Ibuprofen in a Lipid Bilayer: Nanoscale Spatial Arrangement
Abstract
:1. Introduction
2. Theoretical Background
3. Experimental
3.1. Materials and Sample Preparation
3.2. EPR Measurements
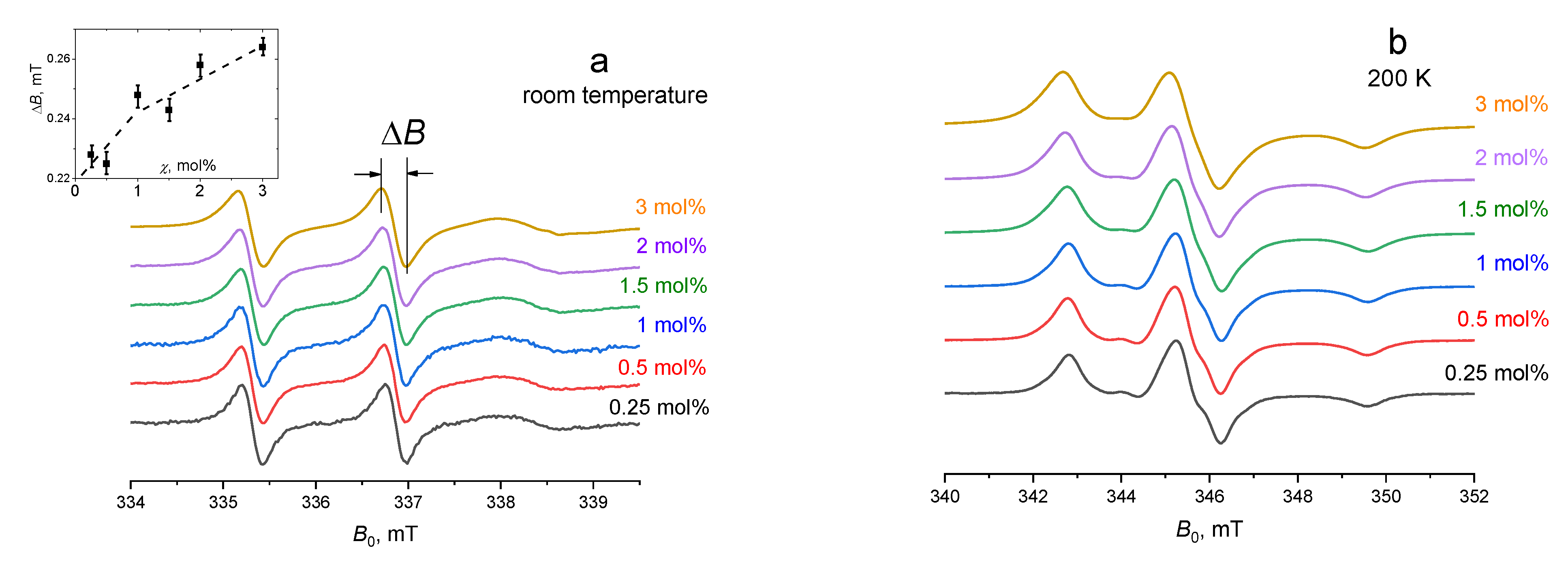
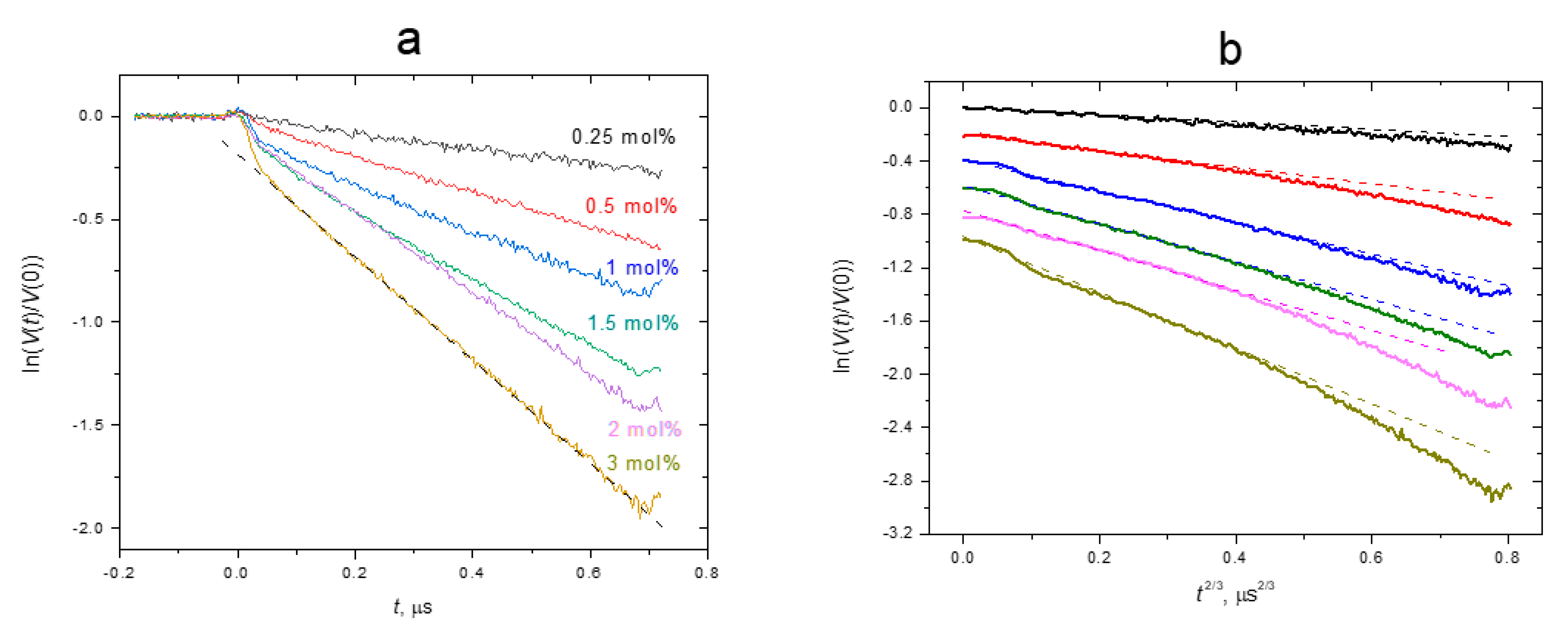
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rainsford, K.D. Ibuprofen: Discovery, Development and Therapeutics; John Wiley & Sons: West Sussex, UK, 2015. [Google Scholar] [CrossRef]
- Schjerning, A.-M.; Mcgettigan, P.; Gislason, G. Cardiovascular effects and safety of (non-aspirin) NSAIDs. Nat. Rev. Cardiol. 2020, 17, 574–584. [Google Scholar] [CrossRef]
- Rainsford, K.D. Ibuprofen: Pharmacology, Therapeutics and Side Effects; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar] [CrossRef]
- Wyss-Coray, T.; Mucke, L. Ibuprofen, inflammation and Alzheimer disease. Nat. Med. 2000, 6, 973–974. [Google Scholar] [CrossRef]
- Gao, X.; Chen, H.; Schwarzschild, M.A.; Ascherio, A. Use of ibuprofen and risk of Parkinson disease. Neurology 2011, 76, 863–869. [Google Scholar] [CrossRef] [Green Version]
- Van Dam, D.; Coen, K.; De Deyn, P. Ibuprofen modifies cognitive disease progression in an Alzheimer’s mouse model. J. Psychopharmacol. 2008, 24, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Khwaja, F.; Allen, J.; Lynch, J.; Andrews, P.; Djakiew, D. Ibuprofen Inhibits Survival of Bladder Cancer Cells by Induced Expression of the p75NTR Tumor Suppressor Protein. Cancer Res. 2004, 64, 6207–6213. [Google Scholar] [CrossRef] [Green Version]
- Zappavigna, S.; Cossu, A.M.; Grimaldi, A.; Bocchetti, M.; Ferraro, G.A.; Nicoletti, G.F.; Filosa, R.; Caraglia, M. Anti-Inflammatory Drugs as Anticancer Agents. Int. J. Mol. Sci. 2020, 21, 2605. [Google Scholar] [CrossRef] [Green Version]
- Endo, H.; Yano, M.; Okumura, Y.; Kido, H. Ibuprofen enhances the anticancer activity of cisplatin in lung cancer cells by inhibiting the heat shock protein 70. Cell Death Dis. 2014, 5, e1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenspan, E.J.; Madigan, J.P.; Boardman, L.A.; Rosenberg, D.W. Ibuprofen Inhibits Activation of Nuclear β-Catenin in Human Colon Adenomas and Induces the Phosphorylation of GSK-3β. Cancer Prev. Res. 2011, 4, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.; Leyva, R.; Kellstein, D.; Arthur, E.; Cruz-Rivera, M. Efficacy of a Fixed-Dose Combination of Ibuprofen and Acetaminophen Compared With Individual Monocomponents in Adult Male Subjects With Endotoxin-Induced Fever: A Randomized Controlled Trial. Clin. Ther. 2021, 43, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Brazier, D.; Perry, R.; Keane, J.; Barrett, K.; Elmaleh, D.R. Pharmacokinetics of Cromolyn and Ibuprofen in Healthy Elderly Volunteers. Clin. Drug Investig. 2017, 37, 1025–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cajaraville, J.P. Ibuprofen Arginate for Rapid-Onset Pain Relief in Daily Practice: A Review of Its Use in Different Pain Conditions. J. Pain Res. 2021, 14, 117–126. [Google Scholar] [CrossRef]
