Oxidative Biomarkers Associated with the Pulmonary Manifestation of Post-COVID-19 Complications
Abstract
:1. Introduction
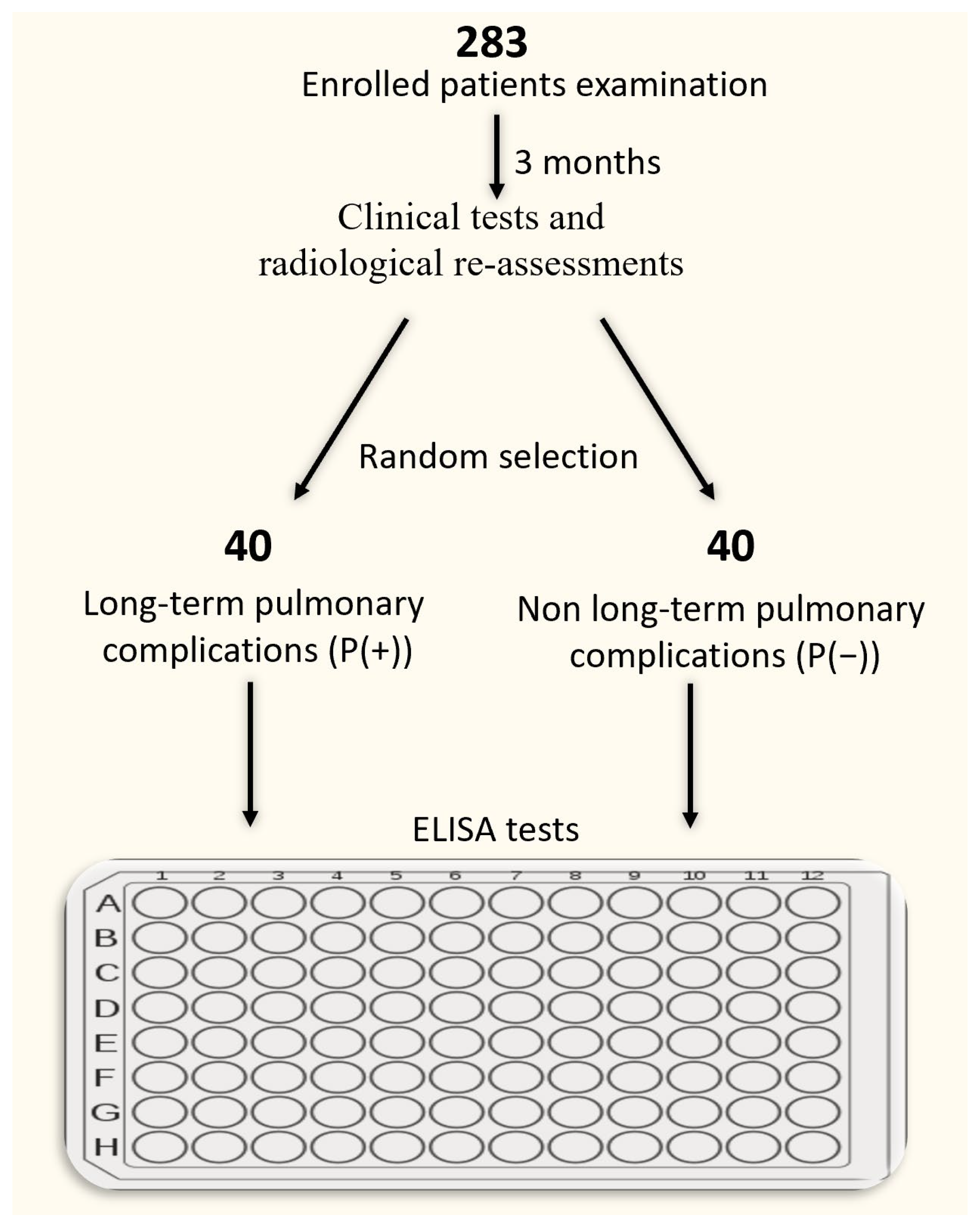
2. Materials and Methods
2.1. Subjects
2.2. Pulmonary Function Tests
2.3. Samples
2.4. ELISA
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Investigated Population
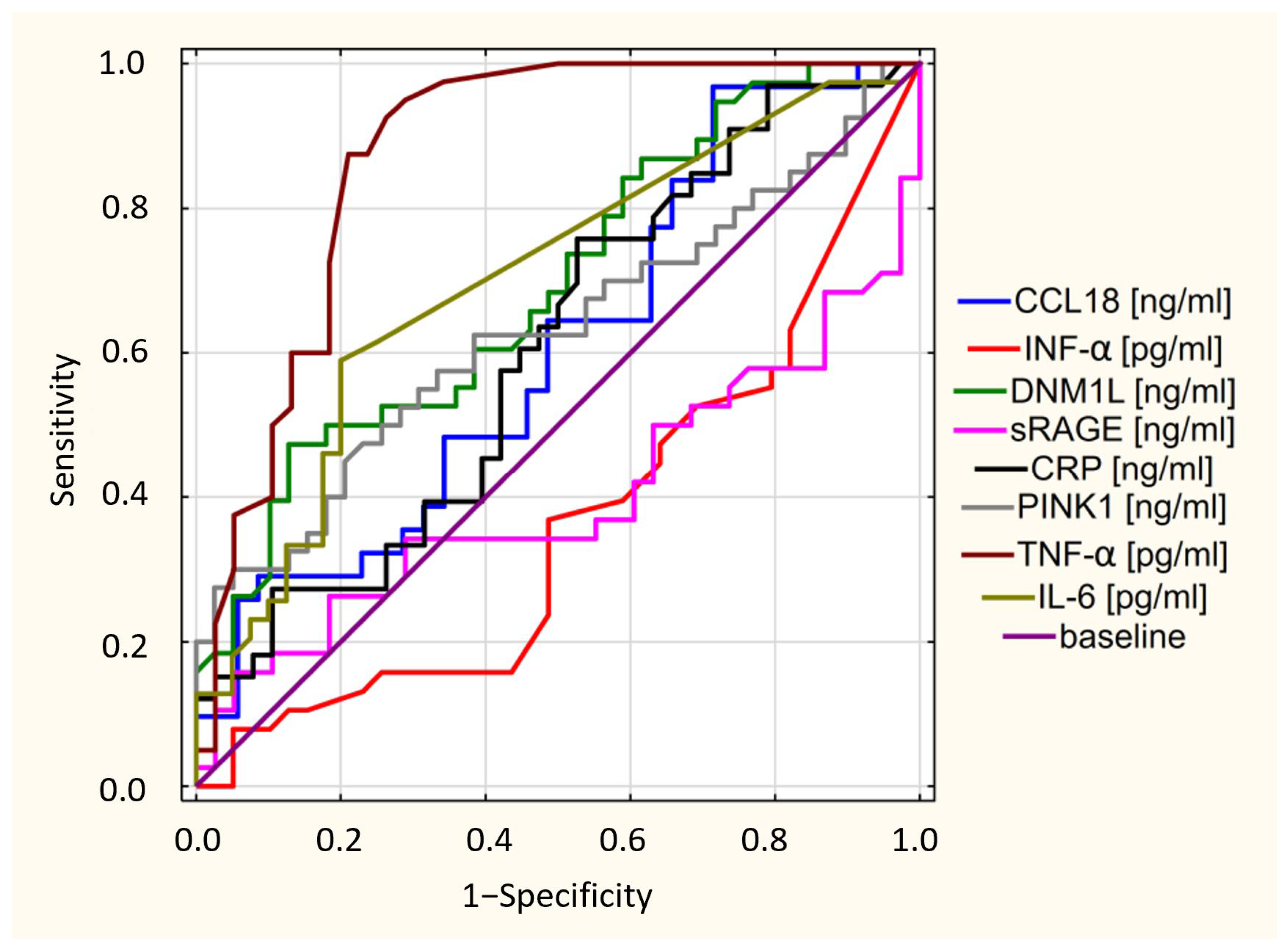
3.2. ELISA Test Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stokes, E.K.; Zambrano, L.D.; Anderson, K.N.; Marder, E.P.; Raz, K.M.; El Burai Felix, S.; Tie, Y.; Fullerton, K.E. Coronavirus Disease 2019 Case Surveillance—United States, January 22–May 30, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 759–765. [Google Scholar] [CrossRef]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the Asymptomatic Proportion of Coronavirus Disease 2019 (COVID-19) Cases on Board the Diamond Princess Cruise Ship, Yokohama, Japan, 2020. Eurosurveillance 2020, 25, 2000180. [Google Scholar] [CrossRef] [Green Version]
- Teuwen, L.A.; Geldhof, V.; Pasut, A.; Carmeliet, P. COVID-19: The Vasculature Unleashed. Nat. Rev. Immunol. 2020, 20, 389–391. [Google Scholar] [CrossRef]
- Zong, M.; Zheng, L.; Zhou, H.; Lu, L.; East, S.; He, H.L.; Wu, H.X.; Chen, H.N.; Wang, L.; Yuan, H.J.; et al. TGF-β and CCL18 as Indicators for Predicting and Monitoring the Development of Pulmonary Fibrosis in Patients with COVID-19. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Desai, A.D.; Lavelle, M.; Boursiquot, B.C.; Wan, E.Y. Long-Term Complications of COVID-19. Am. J. Physiol. Cell Physiol. 2022, 322, C1–C11. [Google Scholar] [CrossRef]
- Niedziela, J.T.; Głowacki, J.; Ochman, M.; Pudlo, R.; Adamczyk-Sowa, M.; Nowowiejska-Wiewióra, A.; Kułaczkowska, Z.; Sobala-Szczygieł, B.; Myrda, K.; Wiewióra, M.; et al. Post-COVID-19 Complications in Hospitalized and Non-Hospitalized Patients: The Silesian Complications of the COVID-19 (SILCOV-19) Database. Pol. Arch. Intern. Med. 2022, 132, 16233. [Google Scholar] [CrossRef]
