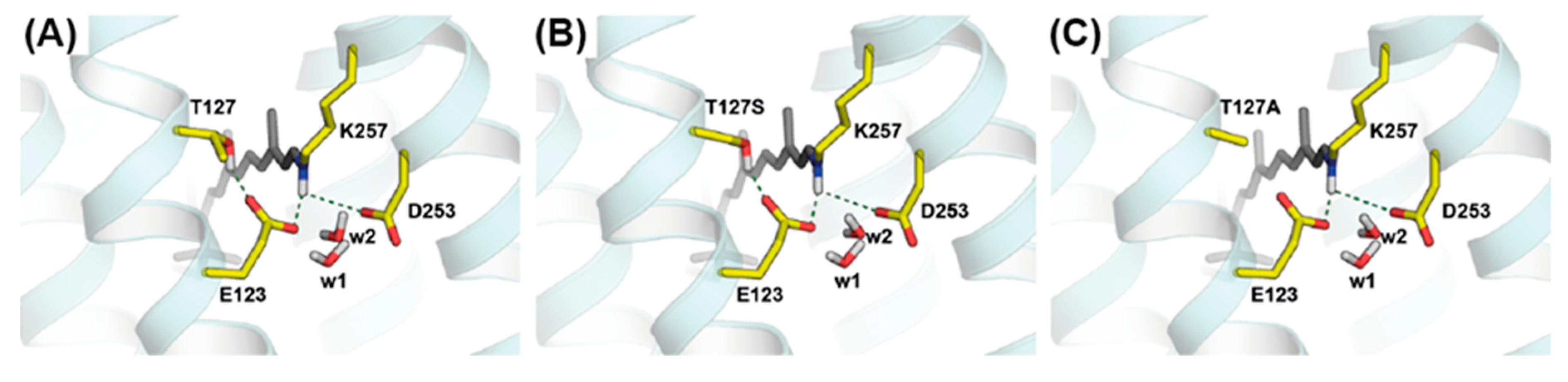
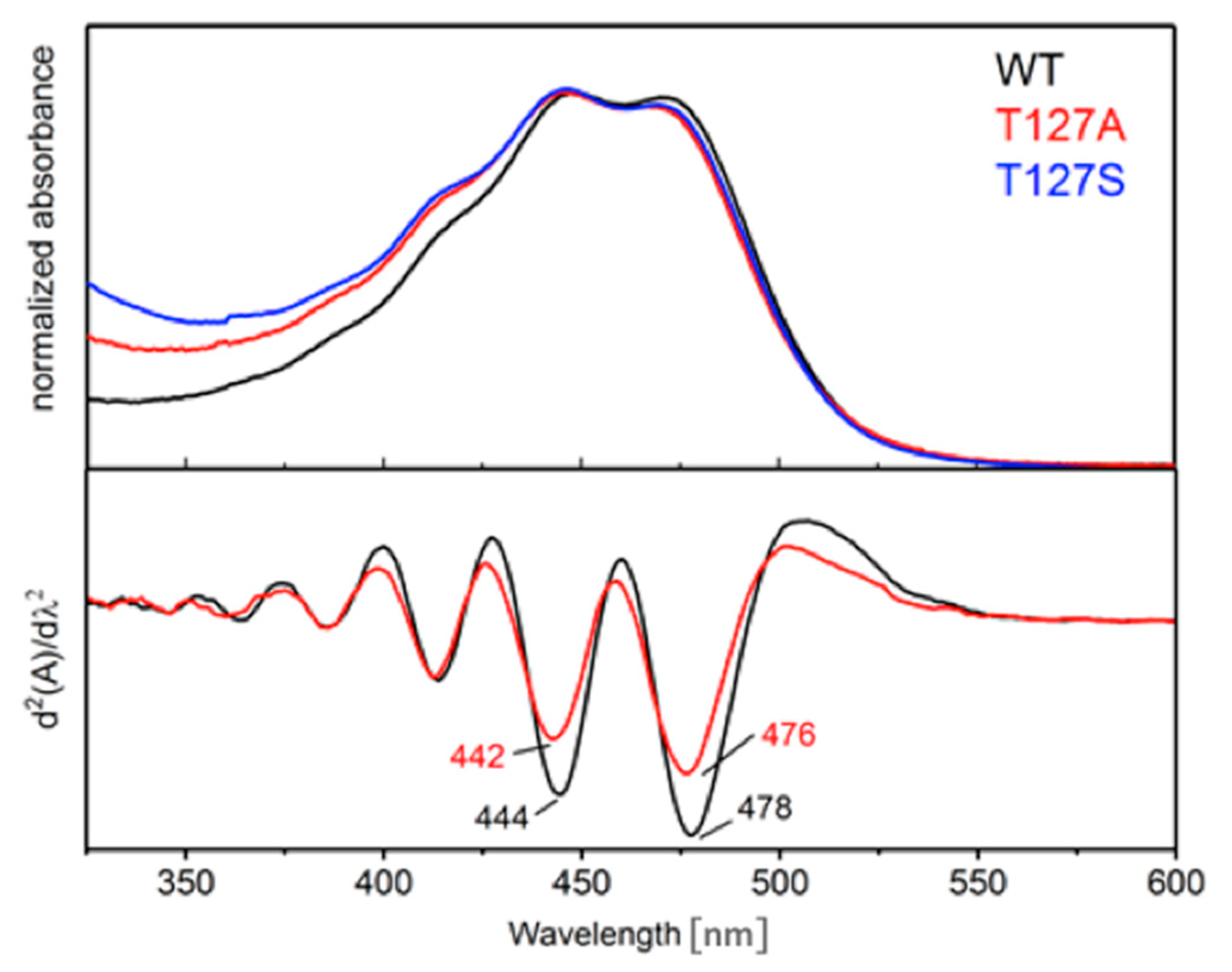
3.1. Spectroscopic Properties of the Dark State of the T127 Variants of CrChR2
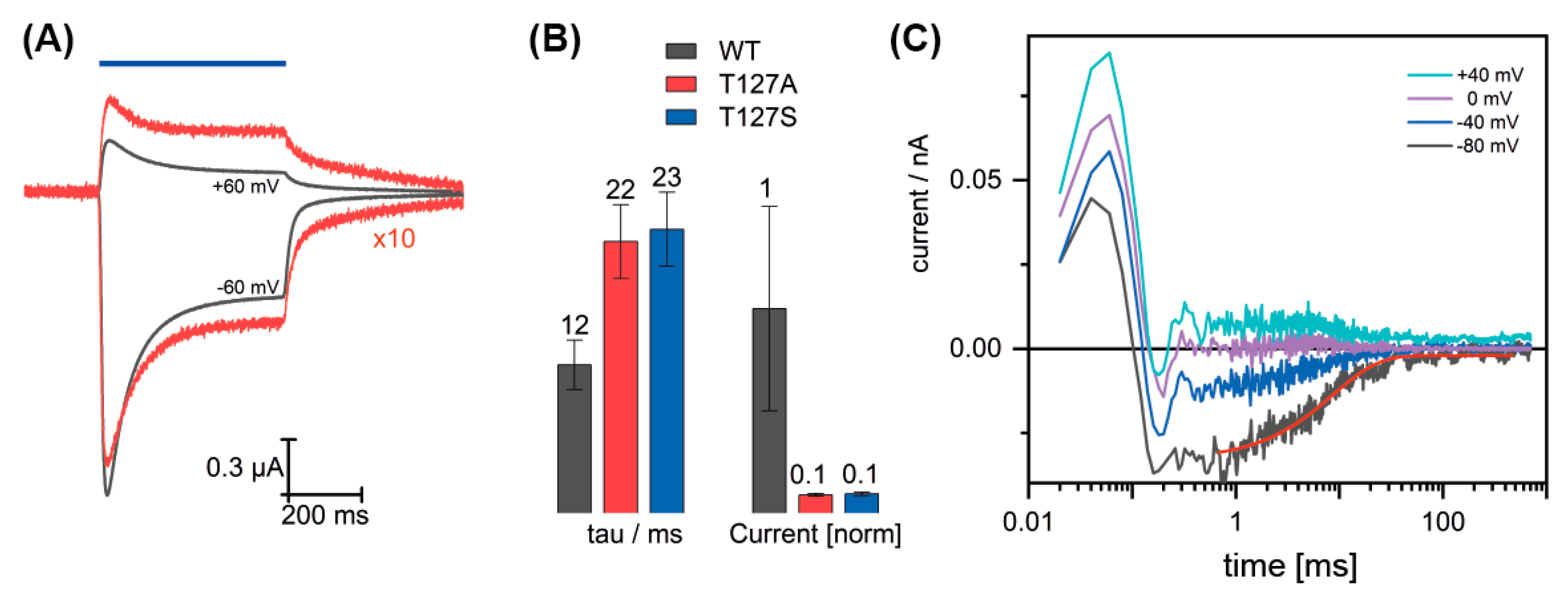
3.2. Channel Conductance of the T127 Variants
3.3. Influence of T127 on the Photocycle Kinetics
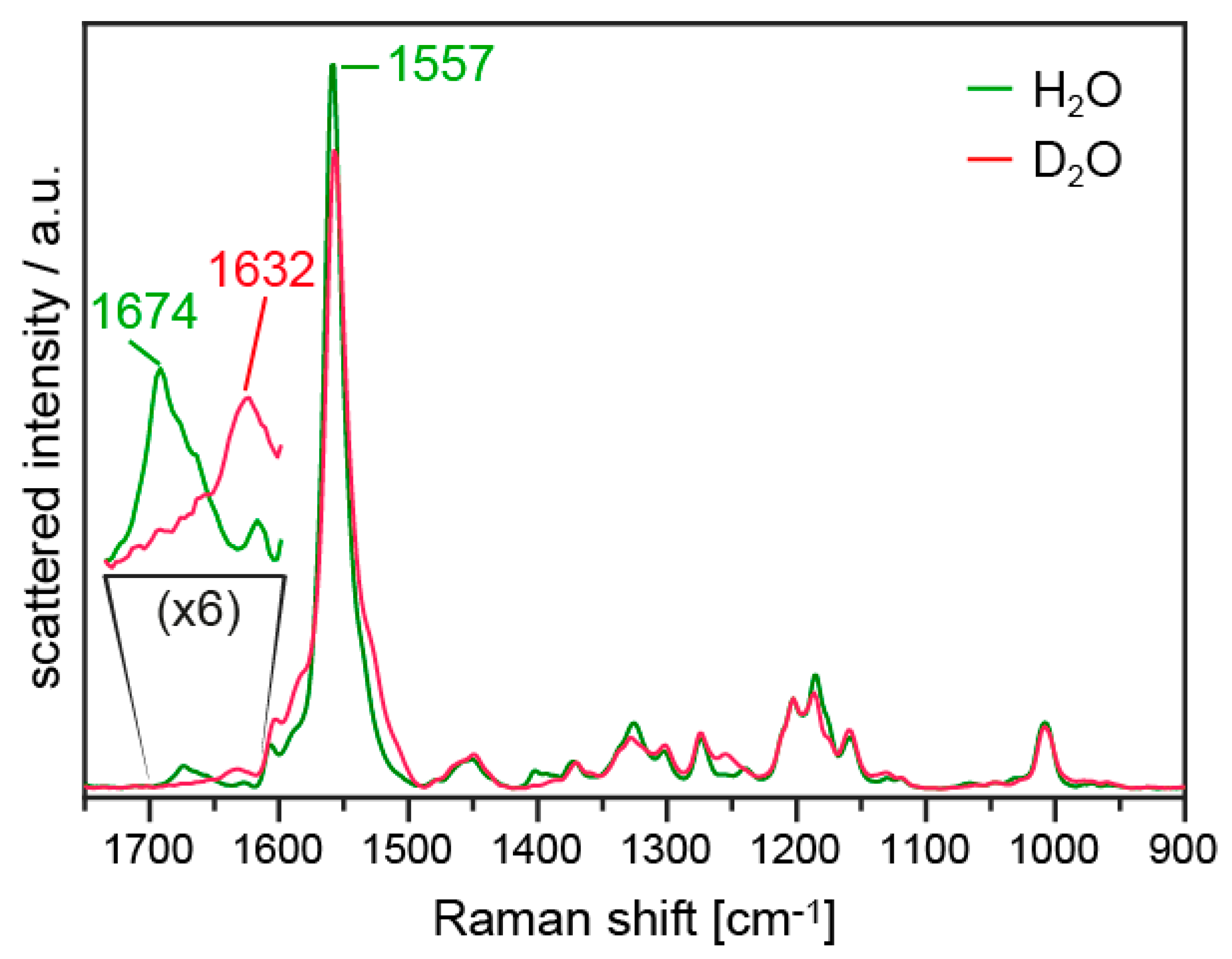
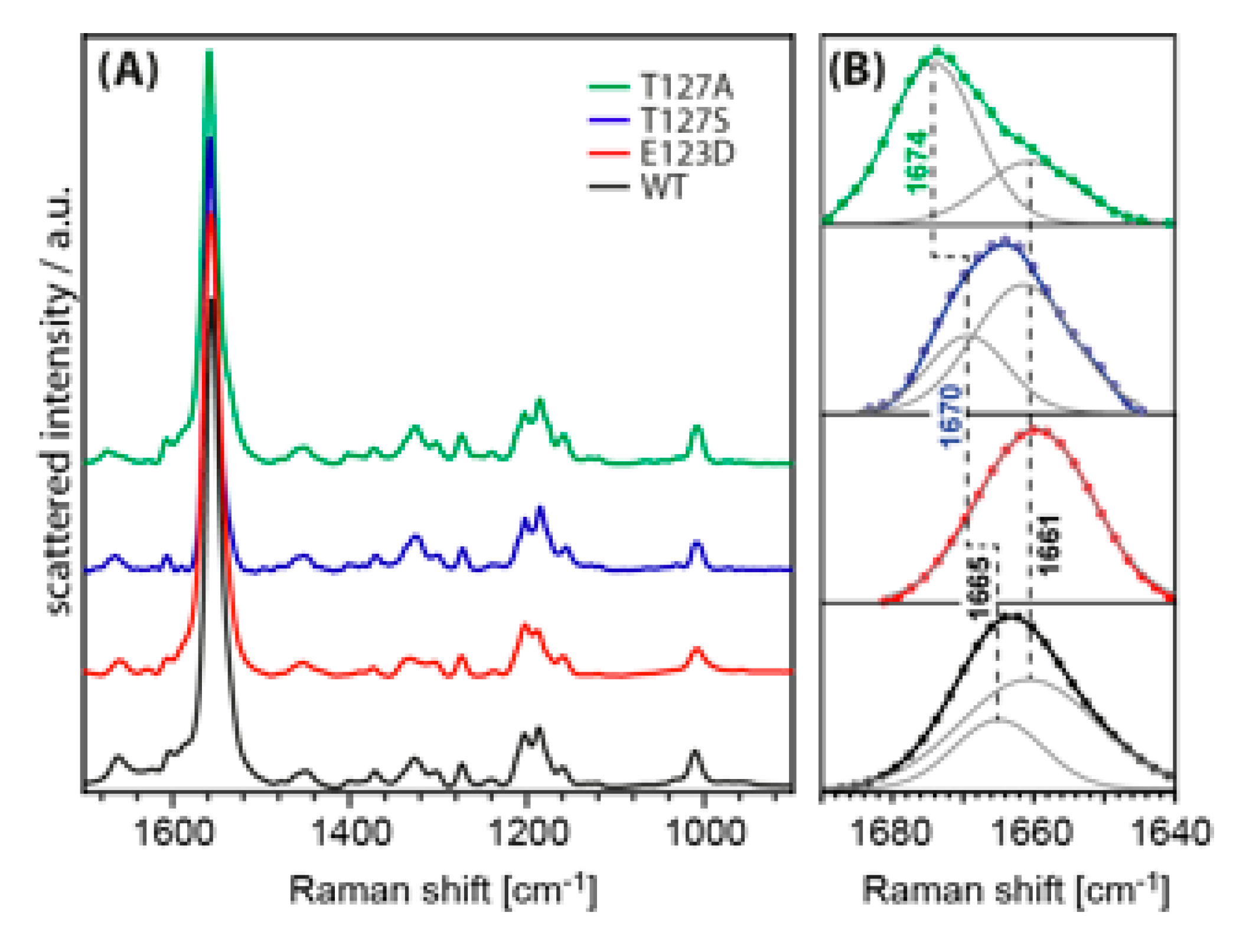
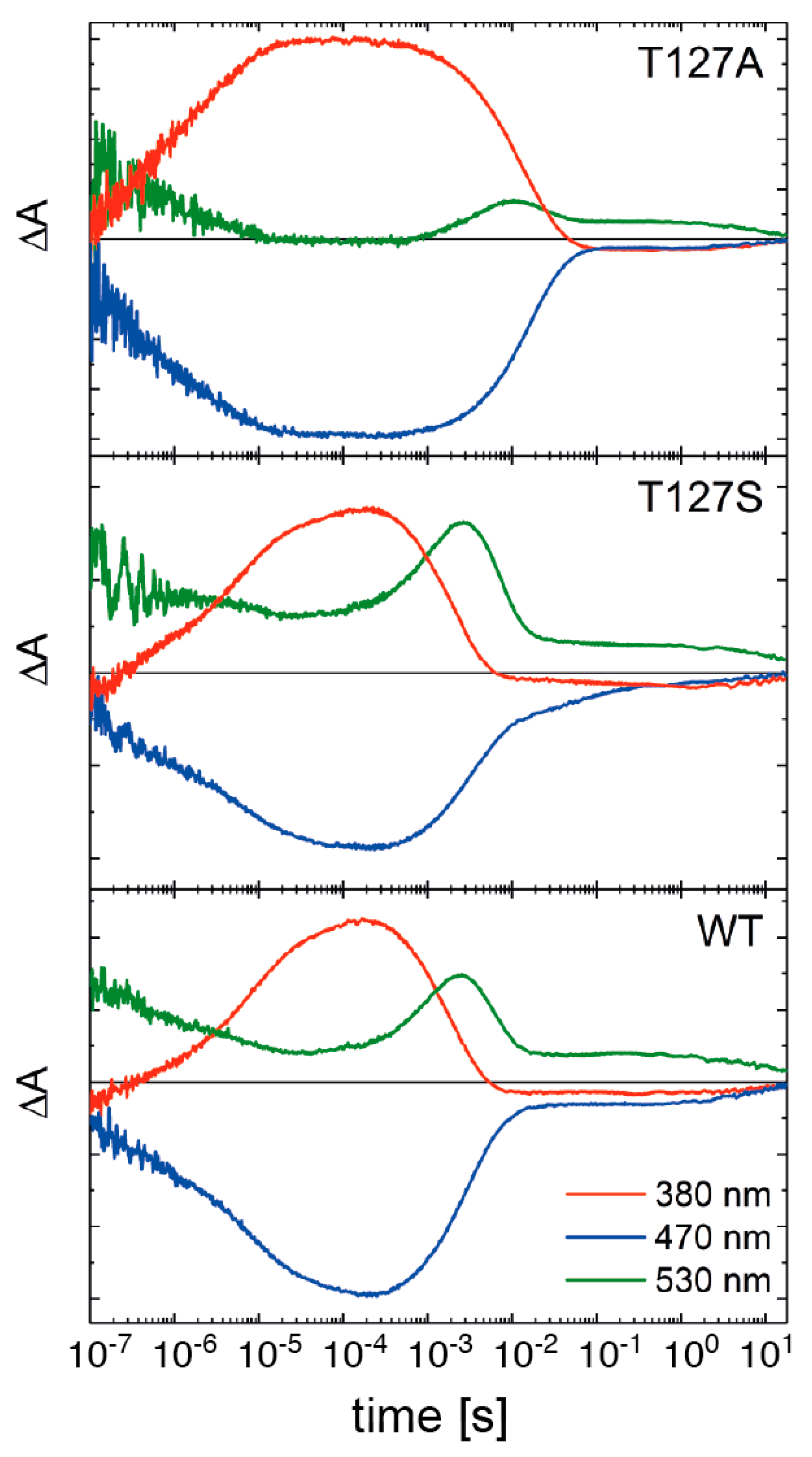
3.4. FTIR Difference Spectroscopy on the T127 Variants
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nagel, G.; Ollig, D.; Fuhrmann, M.; Kateriya, S.; Musti, A.M.; Bamberg, E.; Hegemann, P. Channelrhodopsin-1: A light-gated proton channel in green algae. Science 2002, 296, 2395–2398. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [PubMed]
- Marti, T.; Otto, H.; Mogi, T.; Rosselet, S.J.; Heyn, M.P.; Khorana, H.G. Bacteriorhodopsin mutants containing single substitutions of serine or threonine residues are all active in proton translocation. J. Biol. Chem. 1991, 266, 6919–6927. [Google Scholar]
- Rothschild, K.J.; He, Y.W.; Sonar, S.; Marti, T.; Khorana, H.G. Vibrational spectroscopy of bacteriorhodopsin mutants. Evidence that thr-46 and thr-89 form part of a transient network of hydrogen bonds. J. Biol. Chem. 1992, 267, 1615–1622. [Google Scholar] [PubMed]
