The Transcriptomics to Proteomics of Hair Cell Regeneration: Looking for a Hair Cell in a Haystack
Abstract
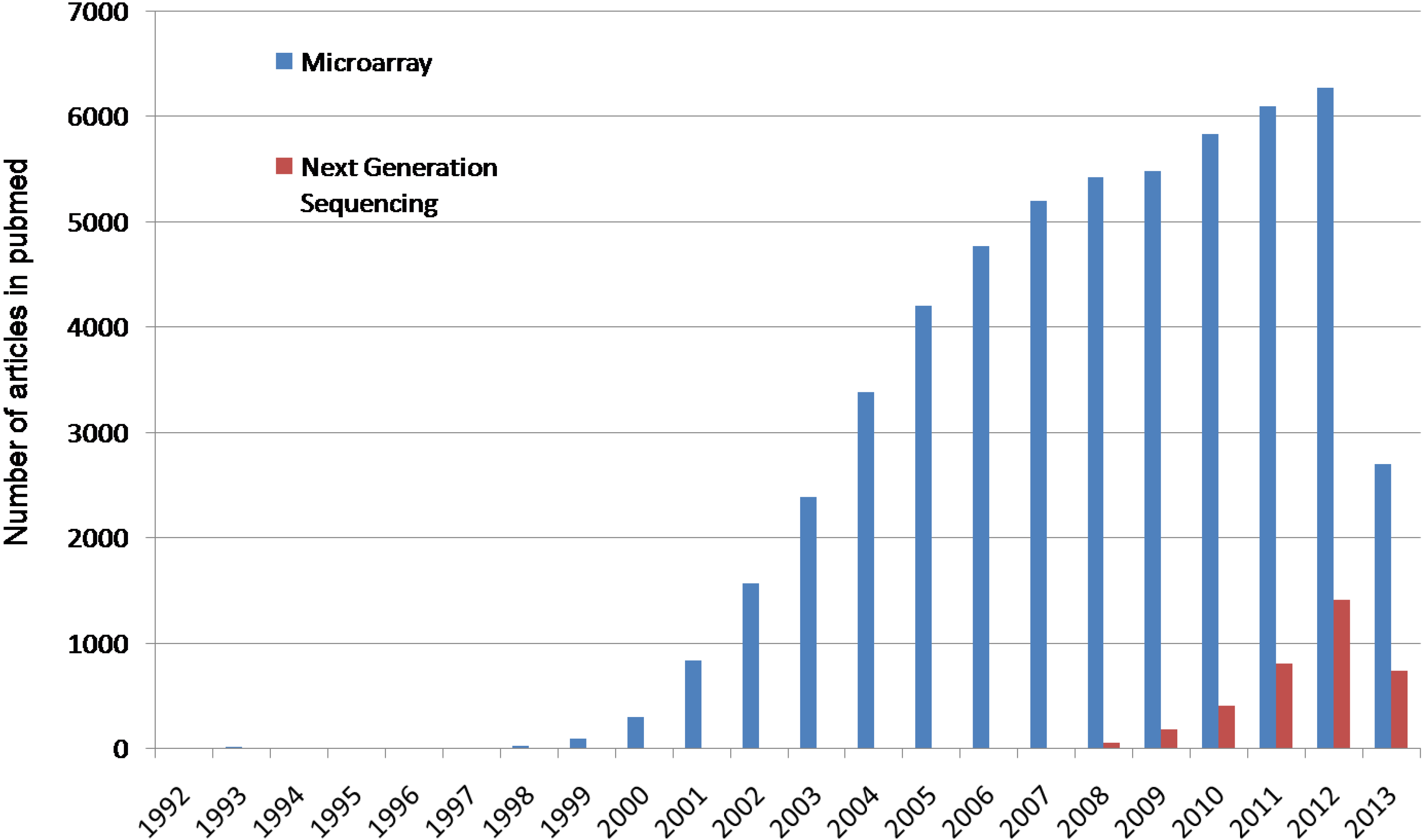
:1. Introduction
2. Experimental Methodologies
| Reference | Organism and organ | Methodology | Developmental stage or treatment |
|---|---|---|---|
| Developmental differences in gene expression | |||
| [38] | Mouse cochlea | Affymetrix GeneChip oligonucleotide array | P2 and P32 |
| [39] | Mouse cochlea | Affymetrix GeneChip oligonucleotide array | P2 and P32 |
| [40] | Mouse cochlea-conditionally immortal cell line | Affymetrix GeneChip oligonucleotide array | 14 days following differentiation |
| [41] | Mouse inner ear | Affymetrix GeneChip oligonucleotide array | E9-E15 |
| [42] | Mouse cochlea | Affymetrix GeneChip oligonucleotide array | P3 and adult |
| Cell/Tissue differences in gene expression | |||
| [27] | Chick cochlea and utricles | Custom built cDNA and oligonucleotide arrays | cochlea versus utricles |
| [28] | Rat cristae ampullaris | Agilent RNA6000 Nano Lab Chip | hair cells versus supporting cells |
| [41] | Mouse inner ear | Affymetrix GeneChip oligonucleotide array | cochlea, utricle, or saccule |
| [24] | Chick inner ear | Custom built TF oligonucleotide array | 30 min and 1, 2, 3 h post-ototoxic or laser trauma |
| [43] | Zebrafish lagena | Affymetrix GeneChip oligonucleotide array | hair cells versus liver hepatocytes |
| Gene expression during regeneration following trauma | |||
| [24] | Chick inner ear | Custom built TF oligonucleotide array | 30 min and 1, 2, 3 h post-ototoxic or laser trauma |
| [25] | Zebrafish inner ear | Agilent Zebrafish oligonucleotide arrays | 2 and 4 days post-acoustic trauma |
| [44] | Zebrafish inner ear | Illumina tag profiling (SAGE) | 0, 1, 2, 4 days post-acoustic trauma |
3. Gene Profiling in the Vertebrate Inner Ear
3.1. Developmental Shifts in Gene Expression
3.2. Cell- and Tissue-Specific Transcript Profiling
3.3. Gene Expression Following Inner Ear Trauma
4. Role of MicroRNAs in the Development, Maturation and Functioning of Hair Cells
4.1. Introduction to the Role of MicroRNAs in the Inner Ear
| Micro RNA | Organism | Reference | Organ of expression |
|---|---|---|---|
| miR182, miR183, miR96 | Zebrafish | [74] | Hair cells of neuromasts and inner ear, nose, cranial ganglia, eye, epiphysis |
| miR183, miR182, miR96 | Mouse | [78] | Inner ear |
| miR141, miR200a, miR200b, miR139 | Zebrafish | [74] | Neuromast, nose, epidermis, taste buds, proctodeum |
| let7g, miR15a, miR17-5p, miR18, miR19a, miR19b, miR20, miR210, miR25, miR26a, miR26b, miR92, miR93 | Zebrafish | [74] | Neuromasts, head, spinal cord, gut, somites |
| miR15a1, miR18a | Zebrafish | [73] | Neuromasts, hair cells, otocyst and head |
| miR199a | Mouse | [73] | Cochlea |
| miR99a, miR15a, miR30b | Mouse | [73] | Cochlear hair and supporting cells, vestibular hair cells, spiral ganglion neurons and basilar membrane |