- Ahmad Alhiyari, M.; Ata, F.; Islam Alghizzawi, M.; Bint I Bilal, A.; Salih Abdulhadi, A.; Yousaf, Z. Post COVID-19 Fibrosis, an Emerging Complicationof SARS-CoV-2 Infection. IDCases 2021, 23, e01041. [Google Scholar] [CrossRef]
- Bridi, G.d.P.; Tanni, S.E.; Baldi, B.G. Current Understanding of Post-COVID Pulmonary Fibrosis: Where Are We? Arch. Bronconeumol. 2023, 59, 69–70. [Google Scholar] [CrossRef]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF Cytokine Triad Is Associated with Post-Acute Sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; das Neves, P.F.M.; Lima, S.S.; Lopes, J.D.C.; Torres, M.K.D.S.; Vallinoto, I.M.V.C.; Bichara, C.D.A.; dos Santos, E.F.; de Brito, M.T.F.M.; da Silva, A.L.S.; et al. Cytokine Profiles Associated with Acute COVID-19 and Long COVID-19 Syndrome. Front. Cell. Infect. Microbiol. 2022, 12, 931. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; El Idrissi Saik, I.; Guessous, F.; Akarid, K. The Pro-Inflammatory Cytokines in COVID-19 Pathogenesis: What Goes Wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Hama Amin, B.J.; Kakamad, F.H.; Ahmed, G.S.; Ahmed, S.F.; Abdulla, B.A.; Mohammed, S.H.; Mikael, T.M.; Salih, R.Q.; Ali, R.K.; Salh, A.M.; et al. Post COVID-19 Pulmonary Fibrosis; a Meta-Analysis Study. Ann. Med. Surg. 2022, 77, 103590. [Google Scholar] [CrossRef]
- Antoniou, K.M.; Vasarmidi, E.; Russell, A.-M.; Andrejak, C.; Crestani, B.; Delcroix, M.; Dinh-Xuan, A.T.; Poletti, V.; Sverzellati, N.; Vitacca, M.; et al. European Respiratory Society Statement on Long COVID-19 Follow-Up. Eur. Respir. J. 2022, 60, 2102174. [Google Scholar] [CrossRef]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol. 2022, 12, 4159. [Google Scholar] [CrossRef]
- Nunn, A.V.W.; Guy, G.W.; Brysch, W.; Bell, J.D. Understanding Long COVID.; Mitochondrial Health and Adaptation— Old Pathways, New Problems. Biomedicines 2022, 10, 3113. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of Mitophagy in Cellular Homeostasis, Physiology and Pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Jarrott, B.; Head, R.; Pringle, K.G.; Lumbers, E.R.; Martin, J.H. “LONG COVID”—A Hypothesis for Understanding the Biological Basis and Pharmacological Treatment Strategy. Pharmacol. Res. Perspect. 2022, 10, e00911. [Google Scholar] [CrossRef]