- Russell, T.S.; Coleman, M.; Rath, P.; Nilsson, A.; Rothschild, K.J. Threonine-89 participates in the active site of bacteriorhodopsin: Evidence for a role in color regulation and schiff base proton transfer. Biochemistry 1997, 36, 7490–7497. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Rummel, G.; Rosenbusch, J.P.; Landau, E.M. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science 1997, 277, 1676–1681. [Google Scholar] [CrossRef]
- Luecke, H.; Richter, H.T.; Lanyi, J.K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science 1998, 280, 1934–1937. [Google Scholar] [CrossRef]
- Kandori, H.; Kinoshita, N.; Yamazaki, Y.; Maeda, A.; Shichida, Y.; Needleman, R.; Lanyi, J.K.; Bizounok, M.; Herzfeld, J.; Raap, J.; et al. Structural change of threonine 89 upon photoisomerization in bacteriorhodopsin as revealed by polarized ftir spectroscopy. Biochemistry 1999, 38, 9676–9683. [Google Scholar] [CrossRef]
- Kandori, H.; Yamazaki, Y.; Shichida, Y.; Raap, J.; Lugtenburg, J.; Belenky, M.; Herzfeld, J. Tight asp-85-thr-89 association during the pump switch of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 2001, 98, 1571–1576. [Google Scholar] [CrossRef]
- Volkov, O.; Kovalev, K.; Polovinkin, V.; Borshchevskiy, V.; Bamann, C.; Astashkin, R.; Marin, E.; Popov, A.; Balandin, T.; Willbold, D.; et al. Structural insights into ion conduction by channelrhodopsin 2. Science 2017, 358, 8862. [Google Scholar] [CrossRef]