| miR18a | Mouse | [73] | Cochlea, Spiral ganglion neurons, vestibular hair and supporting cells |
| Let7a, let7b, let7c, let7d, let7e, let7f, let7g | Mouse | [73] | Cochlea and vestibule |
| Let7i | Mouse | [73] | Vestibule |
| Let7a, let7b, let7c, let7d, let7e, let7f, let7g, let7i | Newt | [86] | Inner ear |
| miR9, miR124a | Mouse | [78] | Inner ear, brain |
| miR10a, miR107, miR124, miR130b, miR146b, miR183, miR190b, miR200c, miR30d, miR30e, miR325, miR333, miR339-3p, miR381, miR429, miR532-3p, miR674, miR99b, miR194, miR186 and miR331-5p | Rat | [83] | Cochlear epithelia |
| miR182, miR140 | Mouse | [79] | Otocyst, spiral ganglion, inner and outer hair cells, utricle, saccule, crista |
| miR194 | Mouse | [79] | Hair cells of cochlea and vestibule, spiral ganglia |
| miR376a, miR376b | Mouse | [80] | Otic placode, organ of Corti, spiral ganglia, stria vascularis, ampulla of vestibular organs |
| miR135 | Mouse | [87] | Hair cells in vestibule, vestibular neurons and spiral ganglia |
| miR205 | Mouse | [87] | Cochlea, spiral ligament, basilar membrane, apical surface of the spiral limbus |
4.2. Important miRNA Clusters in the Inner Ear
5. Proteomic Analysis of the Inner Ear
5.1. Antibody Microarrays
5.2. Liquid Chromatography Coupled with Mass Spectrometry
6. Conclusions and Perspectives
Acknowledgments
Conflict of Interest
References
- Lim, D.J. Ultrastructural cochlear changes following acoustic hyperstimulation and ototoxicity. Ann. Otol. Rhinol. Laryngol. 1976, 85, 740–751. [Google Scholar]
- Lindeman, H.H.; Bredberg, G. Scanning electron microscopy of the organ of Corti after intense auditory stimulation: Effects on stereocilia and cuticular surface of hair cells. Arch. Ohren Nasen Kehlkopfheilkd. 1972, 203, 1–15. [Google Scholar]
- Stockwell, C.W.; Ades, H.W.; Engström, H. Patterns of hair cell damage after intense auditory stimulation. Ann. Otol. Rhinol. Laryngol. 1969, 78, 1144–1168. [Google Scholar]
- Theopold, H.M. Comparative surface studies of ototoxic effects of various aminoglycoside antibiotics on the organ of Corti in the guinea pig. A scanning electron microscopic study. Acta Otolaryngol. 1977, 84, 57–64. [Google Scholar] [CrossRef]
- Keithley, E.M.; Feldman, M.L. Hair cell counts in an age-graded series of rat cochleas. Hear. Res. 1982, 8, 249–262. [Google Scholar] [CrossRef]
- Lombarte, A.; Yan, H.Y.; Popper, A.N.; Chang, J.S.; Platt, C. Damage and regeneration of the hair cell ciliary bundles in a fish ear following treatment with gentamicin. Hear. Res. 1993, 64, 166–174. [Google Scholar] [CrossRef]
- Harris, J.A.; Cheng, A.G.; Cunningham, L.L.; MacDonald, G.; Raible, D.W.; Rubel, E.W. Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio). J. Assoc. Res. Otolaryngol. 2003, 4, 219–234. [Google Scholar] [CrossRef]
- Smith, M.E.; Coffin, A.B.; Miller, D.L.; Popper, A.N. Anatomical and functional recovery of the goldfish (Carassius auratus) ear following noise exposure. J. Exp. Biol. 2006, 209, 4193–4202. [Google Scholar]