- Silvagno, F.; Vernone, A.; Pescarmona, G.P. The Role of Glutathione in Protecting against the Severe Inflammatory Response Triggered by COVID-19. Antioxidants 2020, 9, 624. [Google Scholar] [CrossRef]
- Vollbracht, C.; Kraft, K. Oxidative Stress and Hyper-Inflammation as Major Drivers of Severe COVID-19 and Long COVID: Implications for the Benefit of High-Dose Intravenous Vitamin C. Front. Pharmacol. 2022, 13, 899198. [Google Scholar] [CrossRef]
- Duca, L.; Ottolenghi, S.; Coppola, S.; Rinaldo, R.; Dei Cas, M.; Rubino, F.M.; Paroni, R.; Samaja, M.; Chiumello, D.A.; Motta, I. Differential Redox State and Iron Regulation in Chronic Obstructive Pulmonary Disease, Acute Respiratory Distress Syndrome and Coronavirus Disease 2019. Antioxidants 2021, 10, 1460. [Google Scholar] [CrossRef]
- Wick, K.D.; Siegel, L.; Neaton, J.D.; Oldmixon, C.; Lundgren, J.; Dewar, R.L.; Lane, H.C.; Thompson, B.T.; Matthay, M.A. RAGE Has Potential Pathogenetic and Prognostic Value in Nonintubated Hospitalized Patients with COVID-19. JCI Insight 2022, 7, e157499. [Google Scholar] [CrossRef]
- Queisser, M.A.; Kouri, F.M.; Königshoff, M.; Wygrecka, M.; Schubert, U.; Eickelberg, O.; Preissner, K.T. Loss of RAGE in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2012, 39, 337–345. [Google Scholar] [CrossRef]
- Fritz, G. RAGE: A Single Receptor Fits Multiple Ligands. Trends Biochem. Sci. 2011, 36, 625–632. [Google Scholar] [CrossRef]
- Kierdorf, K.; Fritz, G. RAGE Regulation and Signaling in Inflammation and Beyond. J. Leukoc. Biol. 2013, 94, 55–68. [Google Scholar] [CrossRef]
- NHIS—Adult Tobacco Use—Glossary. Available online: https://www.cdc.gov/nchs/nhis/tobacco/tobacco_glossary.htm (accessed on 5 October 2022).
