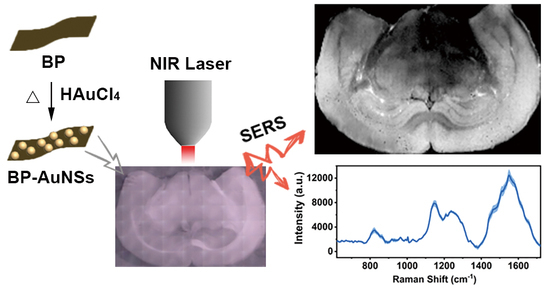
Full-Scale Label-Free Surface-Enhanced Raman Scattering Analysis of Mouse Brain Using a Black Phosphorus-Based Two-Dimensional Nanoprobe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of 2D SERS Substrate
2.3. Characterization
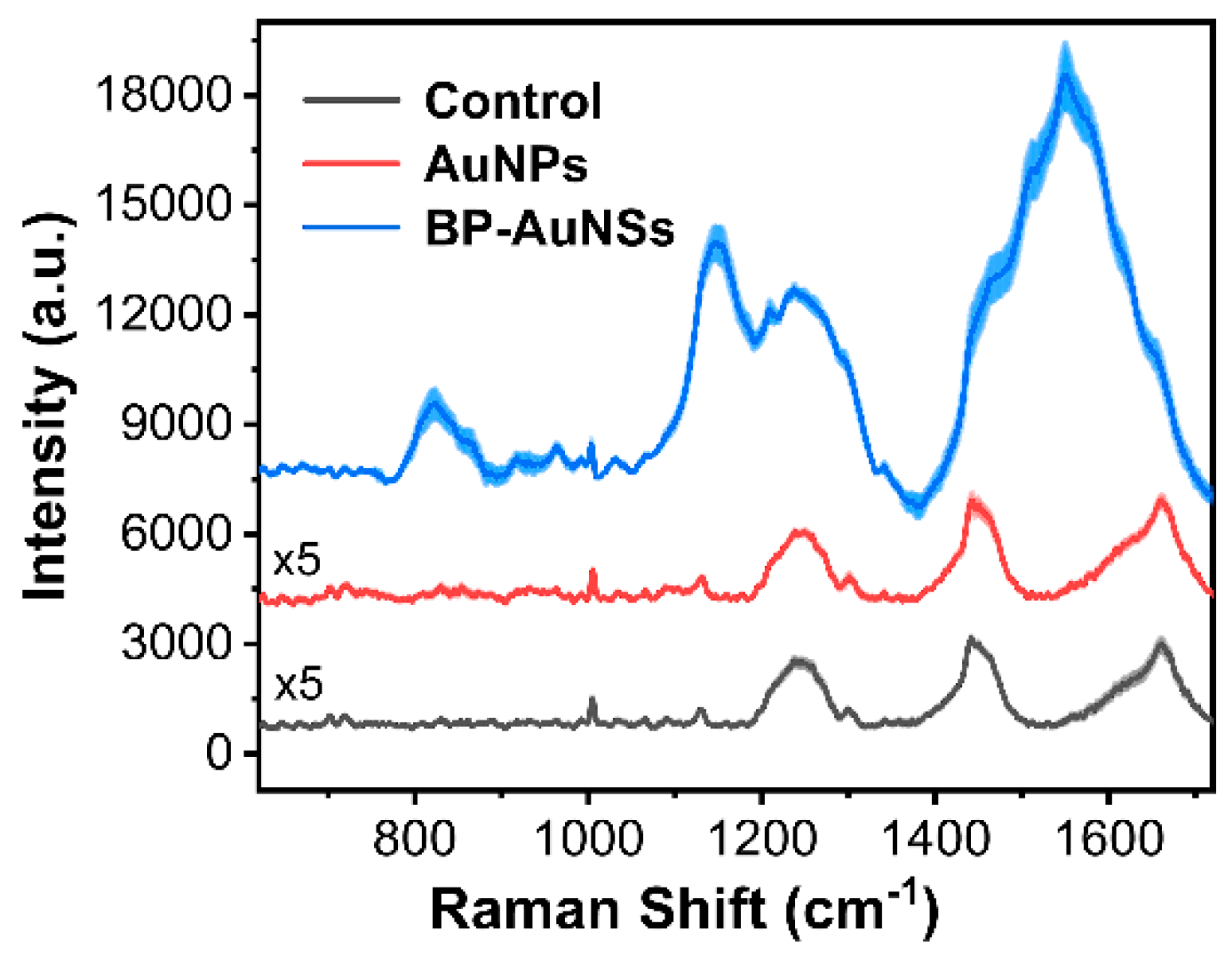
2.4. SERS Activity of BP-AuNSs
2.5. SERS Analysis of Brain Tissue
3. Results and Discussion
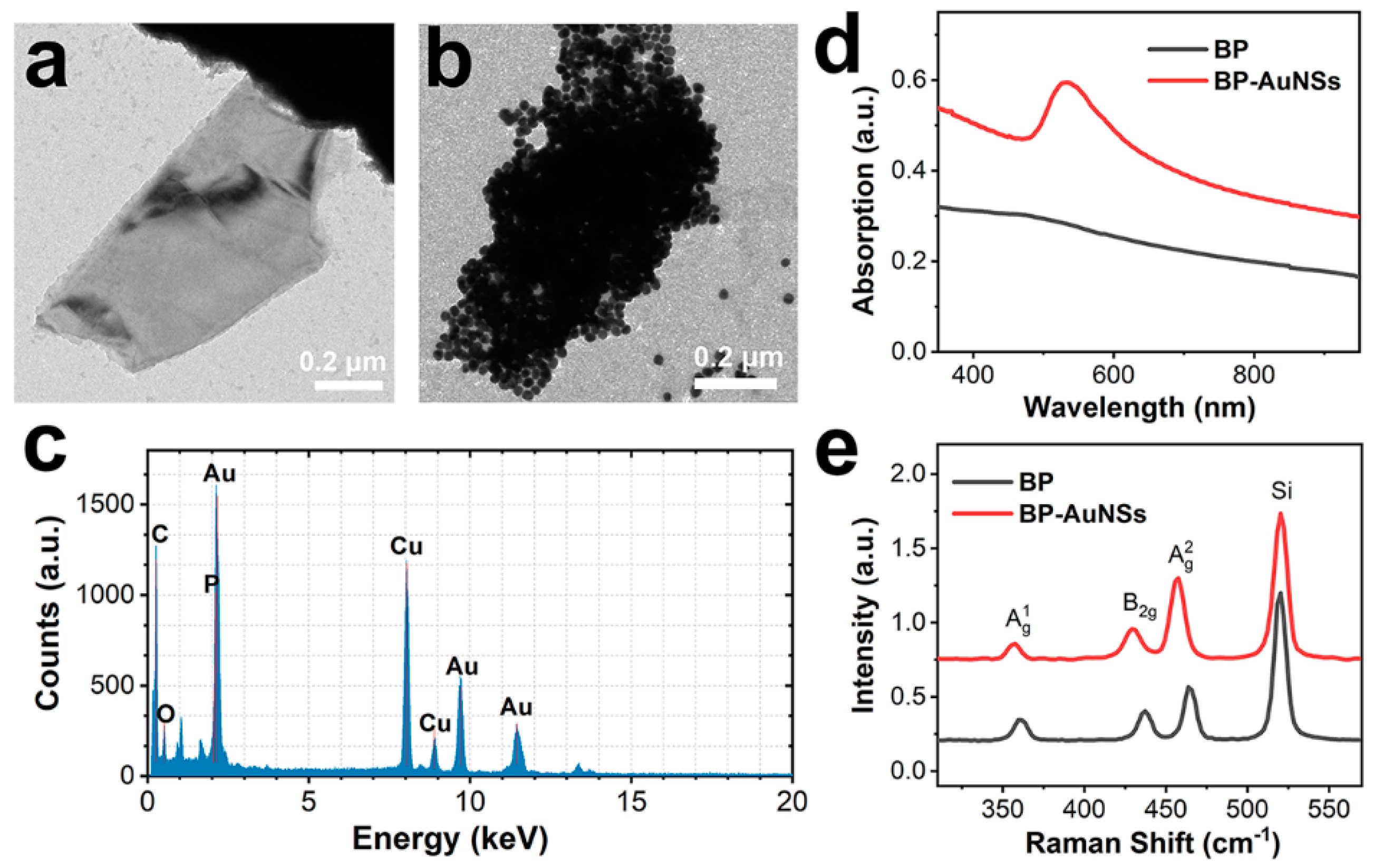
3.1. Characterization of 2D SERS Substrate
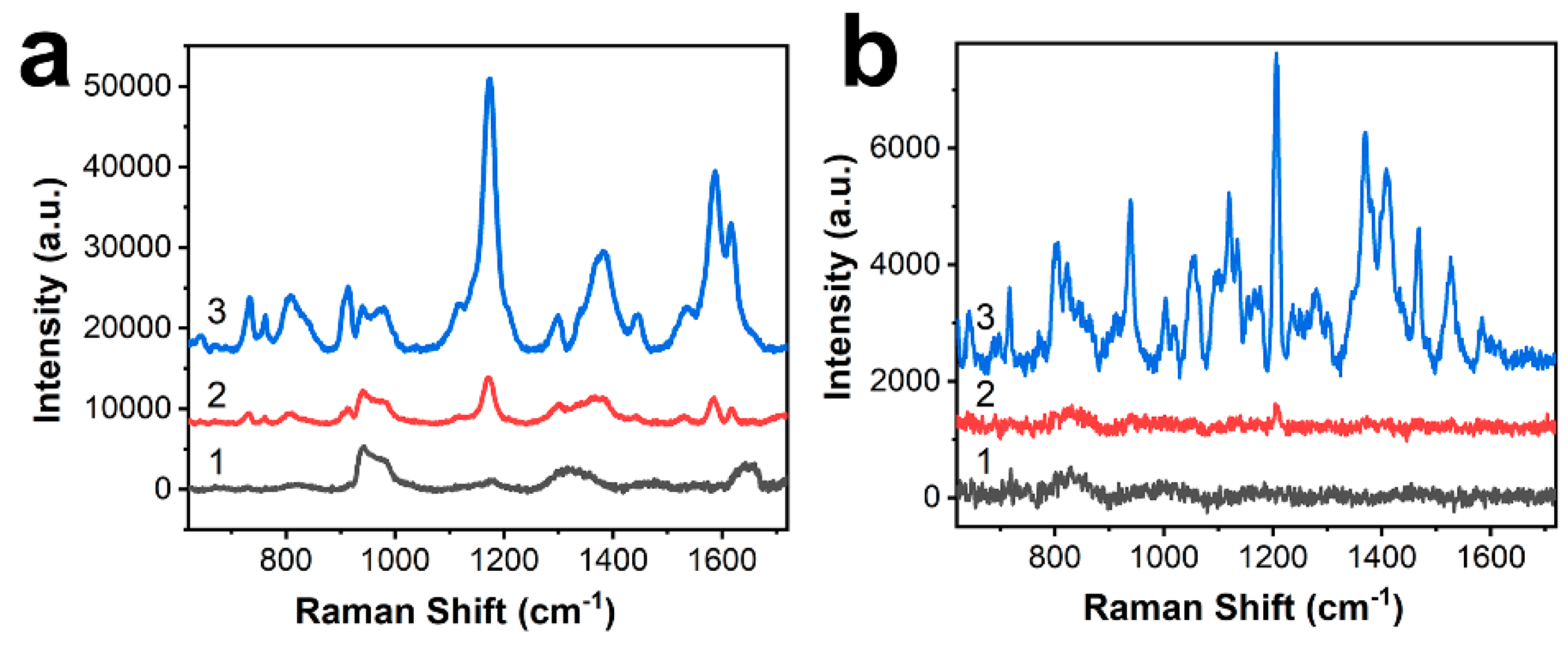
3.2. SERS Performance of BP-AuNSs
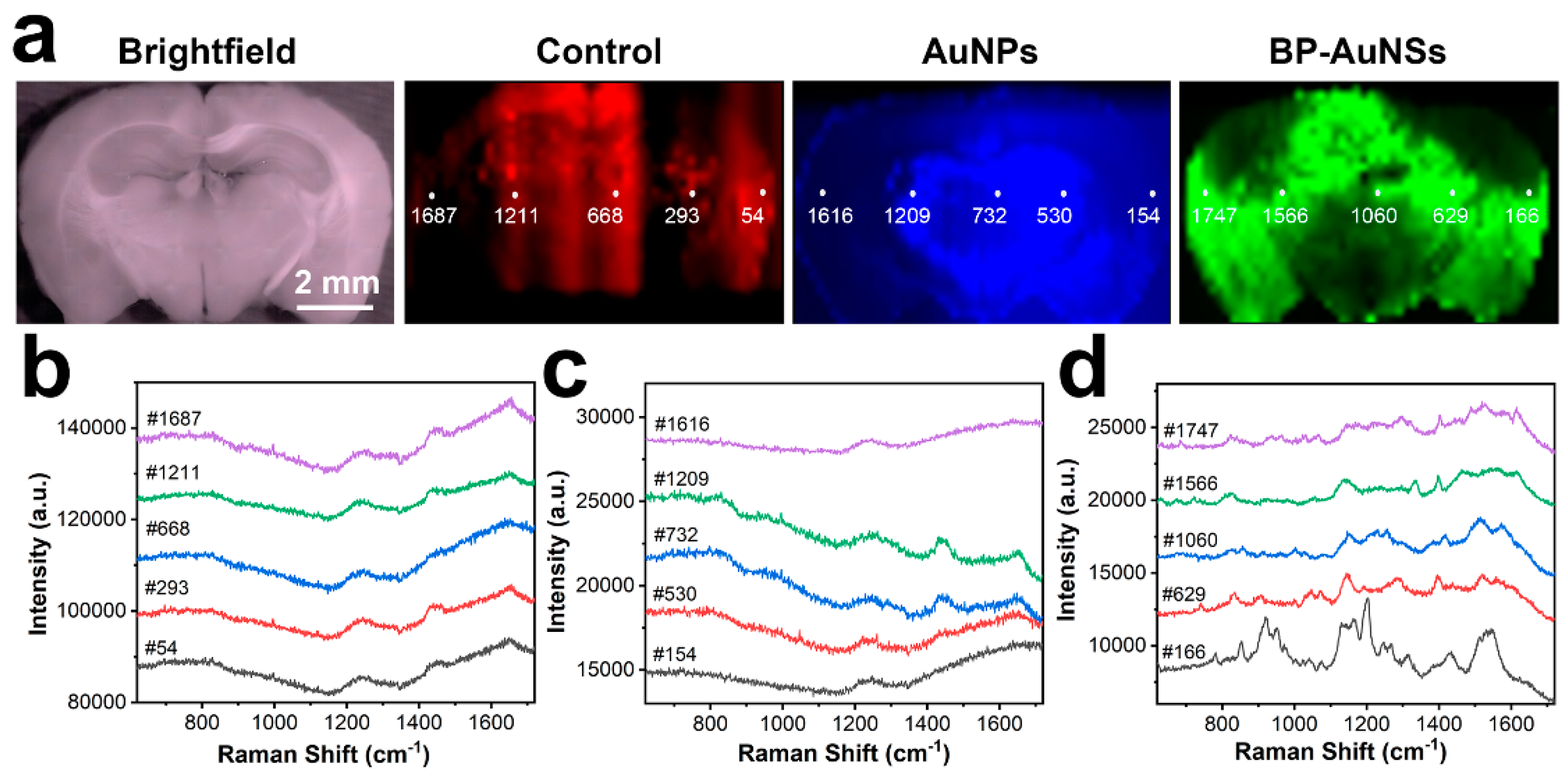
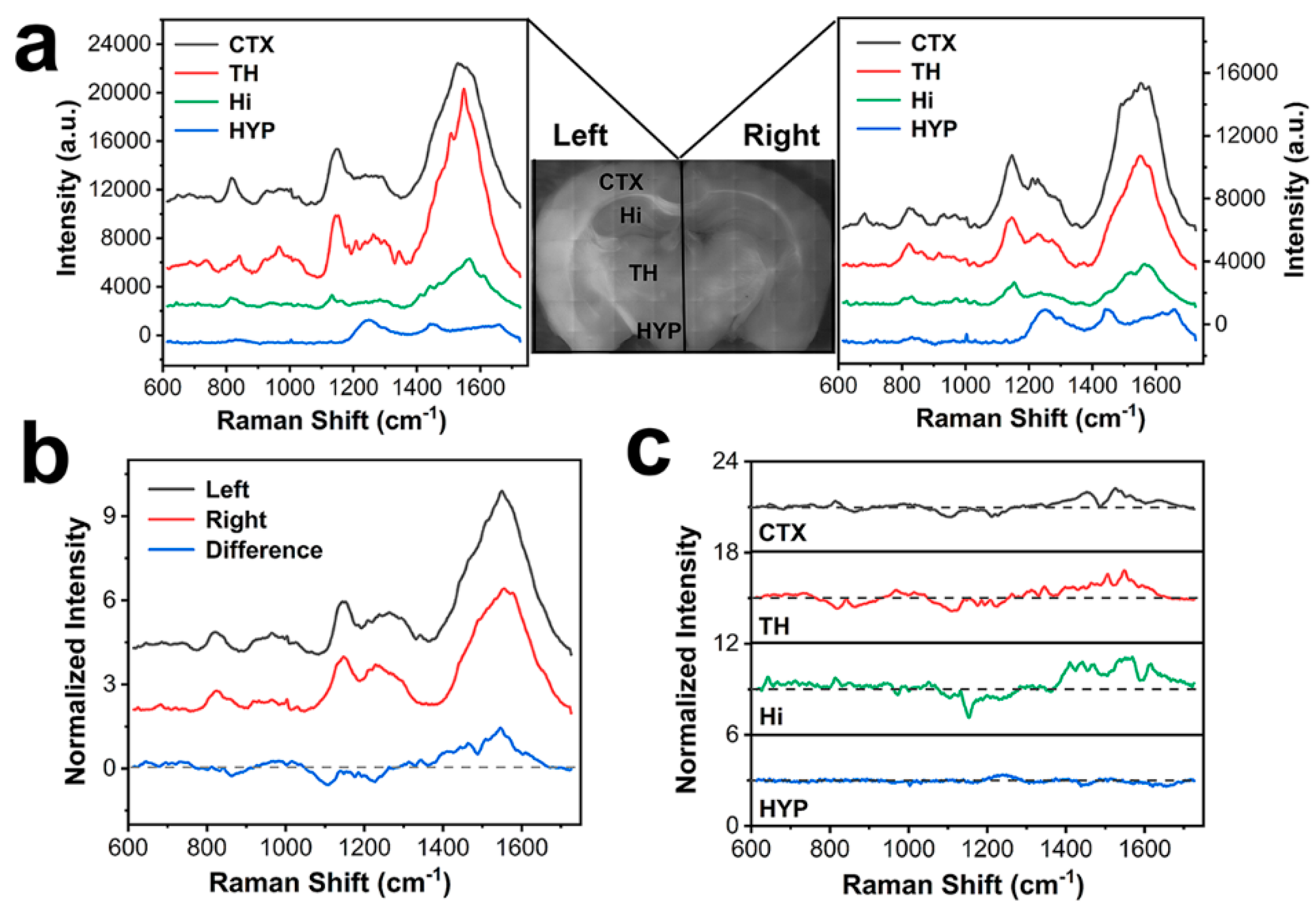
3.3. Full-scale SERS Mapping of Brain Tissues
3.4. SERS Texture Analysis of Brain Tissue
3.5. SERS Spectral Difference Among Various Encephalic Regions
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Risacher, S.L.; Anderson, W.H.; Charil, A.; Castelluccio, P.F.; Shcherbinin, S.; Saykin, A.J.; Schwarz, A.J. Alzheimer disease brain atrophy subtypes are associated with cognition and rate of decline. Neurology 2017, 89, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Fjalldal, S.; Follin, C.; Svard, D.; Rylander, L.; Gabery, S.; Petersen, A.; van Westen, D.; Sundgren, P.C.; Bjorkman-Burtscher, I.M.; Latt, J.; et al. Microstructural white matter alterations and hippocampal volumes are associated with cognitive deficits in craniopharyngioma. Eur. J. Endocrinol. 