- Hou, S.Y.; Govorunova, E.G.; Ntefidou, M.; Lane, C.E.; Spudich, E.N.; Sineshchekov, O.A.; Spudich, J.L. Diversity of chlamydomonas channelrhodopsins. Photochem. Photobiol. 2012, 88, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Govorunova, E.G.; Sineshchekov, O.A.; Rodarte, E.M.; Janz, R.; Morelle, O.; Melkonian, M.; Wong, G.K.-S.; Spudich, J.L. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 2017, 7, 43358. [Google Scholar] [CrossRef] [PubMed]
- Lorenz-Fonfria, V.A.; Resler, T.; Krause, N.; Nack, M.; Gossing, M.; Fischer von Mollard, G.; Bamann, C.; Bamberg, E.; Schlesinger, R.; Heberle, J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA 2013, 110, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Nack, M.; Radu, I.; Gossing, M.; Bamann, C.; Bamberg, E.; von Mollard, G.F.; Heberle, J. The dc gate in channelrhodopsin-2: Crucial hydrogen bonding interaction between c128 and d156. Photochem. Photobiol. Sci. 2010, 9, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.C.; Welke, K.; Sindhikara, D.J.; Hegemann, P.; Elstner, M. Towards an understanding of channelrhodopsin function: Simulations lead to novel insights of the channel mechanism. J. Mol. Biol. 2013, 425, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Krause, N.; Engelhard, C.; Heberle, J.; Schlesinger, R.; Bittl, R. Structural differences between the closed and open states of channelrhodopsin-2 as observed by epr spectroscopy. FEBS Lett. 2013, 587, 3309–3313. [Google Scholar] [CrossRef]