- Schuck, J.B.; Smith, M.E. Cell proliferation follows acoustically-induced hair cell bundle loss in the zebrafish saccule. Hear. Res. 2009, 253, 67–76. [Google Scholar]
- Jones, J.E.; Corwin, J.T. Regeneration of sensory cells after laser ablation in the lateral line system: Hair cell lineage and macrophage behavior revealed by time-lapse video microscopy. J. Neurosci. 1996, 16, 649–662. [Google Scholar]
- Taylor, R.R.; Forge, A. Hair cell regeneration in sensory epithelia from the inner ear of a urodele amphibian. J. Comp. Neurol. 2005, 484, 105–120. [Google Scholar] [CrossRef]
- Avallone, B.; Porritiello, M.; Esposito, D.; Mutone, R.; Balsamo, G.; Marmo, F. Evidence of hair cell regeneration in the crista ampullaris of the lizard Podarcis sicula. Hear. Res. 2003, 178, 79–88. [Google Scholar]
- Cotanche, D.A. Regeneration of hair cell stereociliary bundles in the chick cochlea following severe acoustic trauma. Hear. Res. 1987, 30, 181–195. [Google Scholar] [CrossRef]
- Corwin, J.T.; Cotanche, D.A. Regeneration of sensory hair cells after acoustic trauma. Science 1988, 240, 1772–1774. [Google Scholar]
- Ryals, B.M.; Rubel, E.W. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 1988, 240, 1774–1776. [Google Scholar]
- Weisleder, M.E.; Rubel, E.W. Hair cell regeneration after streptomycin toxicity in the avian vestibular epithelium. J. Comp. Neurol. 1993, 331, 97–110. [Google Scholar] [CrossRef]
- Oesterle, E.C.; Stone, J.S. Hair Cell Regeneration: Mechanisms Guiding Cellular Proliferation and Differention. In Hair Cell Regeneration, Repair, and Protection; Salvi, R.J., Popper, A.N., Fay, R.R., Eds.; Springer: New York, NY, USA, 2008; pp. 141–197. [Google Scholar]
- Stone, L. The development of the lateral line sense organs in the amphibian observed in living and vital-stained preparations. J. Comp. Neurol. 1933, 57, 507–540. [Google Scholar]
- Stone, L. Further experimental studies of the development of lateral-line sense organs in the amphibians observed in living preparations. J. Comp. Neurol. 1937, 68, 83–115. [Google Scholar] [CrossRef]
- Corwin, J.T. Postembryonic production and aging in inner ear hair cells in sharks. J. Comp. Neurol. 1981, 201, 541–553. [Google Scholar]
- Corwin, J.T. Postembryonic growth of the macular neglecta auditory detector in the ray, Raja clavata; continued increases in hair cell number, neural convergence, and physiological sensitivity. J. Comp. Neurol. 1983, 217, 345–356. [Google Scholar] [CrossRef]
- Popper, A.N.; Hoxter, B. Growth of a fish ear: Quantitative analysis of hair cell and ganglion cell proliferation. Hear. Res. 1984, 15, 133–142. [Google Scholar]
- Cruz, R.M.; Lambert, P.R.; Rubel, E.W. Light microscopic evidence of hair cell regeneration after gentamicin toxicity in chick cochlea. Arch. Otolaryngol. Head Neck Surg. 1987, 113, 1058–1062. [Google Scholar] [CrossRef]