2018, 178, 577–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhou, L.; Cui, C.; Liu, Z.; Lu, J. Gray matter morphological anomalies in the cerebellar vermis in first-episode schizophrenia patients with cognitive deficits. BMC Psychiatry 2017, 17, 374. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef] [PubMed]
- Kalkanis, S.N.; Kast, R.E.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Nelson, K.M.; Raghunathan, A.; Poisson, L.M.; Auner, G.W. Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J. Neurooncol. 2014, 116, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Gautam, R.; Chatterjee, S.; Ariese, F.; Sikdar, S.K.; Umapathy, S. Ascorbate protects neurons against oxidative stress: A Raman microspectroscopic study. ACS Chem. Neurosci. 2015, 6, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Zhang, D.; Jiang, Y.; Wu, X.; Shih, P.Y.; Liao, C.S.; Bungart, B.; Xu, X.M.; Drenan, R.; Bartlett, E.; et al. Label-Free Vibrational Spectroscopic Imaging of Neuronal Membrane Potential. J. Phys. Chem. Lett. 2017, 8, 1932–1936. [Google Scholar] [CrossRef]
- Huang, R.; Harmsen, S.; Samii, J.M.; Karabeber, H.; Pitter, K.L.; Holland, E.C.; Kircher, M.F. High Precision Imaging of Microscopic Spread of Glioblastoma with a Targeted Ultrasensitive SERRS Molecular Imaging Probe. Theranostics 2016, 6, 1075–1084. [Google Scholar] [CrossRef] [Green Version]
