Evolving the Era of 5D Ultrasound? A Systematic Literature Review on the Applications for Artificial Intelligence Ultrasound Imaging in Obstetrics and Gynecology
Abstract
:1. Introduction
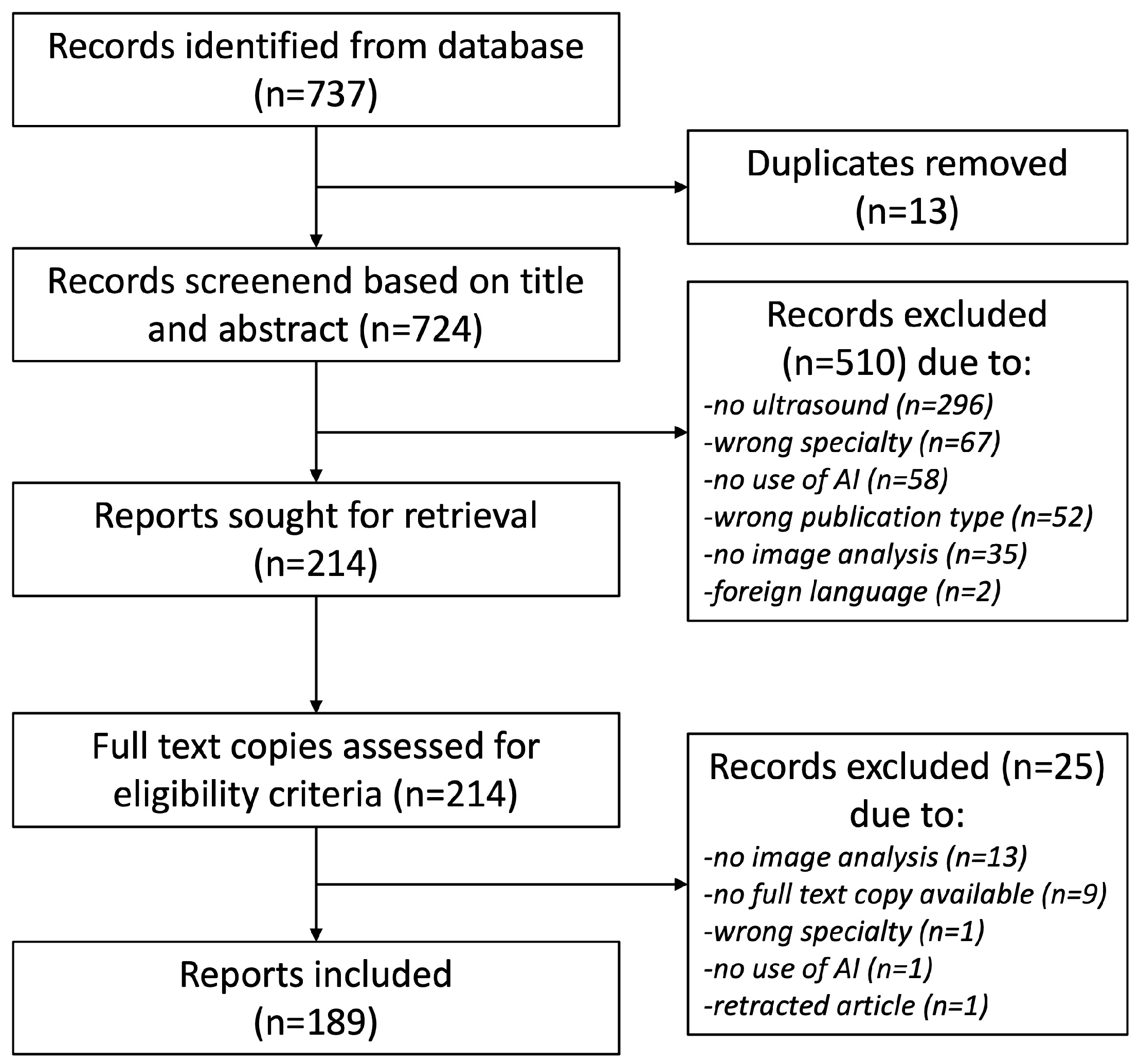
2. Materials and Methods
3. Results
3.1. Included Literature
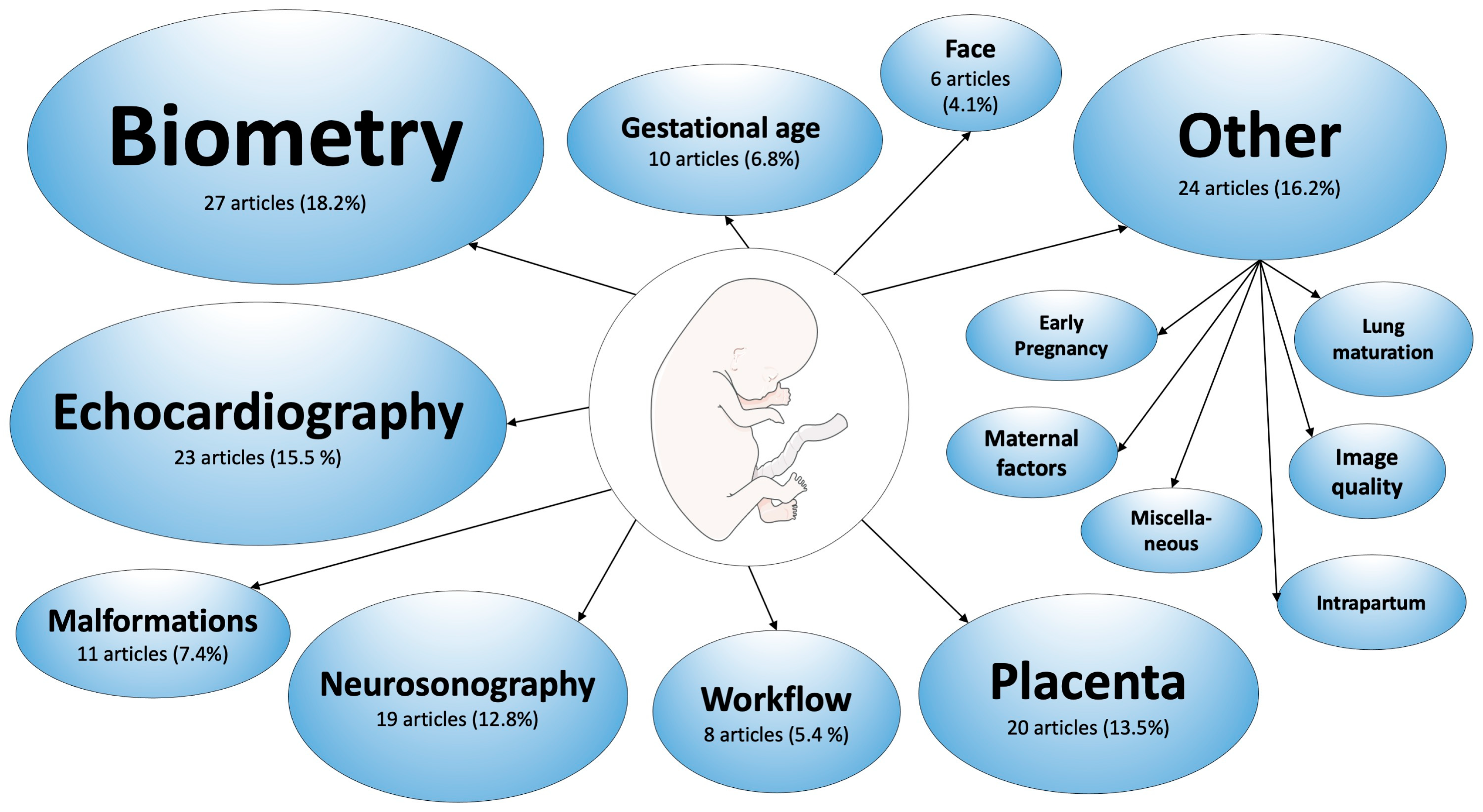
3.2. Applications in Obstetrics
3.2.1. Fetal Biometry
- Equipment-related factors: Especially in hospital settings, repeated examinations may be performed with different US machines, resulting in heterogenous data. Furthermore, image quality depends on resource availability and access to high-end US devices [17], or the use of point-of-care devices [18].
- Patient-related factors: Maternal obesity is known to have an impact on image quality and visualization of the fetus, and thus limits the accuracy of obstetric US examinations [19].