- Graham, B.L.; Steenbruggen, I.; Miller, M.R.; Barjaktarevic, I.Z.; Cooper, B.G.; Hall, G.L.; Hallstrand, T.S.; Kaminsky, D.A.; Mccarthy, K.; Mccormack, M.C.; et al. Standardization of Spirometry 2019 Update. An Official American Thoracic Society and European Respiratory Society Technical Statement. Am. J. Respir. Crit. Care Med. 2019, 200, e70–e88. [Google Scholar] [CrossRef]
- Croghan, C.; Egeghy, P.P. Methods of dealing with values below the limit of detection using SAS. South. SAS User Group 2003, 22, 24. [Google Scholar]
- Asadi-Pooya, A.A.; Akbari, A.; Emami, A.; Lotfi, M.; Rostamihosseinkhani, M.; Nemati, H.; Barzegar, Z.; Kabiri, M.; Zeraatpisheh, Z.; Farjoud-Kouhanjani, M.; et al. Risk Factors Associated with Long COVID Syndrome: A Retrospective Study. Iran. J. Med. Sci. 2021, 46, 428. [Google Scholar] [CrossRef]
- Aul, D.R.; Gates, D.J.; Draper, D.A.; Dunleavy, D.A.; Ruickbie, D.S.; Meredith, D.H.; Walters, D.N.; van Zeller, D.C.; Taylor, D.V.; Bridgett, D.M.; et al. Complications after Discharge with COVID-19 Infection and Risk Factors Associated with Development of Post-COVID Pulmonary Fibrosis. Respir. Med. 2021, 188, 106602. [Google Scholar] [CrossRef]
- Marvisi, M.; Ferrozzi, F.; Balzarini, L.; Mancini, C.; Ramponi, S.; Uccelli, M. First Report on Clinical and Radiological Features of COVID-19 Pneumonitis in a Caucasian Population: Factors Predicting Fibrotic Evolution. Int. J. Infect. Dis. 2020, 99, 485–488. [Google Scholar] [CrossRef]
- Bhowal, C.; Ghosh, S.; Ghatak, D.; De, R. Pathophysiological Involvement of Host Mitochondria in SARS-CoV-2 Infection That Causes COVID-19: A Comprehensive Evidential Insight. Mol. Cell. Biochem. 2023, 478, 1325–1343. [Google Scholar] [CrossRef]
- Prasada Kabekkodu, S.; Chakrabarty, S.; Jayaram, P.; Mallya, S.; Thangaraj, K.; Singh, K.K.; Satyamoorthy, K. Severe Acute Respiratory Syndrome Coronaviruses Contributing to Mitochondrial Dysfunction: Implications for Post-COVID Complications. Mitochondrion 2023, 69, 43–56. [Google Scholar] [CrossRef]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 Hijacking of Host Mitochondria in COVID-19 Pathogenesis. Am. J. Physiol. Cell Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef]