- Laine, L.; Kivitz, A.J.; E Bello, A.; Grahn, A.Y.; Schiff, M.H.; Taha, A.S. Double-Blind Randomized Trials of Single-Tablet Ibuprofen/High-Dose Famotidine vs. Ibuprofen Alone for Reduction of Gastric and Duodenal Ulcers. Am. J. Gastroenterol. 2012, 107, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Hartlieb, K.J.; Ferris, D.P.; Holcroft, J.M.; Kandela, I.; Stern, C.L.; Nassar, M.S.; Botros, Y.Y.; Stoddart, J.F. Encapsulation of Ibuprofen in CD-MOF and Related Bioavailability Studies. Mol. Pharm. 2017, 14, 1831–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javanbakht, S.; Nezhad-Mokhtari, P.; Shaabani, A.; Arsalani, N.; Ghorbani, M. Incorporating Cu-based metal-organic framework/drug nanohybrids into gelatin microsphere for ibuprofen oral delivery. Mater. Sci. Eng. C 2018, 96, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Alsop, R.J.; Armstrong, C.L.; Maqbool, A.; Toppozini, L.; Dies, H.; Rheinstädter, M.C. Cholesterol expels ibuprofen from the hydrophobic membrane core and stabilizes lamellar phases in lipid membranes containing ibuprofen. Soft Matter 2015, 11, 4756–4767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.; Mamontov, E.; Tyagi, M. Effects of NSAIDs on the nanoscopic dynamics of lipid membrane. Biochim. et Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183100. [Google Scholar] [CrossRef]
- Yefimova, S.L.; Tkacheva, T.N.; Kasian, N.A. Study of the Combined Effect of Ibuprofen and Cholesterol on the Microviscosity and Ordering of Model Lipid Membranes by Timeresolved Measurement of Fluorescence Anisotropy Decay. J. Appl. Spectrosc. 2017, 84, 284–290. [Google Scholar] [CrossRef]
- Sharma, V.K.; Nagao, M.; Rai, D.K.; Mamontov, E. Membrane softening by nonsteroidal anti-inflammatory drugs investigated by neutron spin echo. Phys. Chem. Chem. Phys. 2019, 21, 20211–20218. [Google Scholar] [CrossRef]
- Ramadurai, S.; Sarangi, N.K.; Maher, S.; MacConnell, N.; Bond, A.M.; McDaid, D.; Flynn, D.; Keyes, T.E. Microcavity-Supported Lipid Bilayers; Evaluation of Drug–Lipid Membrane Interactions by Electrochemical Impedance and Fluorescence Correlation Spectroscopy. Langmuir 2019, 35, 8095–8109. [Google Scholar] [CrossRef]