- Resler, T.; Schultz, B.J.; Lorenz-Fonfria, V.A.; Schlesinger, R.; Heberle, J. Kinetic and vibrational isotope effects of proton transfer reactions in channelrhodopsin-2. Biophys. J. 2015, 109, 287–297. [Google Scholar] [CrossRef]
- Noguchi, T.; Sugiura, M. Flash-induced ftir difference spectra of the water oxidizing complex in moderately hydrated photosystem ii core films: Effect of hydration extent on s-state transitions. Biochemistry 2002, 41, 2322–2330. [Google Scholar] [CrossRef]
- Schnedermann, C.; Muders, V.; Ehrenberg, D.; Schlesinger, R.; Kukura, P.; Heberle, J. Vibronic dynamics of the ultrafast all-trans to 13-cis photoisomerization of retinal in channelrhodopsin-1. J. Am. Chem. Soc. 2016, 138, 4757–4762. [Google Scholar] [CrossRef]
- DeLano, W.L. The Pymol Molecular Graphics System; DeLano Scientific LLC: San Carlos, CA, USA, 2002. [Google Scholar]
- Becke, A. Density-functional thermochemistry: The role of extract exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14sb: Improving the accuracy of protein side chain and backbone parameters from ff99sb. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R. Nist Standard Reference Database Number 101. Available online: http://cccbdb.nist.gov (accessed on 1 October 2015).