- Mehrzadi, S.; Karimi, M.Y.; Fatemi, A.; Reiter, R.J.; Hosseinzadeh, A. SARS-CoV-2 and Other Coronaviruses Negatively Influence Mitochondrial Quality Control: Beneficial Effects of Melatonin. Pharmacol. Ther. 2021, 224, 107825. [Google Scholar] [CrossRef]
- Elesela, S.; Lukacs, N.W. Role of Mitochondria in Viral Infections. Life 2021, 11, 232. [Google Scholar] [CrossRef]
- de la Cruz-Enríquez, J.; Rojas-Morales, E.; Ruíz-García, M.G.; Tobón-Velasco, J.C.; Jiménez-Ortega, J.C. SARS-CoV-2 Induces Mitochondrial Dysfunction and Cell Death by Oxidative Stress/Inflammation in Leukocytes of COVID-19 Patients. Free Radic. Res. 2021, 55, 982–995. [Google Scholar] [CrossRef]
- Zhu, L.; Mou, C.; Yang, X.; Lin, J.; Yang, Q. Mitophagy in TGEV Infection Counteracts Oxidative Stress and Apoptosis. Oncotarget 2016, 7, 27122. [Google Scholar] [CrossRef] [Green Version]
- Costa, T.J.; Potje, S.R.; Fraga-Silva, T.F.C.; da Silva-Neto, J.A.; Barros, P.R.; Rodrigues, D.; Machado, M.R.; Martins, R.B.; Santos-Eichler, R.A.; Benatti, M.N.; et al. Mitochondrial DNA and TLR9 Activation Contribute to SARS-CoV-2-Induced Endothelial Cell Damage. Vascul. Pharmacol. 2022, 142, 106946. [Google Scholar] [CrossRef]
- Eiyama, A.; Okamoto, K. PINK1/Parkin-Mediated Mitophagy in Mammalian Cells. Curr. Opin. Cell Biol. 2015, 33, 95–101. [Google Scholar] [CrossRef]
- Fu, C.; Cao, N.; Liu, W.; Zhang, Z.; Yang, Z.; Zhu, W.; Fan, S. Crosstalk between Mitophagy and Innate Immunity in Viral Infection. Front. Microbiol. 2022, 13, 1064045. [Google Scholar] [CrossRef]
- Pozzi, A. COVID-19 and Mitochondrial Non-Coding RNAs: New Insights from Published Data. Front. Physiol. 2022, 12, 805005. [Google Scholar] [CrossRef]
- Patrucco, F.; Solidoro, P.; Gavelli, F.; Apostolo, D.; Bellan, M. Idiopathic Pulmonary Fibrosis and Post-COVID-19 Lung Fibrosis: Links and Risks. Microorganisms 2023, 11, 895. [Google Scholar] [CrossRef]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological Dysfunction Persists for 8 Months Following Initial Mild-to-Moderate SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef]
- Ospelnikova, T.P.; Levitskaya, D.S.; Kolodyazhnaya, L.V.; Shitova, A.D.; Osiptsov, V.N.; Kryukova, N.O.; Pakhomov, D.v.; Khromova, E.A.; Arifullina, L.; Baranova, I.A.; et al. The Biological Activity of Interferons in Post-COVID Syndrome. ERJ Open Res. 2022, 8, 240. [Google Scholar] [CrossRef]