- Yamazoe, S.; Naya, M.; Shiota, M.; Morikawa, T.; Kubo, A.; Tani, T.; Hishiki, T.; Horiuchi, T.; Suematsu, M.; Kajimura, M. Large-area surface-enhanced Raman spectroscopy imaging of brain ischemia by gold nanoparticles grown on random nanoarrays of transparent boehmite. ACS Nano 2014, 8, 5622–5632. [Google Scholar] [CrossRef]
- Amharref, N.; Beljebbar, A.; Dukic, S.; Venteo, L.; Schneider, L.; Pluot, M.; Manfait, M. Discriminating healthy from tumor and necrosis tissue in rat brain tissue samples by Raman spectral imaging. Biochim. Biophys. Acta 2007, 1768, 2605–2615. [Google Scholar] [CrossRef] [Green Version]
- Kast, R.; Auner, G.; Yurgelevic, S.; Broadbent, B.; Raghunathan, A.; Poisson, L.M.; Mikkelsen, T.; Rosenblum, M.L.; Kalkanis, S.N. Identification of regions of normal grey matter and white matter from pathologic glioblastoma and necrosis in frozen sections using Raman imaging. J. Neurooncol. 2015, 125, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Shen, A.; Zhao, W.; Baek, S.J.; Yuan, H.; Hu, J. Raman signature from brain hippocampus could aid Alzheimer’s disease diagnosis. Appl. Opt. 2009, 48, 4743–4748. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; McEwen, G.D.; Harihar, S.; Baker, S.M.; DeWald, D.B.; Zhou, A. BRMS1 expression alters the ultrastructural, biomechanical and biochemical properties of MDA-MB-435 human breast