- Hawkins, R.D.; Bashiardes, S.; Powder, K.E.; Sajan, S.A.; Bhonagiri, V.; Speck, J.; Warchol, M.E.; Lovett, M. Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS One 2007, 2, e525. [Google Scholar] [CrossRef]
- Schuck, J.B.; Sun, H.; Penberthy, W.T.; Cooper, N.G.F.; Li, X.; Smith, M.E. Transcriptomic analysis of the zebrafish inner ear points to growth hormone mediated regeneration following acoustic trauma. BMC Neurosci. 2011, 12, 88. [Google Scholar] [CrossRef]
- Alvarado, D.M.; Hawkins, D.R.; Bashiardes, S.; Veile, R.A.; Ku, Y-C.K.; Powder, K.E.; Spriggs, M.K.; Speck, J.D.; Warchol, M.E.; Lovett, M. An RNA interference-based screen of transcription factor genes identifies pathways necessary for sensory regeneration in the avian inner ear. J. Neurosci. 2011, 31, 4535–4543. [Google Scholar]
- Hawkins, R.D.; Bashiardes, S.; Helms, C.A.; Hu, L.; Saccone, N.L.; Warchol, M.E.; Lovett, M. Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: Implications for human hearing and balance disorders. Hum. Mol. Genet. 2003, 12, 1261–1272. [Google Scholar]
- Cristobal, R.; Wackym, P.A.; Cioffi, J.A.; Erbe, C.B.; Roche, J.P.; Popper, P. Assessment of differential gene expression in vestibular epithelia cell types using microarray analysis. Mol. Brain Res. 2005, 133, 19–36. [Google Scholar] [CrossRef]
- Hertzano, R.; Elkon, R. High throughput gene expression analysis of the inner ear. Hear. Res. 2012, 288, 77–88. [Google Scholar] [CrossRef]
- Thalmann, I. Inner ear proteomics: A fad or hear to stay. Brain Res. 2006, 1091, 103–112. [Google Scholar]
- Zhou, J.; Thompson, D.K. Microarray technology and applications in environmental microbiology. Adv. Agron. 2004, 82, 183–270. [Google Scholar]
- Rivolta, M.N.; Holley, M.C. Gene Arrays, Cell Lines, Stem Cells, and Sensory Regeneration in Mammalian Ears. In Hair Cell Regeneration, Repair, and Protection; Salvi, R.J., Popper, A.N., Fay, R.R., Eds.; Springer: New York, NY, USA, 2008; Volume 33, pp. 257–307. [Google Scholar]
- Goulter, A.B.; Harmer, D.W.; Clark, K.L. Evaluation of low density array technology for quantitative parallel measurement of multiple genes in human tissue. BMC Genomics 2006, 7, 1–10. [Google Scholar] [CrossRef]
- Horan, M.P. Application of serial analysis of gene expression to the study of human genetic disease. Hum. Genet. 2009, 126, 605–614. [Google Scholar]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 2012. [Google Scholar] [CrossRef]
- Friedman, L.M.; Avraham, K.B. MicroRNAs and epigenetic regulation in the mammalian ear: Implications for deafness. Mamm. Genome 2009, 20, 581–603. [Google Scholar]
- Chen, Z.-Y.; Corey, D.P. An inner ear gene expression database. JARO 2001, 3, 140–148. [Google Scholar] [CrossRef]
- Chen, Z-Y.; Corey, D.P. Understanding inner ear development with gene expression profiling. J. Neurobiol. 2002, 53, 276–285. [Google Scholar] [CrossRef]