- Bannwarth, C.; Grimme, S. A simplified time-dependent density functional theory approach for electronic ultraviolet and circular dichroism spectra of very large molecules. Comput. Theory Chem. 2014, 1040, 45–53. [Google Scholar] [CrossRef]
- Metz, S.; Kästner, J.; Sokol, A.A.; Keal, T.W.; Sherwood, P. C hem s hell—A modular software package for qm/mm simulations. Wires Comput. Mol. Sci. 2014, 4, 101–110. [Google Scholar] [CrossRef]
- Radu, I.; Bamann, C.; Nack, M.; Nagel, G.; Bamberg, E.; Heberle, J. Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 2009, 131, 7313–7319. [Google Scholar] [CrossRef] [PubMed]
- Aton, B.; Doukas, A.G.; Callender, R.H.; Becher, B.; Ebrey, T.G. Resonance raman studies of the purple membrane. Biochemistry 1977, 16, 2995–2999. [Google Scholar] [CrossRef]
- Nack, M.; Radu, I.; Schultz, B.J.; Resler, T.; Schlesinger, R.; Bondar, A.N.; del Val, C.; Abbruzzetti, S.; Viappiani, C.; Bamann, C.; et al. Kinetics of proton release and uptake by channelrhodopsin-2. FEBS Lett. 2012, 586, 1344–1348. [Google Scholar] [CrossRef]
- Narva, D.; Callender, R.H. On the state of chromophore protonation in rhodopsin: Implication for primary photochemistry in visual pigments. Photochem. Photobiol. 1980, 32, 273–276. [Google Scholar] [CrossRef]