carcinoma cells: An AFM and Raman microspectroscopy study. Cancer Lett. 2010, 293, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Pascut, F.C.; Goh, H.T.; Welch, N.; Buttery, L.D.; Denning, C.; Notingher, I. Noninvasive detection and imaging of molecular markers in live cardiomyocytes derived from human embryonic stem cells. Biophys. J. 2011, 100, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Cialla-May, D.; Zheng, X.S.; Weber, K.; Popp, J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: From cells to clinics. Chem. Soc. Rev. 2017, 46, 3945–3961. [Google Scholar] [CrossRef] [PubMed]
- Aydin, O.; Altas, M.; Kahraman, M.; Bayrak, O.F.; Culha, M. Differentiation of healthy brain tissue and tumors using surface-enhanced Raman scattering. Appl. Spectrosc. 2009, 63, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hu, C.; Li, S.; Zhang, W.; Guo, Z. Rapid intracellular growth of gold nanostructures assisted by functionalized graphene oxide and its application for surface-enhanced Raman spectroscopy. Anal. Chem. 2012, 84, 10338–10344. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, Z.; Si, S.; Zhang, X.; Wu, W.; Deng, H.; Wang, F.; Xiao, X.; Jiang, C. Ultrasensitive SERS Substrate Integrated with Uniform Subnanometer Scale “Hot Spots” Created by a Graphene Spacer for the Detection of Mercury Ions. Small 2017, 13, 1603347. [Google Scholar] [CrossRef]