- Wood, M.; Morales, M.; Miller, E.; Braziel, S.; Giancaspro, J.; Scollan, P.; Rosario, J.; Gayapa, A.; Krmic, M.; Lee, S. Ibuprofen and the Phosphatidylcholine Bilayer: Membrane Water Permeability in the Presence and Absence of Cholesterol. Langmuir 2021, 37, 4468–4680. [Google Scholar] [CrossRef]
- Sun, S.; Sendecki, A.M.; Pullanchery, S.; Huang, D.; Yang, T.; Cremer, P.S. Multistep Interactions between Ibuprofen and Lipid Membranes. Langmuir 2018, 34, 10782–10792. [Google Scholar] [CrossRef] [PubMed]
- Kremkow, J.; Luck, M.; Huster, D.; Müller, P.; Scheidt, H.A. Membrane Interaction of Ibuprofen with Cholesterol-Containing Lipid Membranes. Biomolecules 2020, 10, 1384. [Google Scholar] [CrossRef]
- Teixeira, L.D.S.; Chagas, T.V.; Alonso, A.; Gonzalez-Alvarez, I.; Bermejo, M.; Polli, J.; Rezende, K.R. Biomimetic Artificial Membrane Permeability Assay over Franz Cell Apparatus Using BCS Model Drugs. Pharmaceutics 2020, 12, 988. [Google Scholar] [CrossRef] [PubMed]
- Aloi, E.; Rizzuti, B.; Guzzi, R.; Bartucci, R. Association of ibuprofen at the polar/apolar interface of lipid membranes. Arch. Biochem. Biophys. 2018, 654, 77–84. [Google Scholar] [CrossRef]
- Baranov, D.S.; Smorygina, A.S.; Dzuba, S.A. Synthesis of Spin-Labeled Ibuprofen and Its Interaction with Lipid Membranes. Molecules 2022, 27, 4127. [Google Scholar] [CrossRef] [PubMed]
- Khajeh, A.; Modarress, H. The influence of cholesterol on interactions and dynamics of ibuprofen in a lipid bilayer. Biochim. et Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2431–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jämbeck, J.P.M.; Lyubartsev, A.P. Exploring the Free Energy Landscape of Solutes Embedded in Lipid Bilayers. J. Phys. Chem. Lett. 2013, 4, 1781–1787. [Google Scholar] [CrossRef]
- Milov, A.D.; Salikhov, K.M.; Shirov, M.D. Application of ELDOR in electron-spin echo for paramagnetic center space distribution in solids. Fiz. Tverd. Tela 1981, 23, 975–982. [Google Scholar]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H. Dead-Time Free Measurement of Dipole–Dipole Interactions between Electron Spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef]
- Schiemann, O.; Prisner, T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007, 40, 1–53. [Google Scholar] [CrossRef]
- Fábregas-Ibáñez, L.; Tessmer, M.H.; Jeschke, G.; Stoll, S. Dipolar pathways in dipolar EPR spectroscopy. Phys. Chem. Chem. Phys. 2022, 24, 2504–2520. [Google Scholar] [CrossRef]
- Milov, A.D.; Grishin, Y.A.; Dzuba, S.A.; Tsvetkov, Y.D. Effect of Pumping Pulse Duration on Echo Signal Amplitude in Four-Pulse PELDOR. Appl. Magn. Reson. 2011, 41, 59–67. [Google Scholar] [CrossRef]