- Hu, Z.J.; Xu, J.; Yin, J.M.; Li, L.; Hou, W.; Zhang, L.L.; Zhou, Z.; Yu, Y.Z.; Li, H.J.; Feng, Y.M.; et al. Lower Circulating Interferon-Gamma Is a Risk Factor for Lung Fibrosis in COVID-19 Patients. Front. Immunol. 2020, 11, 2348. [Google Scholar] [CrossRef]
- Colarusso, C.; Maglio, A.; Terlizzi, M.; Vitale, C.; Molino, A.; Pinto, A.; Vatrella, A.; Sorrentino, R. Post-COVID-19 Patients Who Develop Lung Fibrotic-like Changes Have Lower Circulating Levels of IFN-β but Higher Levels of IL-1α and TGF-β. Biomedicines 2021, 9, 1931. [Google Scholar] [CrossRef]
- Jhuti, D.; Rawat, A.; Guo, C.M.; Wilson, L.A.; Mills, E.J.; Forrest, J.I. Interferon Treatments for SARS-CoV-2: Challenges and Opportunities. Infect. Dis. Ther. 2022, 11, 953. [Google Scholar] [CrossRef]
- Wigén, J.; Löfdahl, A.; Bjermer, L.; Elowsson-Rendin, L.; Westergren-Thorsson, G. Converging Pathways in Pulmonary Fibrosis and COVID-19—The Fibrotic Link to Disease Severity. Respir. Med. X 2020, 2, 100023. [Google Scholar] [CrossRef]
- George, P.M.; Wells, A.U.; Jenkins, R.G. Pulmonary Fibrosis and COVID-19: The Potential Role for Antifibrotic Therapy. Lancet Respir. Med. 2020, 8, 807–815. [Google Scholar] [CrossRef]
- Kapandji, N.; Yvin, E.; Devriese, M.; de Margerie-Mellon, C.; Moratelli, G.; Lemiale, V.; Jabaudon, M.; Azoulay, E.; Constantin, J.M.; Dumas, G. Importance of Lung Epithelial Injury in COVID-19- Associated Acute Respiratory Distress Syndrome: Value of Plasma Soluble Receptor for Advanced Glycation End-Products. Am. J. Respir. Crit. Care Med. 2021, 204, 359–362. [Google Scholar] [CrossRef]
- Allen, C.N.S.; Santerre, M.; Arjona, S.P.; Ghaleb, L.J.; Herzi, M.; Llewellyn, M.D.; Shcherbik, N.; Sawaya, B.E. SARS-CoV-2 Causes Lung Inflammation through Metabolic Reprogramming and RAGE. Viruses 2022, 14, 983. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, L.; Delguste, F.; Durand, A.; Guilbaud, A.; Rousselin, C.; Schmidt, A.M.; Tessier, F.; Boulanger, E.; Neviere, R. Advanced Glycation End Products Receptor RAGE Controls Myocardial Dysfunction and Oxidative Stress in High-Fat Fed Mice by Sustaining Mitochondrial Dynamics and Autophagy-Lysosome Pathway. Free Radic. Biol. Med. 2017, 112, 397–410. [Google Scholar] [CrossRef]