- Yin, P.T.; Shah, S.; Chhowalla, M.; Lee, K.-B. Design, Synthesis, and Characterization of Graphene-Nanoparticle Hybrid Materials for Bioapplications. Chem. Rev. 2015, 115, 2483–2531. [Google Scholar] [CrossRef]
- Ling, X.; Xie, L.M.; Fang, Y.; Xu, H.; Zhang, H.L.; Kong, J.; Dresselhaus, M.S.; Zhang, J.; Liu, Z.F. Can Graphene be used as a Substrate for Raman Enhancement? Nano Lett. 2010, 10, 553–561. [Google Scholar] [CrossRef]
- Qiu, M.; Ren, W.X.; Jeong, T.; Won, M.; Park, G.Y.; Sang, D.K.; Liu, L.-P.; Zhang, H.; Kim, J.S. Omnipotent phosphorene: A next-generation, two-dimensional nanoplatform for multidisciplinary biomedical applications. Chem. Soc. Rev. 2018, 47, 5588–5601. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Wang, D.; Liang, W.; Liu, L.; Zhang, Y.; Chen, X.; Sang, D.K.; Xing, C.; Li, Z.; Dong, B.; et al. Novel concept of the smart NIR-light-controlled drug release of black phosphorus nanostructure for cancer therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 501–506. [Google Scholar] [CrossRef]
- Choi, J.R.; Yong, K.W.; Choi, J.Y.; Nilghaz, A.; Lin, Y.; Xu, J.; Lu, X. Black Phosphorus and its Biomedical Applications. Theranostics 2018, 8, 1005–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Yin, J.; Chen, Y.; Pan, S.; Yao, H.; Gao, Y.; Shi, J. 2D-Black-Phosphorus-Reinforced 3D-Printed Scaffolds:A Stepwise Countermeasure for Osteosarcoma. Adv. Mater. 2018, 30, 1705611. [Google Scholar] [CrossRef]