- Unguryan, V.V.; Golysheva, E.A.; Dzuba, S.A. Double Electron–Electron Resonance of Spin-Labeled Cholestane in Model Membranes: Evidence for Substructures inside the Lipid Rafts. J. Phys. Chem. B 2021, 125, 9557–9563. [Google Scholar] [CrossRef] [PubMed]
- Smorygina, A.S.; Golysheva, E.A.; Dzuba, S.A. Clustering of Stearic Acids in Model Phospholipid Membranes Revealed by Double Electron–Electron Resonance. Langmuir 2021, 37, 13909–13916. [Google Scholar] [CrossRef]
- Abragam, A. The Principles of Nuclear Magnetism; Chapter IV; Clarendon Press: Oxford, UK, 1961. [Google Scholar]
- Jeschke, G.; Panek, G.; Godt, A.; Bender, A.; Paulsen, H. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 2004, 26, 223–244. [Google Scholar] [CrossRef]
- Kutsovsky, Y.E.; Mariasov, A.G.; Aristov, Y.; Parmon, V.N. Electron spin echo as a tool for investigation of surface structure of finely dispersed fractal solids. React. Kinet. Catal. Lett. 1990, 42, 19–24. [Google Scholar] [CrossRef]
- Kardash, M.E.; Dzuba, S.A. Lipid-Mediated Clusters of Guest Molecules in Model Membranes and Their Dissolving in the Presence of Lipid Rafts. J. Phys. Chem. B 2017, 121, 5209–5217. [Google Scholar] [CrossRef]
- Pabst, G.; Danner, S.; Podgornik, R.; Katsaras, J. Entropy-Driven Softening of Fluid Lipid Bilayers by Alamethicin. Langmuir 2007, 23, 11705–11711. [Google Scholar] [CrossRef]
- Kuznetsov, N.; Milov, A.D.; Koval, V.; Samoilova, R.; Grishin, Y.A.; Knorre, D.G.; Tsvetkov, Y.D.; Fedorova, O.; Dzuba, S.A. PELDOR study of conformations of double-spin-labeled single- and double-stranded DNA with non-nucleotide inserts. Phys. Chem. Chem. Phys. 2009, 11, 6826–6832. [Google Scholar] [CrossRef]
- Marsh, D. Spin-Label Electron Paramagnetic Resonance Spectroscopy; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar] [CrossRef]
- Poger, D.; Mark, A.E. On the Validation of Molecular Dynamics Simulations of Saturated and cis-Monounsaturated Phosphatidylcholine Lipid Bilayers: A Comparison with Experiment. J. Chem. Theory Comput. 2010, 6, 325–336. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashnik, A.S.; Baranov, D.S.; Dzuba, S.A. Ibuprofen in a Lipid Bilayer: Nanoscale Spatial Arrangement. Membranes 2022, 12, 1077. https://doi.org/10.3390/membranes12111077
Kashnik AS, Baranov DS, Dzuba SA. Ibuprofen in a Lipid Bilayer: Nanoscale Spatial Arrangement. Membranes. 2022; 12(11):1077. https://doi.org/10.3390/membranes12111077
Chicago/Turabian StyleKashnik, Anna S., Denis S. Baranov, and Sergei A. Dzuba. 2022. "Ibuprofen in a Lipid Bilayer: Nanoscale Spatial Arrangement" Membranes 12, no. 11: 1077. https://doi.org/10.3390/membranes12111077