3.2.2. Fetal Echocardiography
3.2.3. Fetal Neurosonography
3.2.4. Fetal Face
3.2.5. Placenta and Umbilical Cord
3.2.6. Fetal Malformations
First Trimester Scan
Second Trimester Scan
3.2.7. Prediction of Gestational Age
3.2.8. Workflow Analysis of Obstetric Ultrasound Scans
3.2.9. Other Applications in Obstetrics
Fetal Lung Maturation
Maternal Factors
Early Pregnancy
Intrapartum Sonography
Image Quality
Miscellaneous
3.3. Applications in Gynecology
3.3.1. Adnexal Masses
3.3.2. Breast Masses
3.3.3. Endometrium
3.3.4. Pelvic Floor
3.3.5. Other Applications in Gynecology
Endometriosis
Uterine Fibroids
Premature Ovarian Failure
Follicle Tracking
Ectopic Pregnancy
4. Discussion
4.1. Benefits
4.2. Limitations
4.3. Strengths and Limitations of This Review
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2/3/4/5D | Two/three/four/five-dimensional |
| 4CV | Four-chamber view |
| AC | Abdominal circumference |
| AI | Artificial intelligence |
| CHD | Congenital heart disease |
| CRL | Crown-rump-length |
| CT | Computed tomography |
| FINE | Fetal intelligent navigation echocardiography |
| GA | Gestational age |
| HC | Head circumference |
| ISUOG | International Society of Ultrasound in Obstetrics & Gynecology |
| MRI | Magnetic resonance imaging |
| NT | Nuchal translucency |
| OB/GYN | Obstetrics and gynecology |
| ROI | Region of interest |
| US | Ultrasound |
References
- Shen, Y.T.; Chen, L.; Yue, W.W.; Xu, H.X. Artificial intelligence in ultrasound. Eur. J. Radiol. 2021, 139, 109717. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices (accessed on 21 August 2023).
- Drukker, L.; Noble, J.A.; Papageorghiou, A.T. Introduction to artificial intelligence in ultrasound imaging in obstetrics and gynecology. Ultrasound Obstet. Gynecol. 2020, 56, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Diniz, P.H.B.; Yin, Y.; Collins, S. Deep Learning Strategies for Ultrasound in Pregnancy. EMJ Reprod. Health 2020, 6, 73–80. [Google Scholar] [CrossRef]
- Reddy, C.D.; van den Eynde, J.; Kutty, S. Artificial intelligence in perinatal diagnosis and management of congenital heart disease. Semin. Perinatol. 2022, 46, 151588. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.N.; Wang, N.; He, M.; Zhang, L.H.; Cai, H.M.; Xian, J.B.; Lin, M.F.; Zheng, J.; Yang, Y.Z. Using deep-learning algorithms to classify fetal brain ultrasound images as normal or abnormal. Ultrasound Obstet. Gynecol. 2020, 56, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Komatsu, M.; Komatsu, R.; Matsuoka, R.; Yasutomi, S.; Dozen, A.; Shozu, K.; Arakaki, T.; Machino, H.; Asada, K.; et al. Medical Professional Enhancement Using Explainable Artificial Intelligence in Fetal Cardiac Ultrasound Screening. Biomedicines 2022, 10, 551. [Google Scholar] [CrossRef] [PubMed]
- Sarno, L.; Neola, D.; Carbone, L.; Saccone, G.; Carlea, A.; Miceli, M.; Iorio, G.G.; Mappa, I.; Rizzo, G.; Di Girolamo, R.; et al. Use of artificial intelligence in obstetrics: Not quite ready for prime time. Am. J. Obstet. Gynecol. MFM 2023, 5, 100792. [Google Scholar] [CrossRef]
- Leung, K.-Y. Applications of Advanced Ultrasound Technology in Obstetrics. Diagnostics 2021, 11, 1217. Available online: https://pubmed.ncbi.nlm.nih.gov/34359300/ (accessed on 21 August 2023). [CrossRef]
- Rizzo, G.; Aiello, E.; Elena Pietrolucci, M.; Arduini, D. The feasibility of using 5D CNS software in obtaining standard fetal head measurements from volumes acquired by three-dimensional ultrasonography: Comparison with two-dimensional ultrasound. J. Matern.-Fetal Neonatal Med. 2016, 29, 2217–2222. [Google Scholar] [CrossRef]
- Deshmukh, N.P.; Caban, J.J.; Taylor, R.H.; Hager, G.D.; Boctor, E.M. Five-dimensional ultrasound system for soft tissue visualization. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1927–1939. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Moher, D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 29, n71. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Healthcare Interventions: Explanation and Elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [PubMed]