- Rivolta, M.N.; Halsall, A.; Johnson, C.M.; Tones, M.A.; Holley, M.C. Transcript profiling of functionally related groups of genes during conditional differentiation of a mammalian cochlear cell line. Genome Res. 2002, 12, 1091–1099. [Google Scholar] [CrossRef]
- Sajan, S.A.; Warchol, M.E.; Lovett, M. Towards a systems biology of mouse inner ear organogenesis: Gene expression pathways, patterns and network analysis. Genetics 2007, 177, 631–653. [Google Scholar] [CrossRef]
- Smeti, I.; Assou, S.; Savary, E.; Masmoudi, S.; Zine, A. Transcriptomic analysis of the developing and adult cochlear sensory epithelia. PLoS One 2012, 7, e42987. [Google Scholar] [CrossRef] [Green Version]
- McDermott, B.M.; Baucom, J.M.; Hudspeth, A.J. Analysis and functional evaluation of the hair cell transcriptome. Proc. Natl. Acad. Sci. USA 2007, 104, 11820–11825. [Google Scholar] [CrossRef]
- Liang, J.; Wang, D.; Renaud, G.; Wolfsberg, T.G.; Wilson, A.F.; Burgess, S.M. The stat3/socs3a pathway: Regulator of hair cell regeneration. J. Neurosci. 2012, 32, 10662–10673. [Google Scholar]
- Roberson, D.W.; Rubel, E.W. Cell division in the gerbil cochlea after acoustic trauma. Am. J. Otol. 1994, 15, 28–34. [Google Scholar]
- Sobkowicz, H.M.; August, B.W.; Slapnick, S.M. Cellular interactions as a response to injury in the organ of Corti in culture. Int. J. Dev. Neurosci. 1997, 15, 463–485. [Google Scholar] [CrossRef]
- Warchol, M.E.; Corwin, J.T. Supporting cells in avian vestibular organs proliferate in serum-free culture. Hear. Res. 1993, 71, 28–36. [Google Scholar] [CrossRef]
- Forge, A.; Li, L.; Corwin, J.T.; Nevill, G. Ultrastructural evidence for hair cell regeneration in the mammalian inner ear. Science 1993, 259, 1616–1619. [Google Scholar]
- Forge, A.; Nevill, G. Hair cell recovery in the vestibular sensory epithelia of mature guinea pigs. J. Comp. Neurol. 1998, 397, 69–88. [Google Scholar] [CrossRef]
- Zheng, Q.; Rozanas, C.; Thalmann, I.; Chance, M.; Alagramam, K. Inner ear proteomics of mouse models for deafness, a discovery strategy. Brain Res. 2006, 1091, 113–121. [Google Scholar]
- Glokler, J.; Angenendt, P. Protein and antibody microarray technology. J. Chromatogr. B 2003, 797, 229–240. [Google Scholar] [CrossRef]
- Kusnezow, W.; Hoheisel, J. Antibody microarrays: Promises and problems. Biotechniques 2002, 33, S14–S23. [Google Scholar]
- Jamesdaniel, S.; Ding, D.; Kermany, M.; Davidson, B.; Knight, P., III; Salvi, R.; Coling, D.E. Proteomic analysis of the balance between survival and cell death responses in cisplatin-mediated ototoxicity. J. Proteome Res. 2008, 7, 3516–3524. [Google Scholar]
- Jamesdaniel, S.; Hu, B.; Kermany, M.; Jiang, H.; Ding, D.; Coling, D.; Salvi, R. Noise induced changes in the expression of p38/MAPK signaling proteins in the sensory epithelium of the inner ear. J. Proteomics 2011, 75, 410–424. [Google Scholar] [CrossRef]