- Zhou, W.; Cui, H.; Ying, L.; Yu, X.F. Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing. Angew. Chem. Int. Ed. 2018, 57, 10268–10272. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhao, Y.; Li, Z.; Cui, H.; Zhou, Y.; Li, W.; Tao, W.; Zhang, H.; Wang, H.; Chu, P.K.; et al. TiL4-Coordinated Black Phosphorus Quantum Dots as an Efficient Contrast Agent for In Vivo Photoacoustic Imaging of Cancer. Small 2017, 13, 1602896. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ouyang, J.; Yi, X.; Xu, Y.; Niu, C.; Zhang, W.; Wang, L.; Sheng, J.; Deng, L.; Liu, Y.N.; et al. Black Phosphorus Nanosheets as a Neuroprotective Nanomedicine for Neurodegenerative Disorder Therapy. Adv. Mater. 2018, 30, 1703458. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Liu, Z.; Li, Y.; Hou, Y.; Fei, X.; Su, C.; Wang, S.; Zhuang, Z.; Guo, Z. Facile synthesis of black phosphorus-Au nanocomposites for enhanced photothermal cancer therapy and surface-enhanced Raman scattering analysis. Biomater. Sci. 2017, 5, 2048–2055. [Google Scholar] [CrossRef]
- Mo, J.; Xie, Q.; Wei, W.; Zhao, J. Revealing the immune perturbation of black phosphorus nanomaterials to macrophages by understanding the protein corona. Nat. Commun. 