- Guo, Y.; Beyle, F.E.; Bold, B.M.; Watanabe, H.C.; Koslowski, A.; Thiel, W.; Hegemann, P.; Marazzi, M.; Elstner, M. Active site structure and absorption spectrum of channelrhodopsin-2 wild-type and c128t mutant. Chem. Sci. 2016, 7, 3879–3891. [Google Scholar] [CrossRef]
- Nack, M.; Radu, I.; Bamann, C.; Bamberg, E.; Heberle, J. The retinal structure of channelrhodopsin-2 assessed by resonance raman spectroscopy. FEBS Lett. 2009, 583, 3676–3680. [Google Scholar] [CrossRef]
- Lórenz-Fonfría, V.A.; Schultz, B.-J.; Resler, T.; Schlesinger, R.; Bamann, C.; Bamberg, E.; Heberle, J. Pre-gating conformational changes in the cheta variant of channelrhodopsin-2 monitored by nanosecond ir spectroscopy. J. Am. Chem. Soc. 2015, 137, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Lorenz-Fonfria, V.A.; Bamann, C.; Resler, T.; Schlesinger, R.; Bamberg, E.; Heberle, J. Temporal evolution of helix hydration in a light-gated ion channel correlates with ion conductance. Proc. Natl. Acad. Sci. USA 2015, 112, 5796–5804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saita, M.; Pranga-Sellnau, F.; Resler, T.; Schlesinger, R.; Heberle, J.; Lorenz-Fonfria, V.A. Photoexcitation of the p4(480) state induces a secondary photocycle that potentially desensitizes channelrhodopsin-2. J. Am. Chem. Soc. 2018, 140, 