- Peng, H.; Liu, M.; Pecka, J.; Beisel, K.; Ding, S.-J. Proteomic analysis of the organ of Corti using nanoscale liquid chromatography coupled with tandem mass spectrometry. Int. J. Mol. Sci. 2012, 13, 8171–8188. [Google Scholar] [CrossRef]
- Shin, J.-B.; Streijger, F.; Beynon, A.; Peters, T.; Gadzala, L.; McMillen, D.; Bystrom, C.; van der Zee, C.E.E.M.; Wallimann, T.; Gillespie, P.G. Hair bundles are specialized for ATP delivery via creatine kinase. Neuron 2007, 53, 371–386. [Google Scholar]
- Spinelli, K.; Klimek, J.; Wilmarth, P.; Shin, J.-B.; Choi, D.; David, L.; Gillespie, P.G. Distinct energy metabolism of auditory and vestibular sensory epithelia revealed by quantitative mass spectrometry using MS2 intensity. Proc. Natl. Acad. Sci. USA 2012, 109, E268–E277. [Google Scholar]
- Cho, Y.; Gong, T.-W.L.; Stöver, T.; Lomax, M.I.; Altschuler, R.A. Gene expression profiles of the rat cochlea, cochlear nucleus, and inferior colliculus. JARO 2001, 3, 54–67. [Google Scholar] [CrossRef]
- Kil, J.; Warchol, M.E.; Corwin, J.T. Cell death, cell proliferation, and estimates of hair cell life spans in the vestibular organs of chicks. Hear. Res. 1997, 114, 117–126. [Google Scholar] [CrossRef]
- Stone, J.S.; Oesterle, E.C.; Rubel, E.W. Recent insights into regeneration of auditory and vestibular hair cells. Curr. Opin. Neurol. 1998, 11, 17–24. [Google Scholar] [CrossRef]
- Goodyear, R.J.; Gates, R.; Lukashkin, A.N.; Richardson, G.P. Hair cell numbers continue to increase in utricular macula of the early posthatch chick. J. Neurocytol. 1999, 28, 851–861. [Google Scholar] [CrossRef]
- Wilkins, H.R.; Presson, J.C.; Popper, A.N. Proliferation of vertebrate inner ear supporting cells. J. Neurobiol. 1999, 39, 527–535. [Google Scholar] [CrossRef]
- Oesterle, E.C.; Rubel, E.W. Postnatal production of supporting cells in the chick cochlea. Hear. Res. 1993, 66, 213–224. [Google Scholar] [CrossRef]
- Lawoko-Kerali, G.; Rivolta, M.N.; Holley, M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J. Comp. Neurol. 2002, 442, 378–391. [Google Scholar] [CrossRef]
- Sun, H.; Lin, C.-H.; Smith, M.E. Growth hormone promotes hair cell regeneration in the zebrafish (Danio rerio) inner ear following acoustic trauma. PLoS One 2011, 6, e28372. [Google Scholar] [CrossRef]
- Camarero, G.; Avendano, C.; Fernandez-Moreno, C.; Villar, A.; Contreras, J.; de Pablo, F.; Pichel, J.G.; Varela-Nieto, I. Delayed inner ear maturation and neuronal loss in postnatal IGF-1 deficient mice. J. Neurosci. 2001, 21, 7630–7641. [Google Scholar]
- Reznik, S.I.; Jaramillo, A.; Zhang, L.; Patterson, G.A.; Cooper, J.D.; Monhanakumar, T. Anti-HLA antibody binding to HAL class I molecules induces proliferation of airway epithelial cells: A potential mechanism for brochiolitis obliterans syndrome. J. Thorac. Cariovasc. Surg. 2000, 119, 39–45. [Google Scholar] [CrossRef]