2018, 9, 2480. [Google Scholar] [CrossRef]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Xie, H.; Shao, J.; Ma, Y.; Wang, J.; Huang, H.; Yang, N.; Wang, H.; Ruan, C.; Luo, Y.; Wang, Q.Q.; et al. Biodegradable near-infrared-photoresponsive shape memory implants based on black phosphorus nanofillers. Biomaterials 2018, 164, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liang, L.; Ling, X.; Zhang, S.; Mao, N.; Zhang, N.; Sumpter, B.G.; Meunier, V.; Tong, L.; Zhang, J. Enhanced Raman Scattering on In-Plane Anisotropic Layered Materials. J. Am. Chem. Soc. 2015, 137, 15511–15517. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Ohtsuka, A.; Matsuoka, H.; Taguchi, T.; Murakami, T.; Abe, K.; Ninomiya, Y. Intensely positively charged perineuronal nets in the adult rat brain as detected by staining with anionic iron colloid. Arch. Histol. Cytol. 2001, 64, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.J., Jr.; Prescott, B.; Olins, D.E. Secondary structure of histones and DNA in chromatin. Science 1977, 197, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Kneipp, J.; Kneipp, H.; McLaughlin, M.; Brown, D.; Kneipp, K. In vivo molecular probing of cellular compartments with gold nanoparticles and nanoaggregates. Nano Lett. 2006, 6, 2225–2231. [Google Scholar] [CrossRef] [PubMed]
- Willets, K.A. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 2009, 394, 85–94. [Google Scholar] [CrossRef] [PubMed]

| Raman Shift (cm−1) | Tentative Assignment |
|---|---|