9899–9903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bamann, C.; Kirsch, T.; Nagel, G.; Bamberg, E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008, 375, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Ernst, O.P.; Murcia, P.A.S.; Daldrop, P.; Tsunoda, S.P.; Kateriya, S.; Hegemann, P. Photoactivation of channelrhodopsin. J. Biol. Chem. 2008, 283, 1637–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lórenz-Fonfría, V.A.; Heberle, J. Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 626–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Val, C.; Royuela-Flor, J.; Milenkovic, S.; Bondar, A.N. Channelrhodopsins: A bioinformatics perspective. Biochim. Biophys. Acta 2014, 1837, 643–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodor, S.; Gebhard, R.; Lugtenburg, J.; Bogomolni, R.; Mathies, R. Structure of the retinal chromophore in sensory rhodopsin i from resonance raman spectroscopy. J. Biol. Chem. 1989, 264, 18280–18283. [Google Scholar]
- Heberle, J.; Oesterhelt, D.; Dencher, N.A. Decoupling of photo- and proton cycle in the asp85--> glu mutant of bacteriorhodopsin. EMBO J. 1993, 12, 3721–3727. [Google Scholar] [CrossRef]
- Berndt, A.; Schoenenberger, P.; Mattis, J.; Tye, K.M.; Deisseroth, K.; Hegemann, P.; Oertner, T.G. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. USA 2011, 108, 7595–7600. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| RSBH+ Atom Pairs (Distances in Å) | WT | T127S | T127A |