- Herrington, J.; Carter-Su, C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol. Metabol. 2001, 12, 252–257. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lewis, M.A.; Quint, E.; Glazier, A.M.; Fuchs, H.; de Angelis, M.H.; Langford, C.; van Dongen, S.; Abreu-Goodger, C.; Piipari, M.; Redshaw, N.; et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009, 41, 614–618. [Google Scholar] [CrossRef]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Friedman, L.M.; Dror, A.A.; Mor, E.; Tenne, T.; Toren, G.; Satoh, T.; Biesemeier, D.J.; Shomron, N.; Fekete, D.M.; Hornstein, E.; Avraham, K.B. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl. Acad. Sci. USA 2009, 106, 7915–7920. [Google Scholar] [CrossRef]
- Weinholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H.A. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Weinstein, E.G.; Abdelhakim, A.; Yekta, S.; Rhoades, M.W.; Burge, C.B.; Bartel, D.P. The microRNAs of Caenorhabditis elegans. Gene Dev. 2003, 17, 991–1008. [Google Scholar] [CrossRef]
- Li, H.; Kloosterman, W.; Fekete, D.M. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J. Neurosci. 2010, 30, 3254–3263. [Google Scholar] [CrossRef]
- Weston, M.D.; Pierce, M.L.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111, 95–104. [Google Scholar]
- Wang, X.-R.; Zhang, X.-M.; Zhen, J.; Zhang, P.-X.; Xu, G.; Jiang, H. MicroRNA expression in the embryonic mouse inner ear. Neuroreport 2010, 21, 611–617. [Google Scholar] [CrossRef]
- Yan, D.; Xing, Y.; Ouyang, X.; Zhu, J.; Chen, Z.-Y.; Lang, H.; Liu, X.Z. Analysis of miR-376 RNA cluster members in the mouse inner ear. Int. J. Exp. Pathol. 2012, 93, 450–457. [Google Scholar]
- Soukup, G.A.; Fritzsch, B.; Pierce, M.L.; Weston, M.D.; Jahan, I.; McManus, M.T.; Harfe, B.D. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev. Biol. 2009, 328, 328–341. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H.; McGee, J.; Walsh, E.; Soukup, G.; He, D. Identifying microRNAs involved in degeneration of the organ of Corti during age-related hearing loss. PLoS One 2013, 8, e62786. [Google Scholar] [CrossRef]
- Patel, M.; Cai, Q.; Ding, D.; Salvi, R.; Hu, Z.; Hu, B.H. The miR-183/Taok1 target pair is implicated in cochlear responses to acoustic trauma. PLoS One 2013, 8, e58471. [Google Scholar] [CrossRef]
- Frucht, C.; Uduman, M.; Duke, J.; Kleinstein, S.; Santos-Sacchi, J.; Navaratnam, D. Gene expression analysis of forskolin treated basilar papillae identifies microRNA181a as a mediator of proliferation. PLoS One 2010, 5, e11502. [Google Scholar] [CrossRef]
- Frucht, C.; Santos-Sacchi, J.; Navaratnam, D. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci. Lett. 2011, 493, 44–48. [Google Scholar] [CrossRef]
- Tsonis, P.A.; Call, M.K.; Grogg, M.W.; Sartor, M.A.; Taylor, R.R.; Forge, A.; Fyffe, R.; Goldenberg, R.; Cowper-Sallari, R.; Tomlinson, C.R. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem. Biophys. Res. Comm. 2007, 362, 940–945. [Google Scholar] [CrossRef]
- Elkan-Miller, T.; Ulitsky, I.; Hertzano, R.; Rudnicki, A.; Dror, A.; Lenz, D.; Elkon, R.; Irmler, M.; Beckers, J.; Shamir, R.; et al. Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS One 2011, 6, e18195. [Google Scholar] [CrossRef]