| 645 L | Tyr, COO- bend or C-S str (p) |
| 730 L | A (d) |
| 747 L | T (d); Trp ring breath (p) |
| 860 R | Ribose: C-C str, ring breath, C-O-C str (c) |
| 964 L | C-C bk str (p) |
| 1025 L | C-H in-plane Phe (p); glycogen |
| 1109 R | C-N str (p) |
| 1176 R | T, G, C-N str (d) |
| 1230 R | Amide III (p) |
| 1343 L | CH2 def (p); A,G (d) |
| 1410 L | COO- sym str (p) |
| 1445–1470 L | CH2/CH3 def (p, l) |
| 1508–1530 L | A, C, G (d) |
| 1546 L | Amide II, Trp (p) |
| 1590 L | Amide II, Phe, Tyr (p); G, A (d) |
| 1611 L | Tyr, Phe C=C bend (p) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Ding, F.; Li, D.; Zhang, W.; Cao, L.; Liu, Z. Full-Scale Label-Free Surface-Enhanced Raman Scattering Analysis of Mouse Brain Using a Black Phosphorus-Based Two-Dimensional Nanoprobe. Appl. Sci. 2019, 9, 398. https://doi.org/10.3390/app9030398
Guo T, Ding F, Li D, Zhang W, Cao L, Liu Z. Full-Scale Label-Free Surface-Enhanced Raman Scattering Analysis of Mouse Brain Using a Black Phosphorus-Based Two-Dimensional Nanoprobe. Applied Sciences. 2019; 9(3):398. https://doi.org/10.3390/app9030398
Chicago/Turabian StyleGuo, Tiejun, Fangsheng Ding, Dongling Li, Wen Zhang, Liren Cao, and Zhiming Liu. 2019. "Full-Scale Label-Free Surface-Enhanced Raman Scattering Analysis of Mouse Brain Using a Black Phosphorus-Based Two-Dimensional Nanoprobe" Applied Sciences 9, no. 3: 398. https://doi.org/10.3390/app9030398