- Aquaro, S.; Holms, R.D. Long COVID (PASC) Is Maintained by a Self-Sustaining Pro-Inflammatory TLR4/RAGE-Loop of S100A8/A9 > TLR4/RAGE Signalling, Inducing Chronic Expression of IL-1b, IL-6 and TNFa: Anti-Inflammatory Ezrin Peptides as Potential Therapy. Immuno 2022, 2, 512–533. [Google Scholar] [CrossRef]
- Jankovic, M.; Nikolic, D.; Novakovic, I.; Petrovic, B.; Lackovic, M.; Santric-Milicevic, M. MiRNAs as a Potential Biomarker in the COVID-19 Infection and Complications Course, Severity, and Outcome. Diagnostics 2023, 13, 1091. [Google Scholar] [CrossRef]
- Guiot, J.; Henket, M.; Remacle, C.; Cambier, M.; Struman, I.; Winandy, M.; Moermans, C.; Louis, E.; Malaise, M.; Ribbens, C.; et al. Systematic Review of Overlapping MicroRNA Patterns in COVID-19 and Idiopathic Pulmonary Fibrosis. Respir. Res. 2023, 24, 112. [Google Scholar] [CrossRef]
- Centa, A.; Fonseca, A.S.; Da Silva Ferreira, S.G.; Azevedo, M.L.V.; De Paula, C.B.V.; Nagashima, S.; MacHado-Souz, C.; Dos Santos Miggiolaro, A.F.R.; Baena, C.P.; De Noronha, L.; et al. Deregulated MiRNA Expression Is Associated with Endothelial Dysfunction in Post-Mortem Lung Biopsies of COVID-19 Patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2021, 320, L405–L412. [Google Scholar] [CrossRef]
- Maranini, B.; Ciancio, G.; Ferracin, M.; Cultrera, R.; Negrini, M.; Sabbioni, S.; Govoni, M. MicroRNAs and Inflammatory Immune Response in SARS-CoV-2 Infection: A Narrative Review. Life 2022, 12, 288. [Google Scholar] [CrossRef]

| Parameter | P(+) n = 36 | P(−) n = 37 | |||
|---|---|---|---|---|---|
| Volume (L) | % of Predicted Value * | Volume (L) | % of Predicted Value * | * p-Value | |
| FVC, mean (SD) | 3.2 (1.24) | 74 (20.2) | 3.8 (0.99) | 92 (11.56) | <0.001 |
| FEV1, mean (SD) | 2.6 (0.95) | 76 (18.45) | 3 (0.72) | 90 (12.28) | <0.001 |
| TLCO, mean (SD) | 5.7 (2.56) | 67 (25.67) | 7.9 (2.13) | 97 (12.87) | <0.001 |
| P(+) n = 40 | P(−) n = 40 | p-Value | |
|---|---|---|---|
| Age (years), mean (SD) | 56 (12.21) | 53 (11.3) | 0.210 |
| Male sex, n (%) | 31 (77.5%) | 21 (52%) | 0.019 |
| BMI (kg/m2), mean (SD) | 28 (5.28) | 28 (4.7) | 0.699 |