- Weston, M.; Pierce, M.; Jensen-Smith, H.; Fritzsch, B.; Rocha-Sanchez, S.; Beisel, K.; Soukup, G. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev. Dynam. 2011, 240, 808–819. [Google Scholar] [CrossRef]
- Mencia, A.; Modamio-Hoybjor, S.; Reshaw, N.; Morin, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Solda, G.; Robusto, M.; Primignani, P.; Castorina, P.; Benzoni, E.; Cesarani, A.; Ambrosetti, U.; Asselta, R.; Duga, S. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum. Mol. Genet. 2012. [Google Scholar] [CrossRef]
- Kiernan, A.; Pelling, A.; Leung, K.; Tang, A.; Bell, D.; Tease, C.; Lovell-Badge, R.; Steel, K.P.; Cheah, K.S.E. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 2005, 434, 1031–1035. [Google Scholar] [CrossRef]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Uthaiah, R.; Hudspeth, A. Molecular anatomy of the hair cell’s ribbon synapse. J. Neurosci. 2010, 30, 12387–12399. [Google Scholar] [CrossRef]
- Lee, K.Y.; Nakagawa, T.; Okano, T.; Hori, R.; Ono, K.; Tabata, Y.; Lee, S.H.; Ito, J. Novel therapy for hearing loss: Delivery of insulin-like growth factor 1 to the cochlea using gelatin hydrogel. Otol. Neurotol. 2007, 28, 976–981. [Google Scholar] [CrossRef]
- Nakagawa, T.; Sakamoto, T.; Hiraumi, H.; Kikkawa, Y.S.; Yamamoto, N.; Hamaguchi, K.; Ono, K.; Yamamoto, M.; Tabata, Y.; Teramukai, S.; et al. Topical insulin-like growth factor 1 treatment using gelatin hydrogels for glucocorticoid-resistant sudden sensorineural hearing loss: A prospective clinical trial. BMC Med. 2010, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Yamamoto, N.; Nakagawa, T.; Ito, J. Insulin-like growth factor 1 inhibits hair cell apoptosis and promotes the cell cycle of supporting cells by activating different downstream cascades after pharmacological hair cell injury. Mol. Cell Neurosci. 2013, 56, 29–38. [Google Scholar] [CrossRef] [Green Version]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Smith, M.E.; Rajadinakaran, G. The Transcriptomics to Proteomics of Hair Cell Regeneration: Looking for a Hair Cell in a Haystack. Microarrays 2013, 2, 186-207. https://doi.org/10.3390/microarrays2030186
Smith ME, Rajadinakaran G. The Transcriptomics to Proteomics of Hair Cell Regeneration: Looking for a Hair Cell in a Haystack. Microarrays. 2013; 2(3):186-207. https://doi.org/10.3390/microarrays2030186
Chicago/Turabian StyleSmith, Michael E., and Gopinath Rajadinakaran. 2013. "The Transcriptomics to Proteomics of Hair Cell Regeneration: Looking for a Hair Cell in a Haystack" Microarrays 2, no. 3: 186-207. https://doi.org/10.3390/microarrays2030186
APA StyleSmith, M. E., & Rajadinakaran, G. (2013). The Transcriptomics to Proteomics of Hair Cell Regeneration: Looking for a Hair Cell in a Haystack. Microarrays, 2(3), 186-207. https://doi.org/10.3390/microarrays2030186