|---|---|---|---|
| C=N | 1.310 | 1.308 | 1.309 |
| N–H | 1.076 | 1.072 | 1.092 |
| Model | WT | T127S | T127A |
|---|---|---|---|
| Excitation energy nm (eV) | 389 (3.18) | 392 (3.16) | 388 (3.19) |
| Model | WT | T127S | T127A |
|---|---|---|---|
| Frequencies (cm−1) | 1683 cm−1 | 1683 cm−1 | 1702 cm−1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehrenberg, D.; Krause, N.; Saita, M.; Bamann, C.; Kar, R.K.; Hoffmann, K.; Heinrich, D.; Schapiro, I.; Heberle, J.; Schlesinger, R. Atomistic Insight into the Role of Threonine 127 in the Functional Mechanism of Channelrhodopsin-2. Appl. Sci. 2019, 9, 4905. https://doi.org/10.3390/app9224905
Ehrenberg D, Krause N, Saita M, Bamann C, Kar RK, Hoffmann K, Heinrich D, Schapiro I, Heberle J, Schlesinger R. Atomistic Insight into the Role of Threonine 127 in the Functional Mechanism of Channelrhodopsin-2. Applied Sciences. 2019; 9(22):4905. https://doi.org/10.3390/app9224905
Chicago/Turabian StyleEhrenberg, David, Nils Krause, Mattia Saita, Christian Bamann, Rajiv K. Kar, Kirsten Hoffmann, Dorothea Heinrich, Igor Schapiro, Joachim Heberle, and Ramona Schlesinger. 2019. "Atomistic Insight into the Role of Threonine 127 in the Functional Mechanism of Channelrhodopsin-2" Applied Sciences 9, no. 22: 4905. https://doi.org/10.3390/app9224905