| Comorbidities, n (%) | |||
| Hypertension | 18 (45%) | 14 (35%) | 0.367 |
| Obesity | 10 (25%) | 7 (17.5%) | 0.419 |
| Heart failure | 5 (12.5%) | 1 (2.5%) | 0.094 |
| Type 2 diabetes | 4 (10%) | 8 (20%) | 0.216 |
| Asthma | 4 (10%) | 4 (10%) | 0.992 |
| Coronary heart disease | 4 (10%) | 2 (5%) | 0.404 |
| Pulmonary hypertension | 2 (5%) | 1 (2.5%) | 0.569 |
| Smoking | |||
| Active smokers, n (%) | 2 (5%) | 8 (20%) | 0.024 |
| Pack-years, mean (SD) | 16 (13) | 11 (13) | 0.118 |
| Ex-smokers, n (%) | 22 (55%) | 18 (45%) | 0.420 |
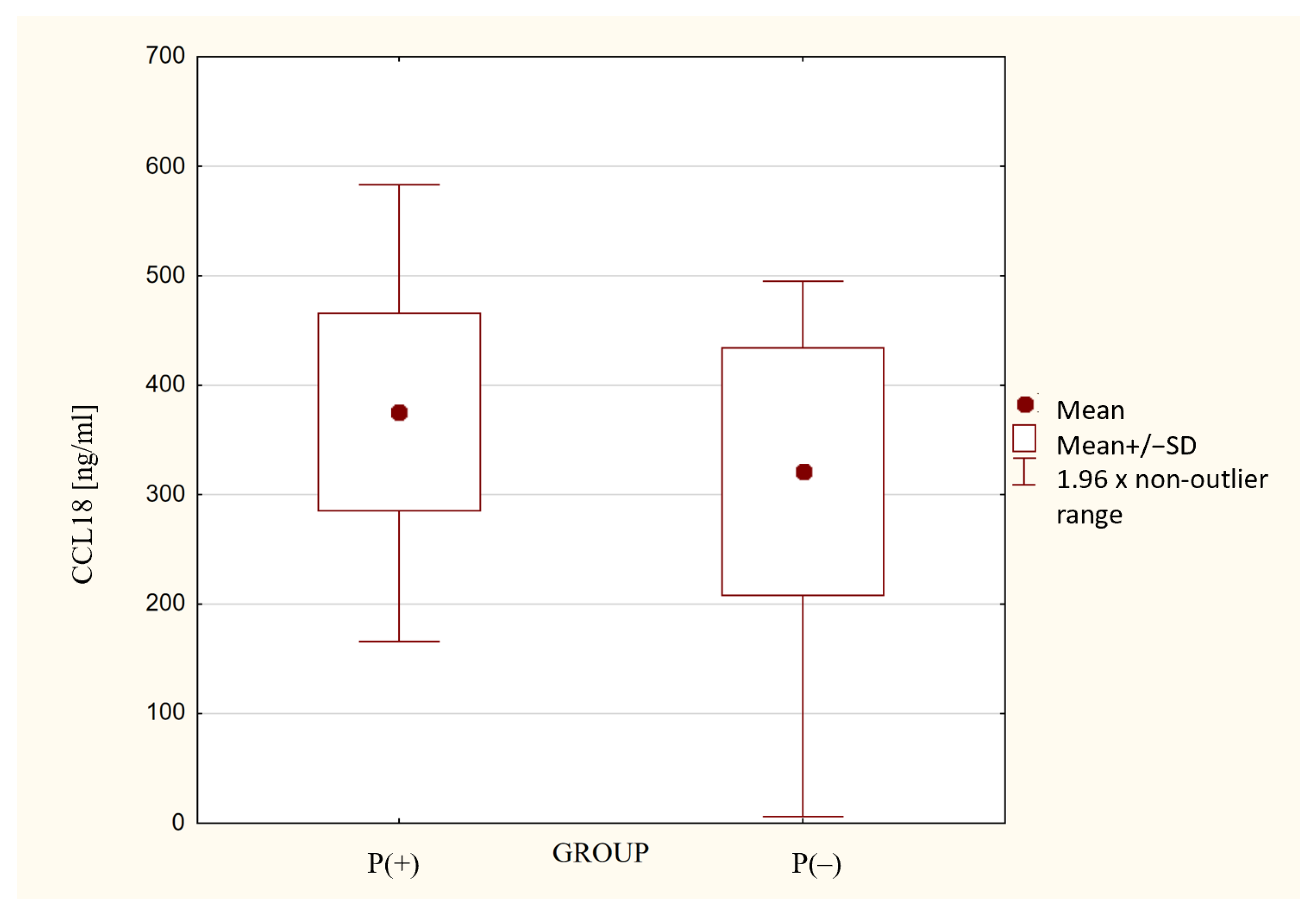
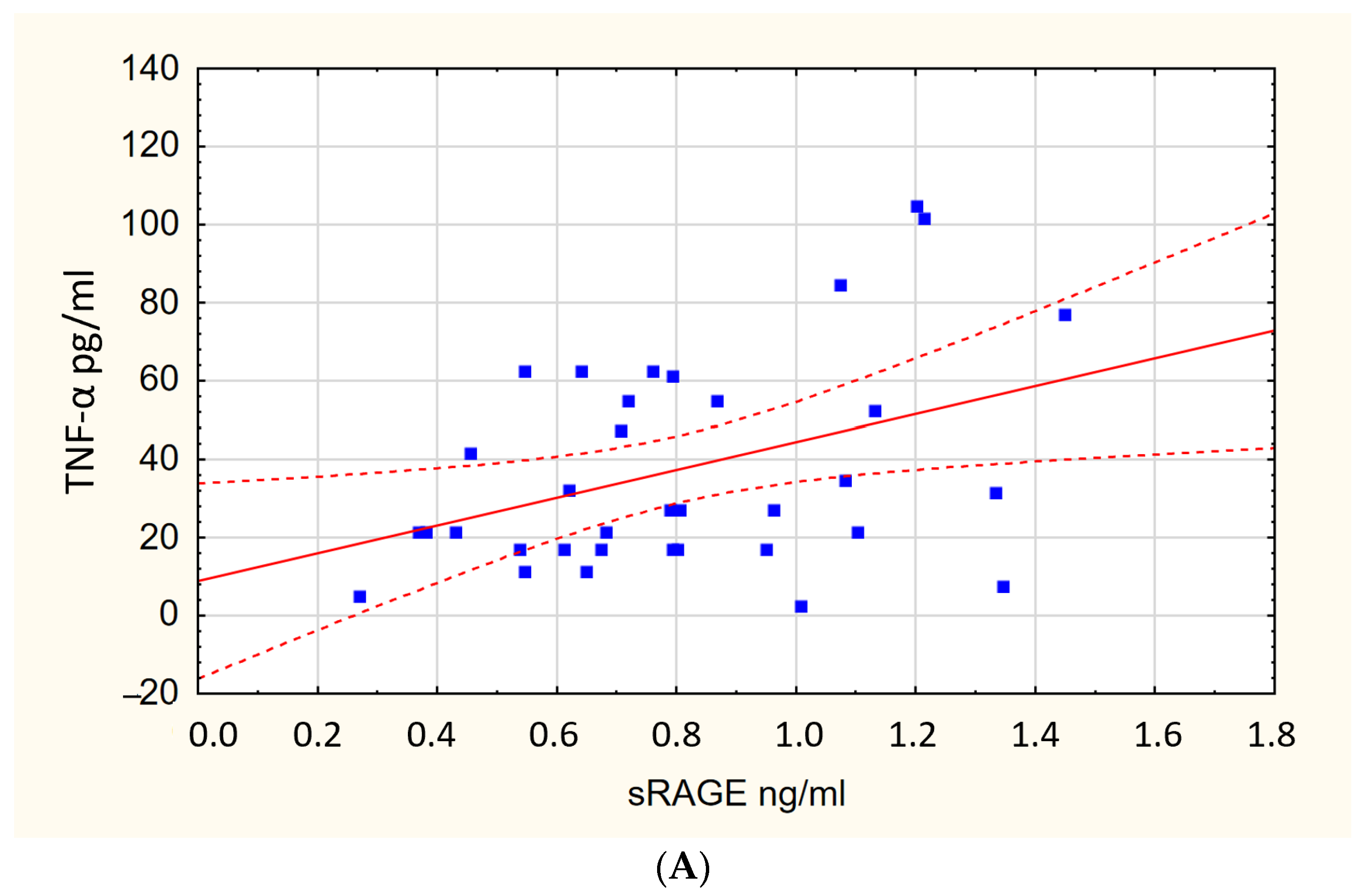
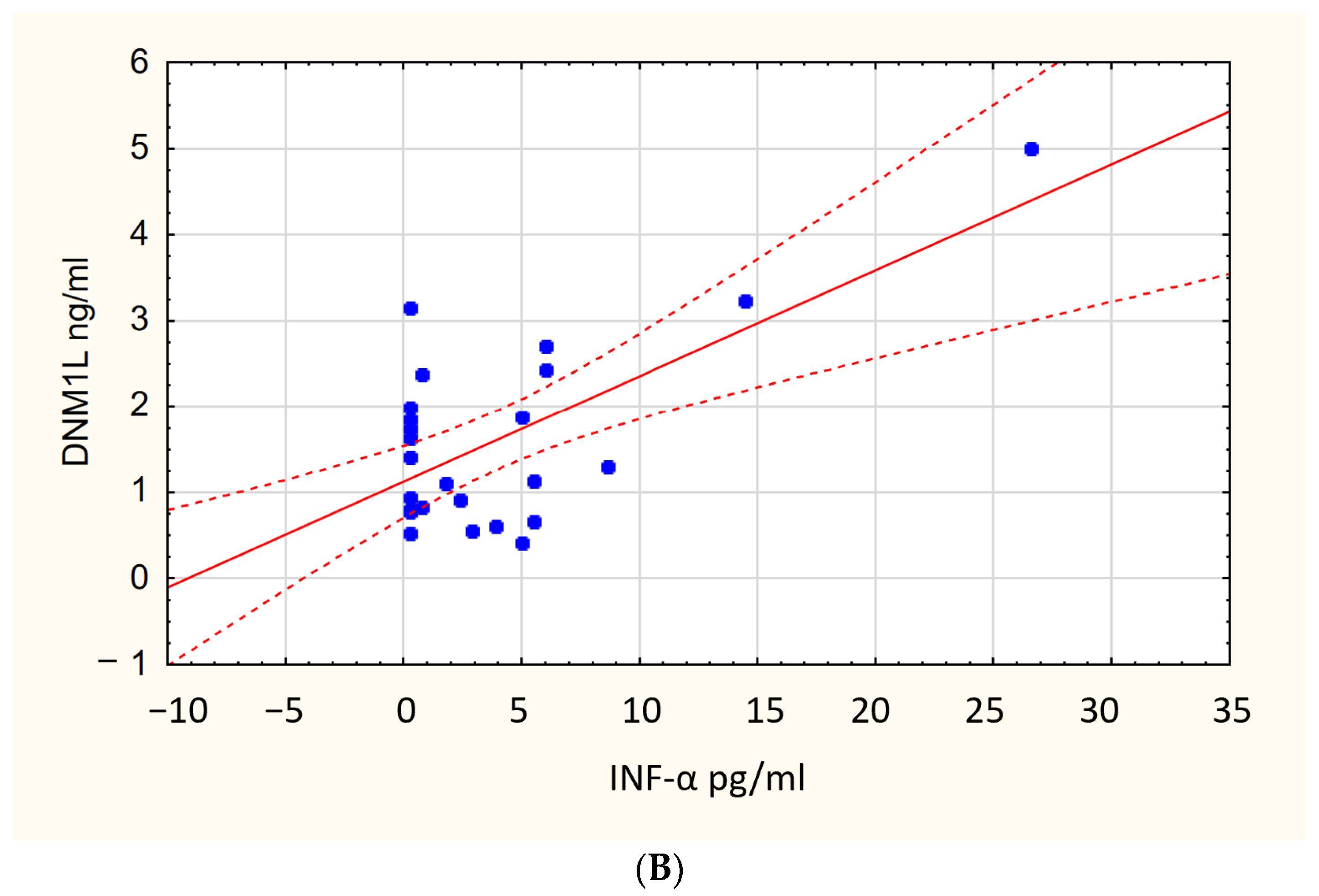
| Biomarker | sRAGE [ng/mL] | IL-6 [pg/mL] | IFN-α [pg/mL] | TNF-α [pg/mL] | PINK1 [ng/mL] | DNM1L [ng/mL] | MFN2 [ng/mL] |
|---|---|---|---|---|---|---|---|
| P(+) | 0.77 [0.61–1.07] | 1.0 [0.1–75.7] | 2.9 [0.2–5.5] | 32.4 [2.8–318] | 1.62 [1.02–2.29] | 1.6 [0.9–2.4] | 0.3 [0.2–0.5] |
| P(−) | 0.88 [0.7–1.03] | 0.5 [0.14–18.1] | 4.4 [2.3–7.6] | 2.4 [2.0–158] | 1.34 [0.94–1.74] | 0.9 [0.5–1.6] | 0.2 [0.1–0.3] |
| p-Value | 0.135 | 0.002 | 0.039 | <0.001 | 0.046 | 0.004 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siekacz, K.; Kumor-Kisielewska, A.; Miłkowska-Dymanowska, J.; Pietrusińska, M.; Bartczak, K.; Majewski, S.; Stańczyk, A.; Piotrowski, W.J.; Białas, A.J. Oxidative Biomarkers Associated with the Pulmonary Manifestation of Post-COVID-19 Complications. J. Clin. Med. 2023, 12, 4253. https://doi.org/10.3390/jcm12134253
Siekacz K, Kumor-Kisielewska A, Miłkowska-Dymanowska J, Pietrusińska M, Bartczak K, Majewski S, Stańczyk A, Piotrowski WJ, Białas AJ. Oxidative Biomarkers Associated with the Pulmonary Manifestation of Post-COVID-19 Complications. Journal of Clinical Medicine. 2023; 12(13):4253. https://doi.org/10.3390/jcm12134253
Chicago/Turabian StyleSiekacz, Kamil, Anna Kumor-Kisielewska, Joanna Miłkowska-Dymanowska, Małgorzata Pietrusińska, Krystian Bartczak, Sebastian Majewski, Adam Stańczyk, Wojciech J. Piotrowski, and Adam J. Białas. 2023. "Oxidative Biomarkers Associated with the Pulmonary Manifestation of Post-COVID-19 Complications" Journal of Clinical Medicine 12, no. 13: 4253. https://doi.org/10.3390/jcm12134253