- Hadlock, F.P.; Harrist, R.B.; Sharman, R.S.; Deter, R.L.; Park, S.K. Estimation of fetal weight with the use of head, body, and femur measurements—A prospective study. Am. J. Obstet. Gynecol. 1985, 151, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Fletcher, T.P.; Smith, S.; Koch, L.M.; Kainz, B.; Rueckert, D. SonoNet: Real-Time Detection and Localisation of Fetal Standard Scan Planes in Freehand Ultrasound. IEEE Trans. Med. Imaging 2017, 36, 2204–2215. [Google Scholar] [CrossRef] [PubMed]
- Sarris, I.; Ioannou, C.; Chamberlain, P.; Ohuma, E.; Roseman, F.; Hoch, L.; Altman, D.G.; Papageorghiou, A.T.; International Fetal and Newborn Growth Consortium for the 21st Century (INTERGROWTH-21st). Intra- and interobserver variability in fetal ultrasound measurements. Ultrasound Obstet. Gynecol. 2012, 39, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Sendra-Balcells, C.; Campello, V.M.; Torrents-Barrena, J.; Ahmed, Y.A.; Elattar, M.; Ohene-Botwe, B.; Nyangulu, P.; Stones, W.; Ammar, M.; Benamer, L.N.; et al. Generalisability of fetal ultrasound deep learning models to low-resource imaging settings in five African countries. Sci. Rep. 2023, 13, 2728. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Luo, J.; Cheng, J.; Lu, Y. Efficient fetal ultrasound image segmentation for automatic head circumference measurement using a lightweight deep convolutional neural network. Med. Phys. 2022, 49, 5081–5092. [Google Scholar] [CrossRef] [PubMed]
- Dashe, J.S.; McIntire, D.D.; Twickler, D.M. Effect of Maternal Obesity on the Ultrasound Detection of Anomalous Fetuses. Obstet. Gynecol. 2009, 113, 1001–1007. [Google Scholar] [CrossRef]
- Song, J.; Liu, J.; Liu, L.; Jiang, Y.; Zheng, H.; Ke, H.; Yang, L.; Zhang, Z. The birth weight of macrosomia influence the accuracy of ultrasound estimation of fetal weight at term. J. Clin. Ultrasound 2022, 50, 967–973. [Google Scholar] [CrossRef]
- Jang, J.; Park, Y.; Kim, B.; Lee, S.M.; Kwon, J.-Y.; Seo, J.K. Automatic Estimation of Fetal Abdominal Circumference from Ultrasound Images. IEEE J. Biomed. Health Inform. 2018, 22, 1512–1520. [Google Scholar] [CrossRef]
- Grandjean, G.A.; Hossu, G.; Bertholdt, C.; Noble, P.; Morel, O.; Grangé, G. Artificial intelligence assistance for fetal head biometry: Assessment of automated measurement software. Diagn. Interv. Imaging 2018, 99, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Pluym, I.D.; Afshar, Y.; Holliman, K.; Kwan, L.; Bolagani, A.; Mok, T.; Silver, B.; Ramirez, E.; Han, C.S.; Platt, L.D. Accuracy of automated three-dimensional ultrasound imaging technique for fetal head biometry. Ultrasound Obstet. Gynecol. 2021, 57, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dou, H.; Huang, R.; Xue, W.; Huang, Y.; Qian, J.; Zhang, Y.; Luo, H.; Guo, H.; Wang, T.; et al. Agent with Warm Start and Adaptive Dynamic Termination for Plane Localization in 3D Ultrasound. IEEE Trans. Med. Imaging 2021, 40, 1950–1961. [Google Scholar] [CrossRef] [PubMed]
- Płotka, S.; Klasa, A.; Lisowska, A.; Seliga-Siwecka, J.; Lipa, M.; Trzciński, T.; Sitek, A. Deep learning fetal ultrasound video model match human observers in biometric measurements. Phys. Med. Biol. 2022, 67, 045013. [Google Scholar] [CrossRef] [PubMed]
- Sridar, P.; Kumar, A.; Quinton, A.; Nanan, R.; Kim, J.; Krishnakumar, R. Decision Fusion-Based Fetal Ultrasound Image Plane Classification Using Convolutional Neural Networks. Ultrasound Med. Biol. 2019, 45, 1259–1273. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Bonet-Carne, E.; Eixarch, E.; Crispi, F.; Gratacos, E. Evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes. Sci. Rep. 2020, 10, 10200. [Google Scholar] [CrossRef]
- Chen, H.; Wu, L.; Dou, Q.; Qin, J.; Li, S.; Cheng, J.-Z.; Ni, D.; Heng, P.-A. Ultrasound Standard Plane Detection Using a Composite Neural Network Framework. IEEE Trans. Cybern. 2017, 47, 1576–1586. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, H.; Luo, H.; Li, K. Automatic quality assessment for 2D fetal sonographic standard plane based on multitask learning. Medicine 2021, 100, e24427. [Google Scholar] [CrossRef]
- Rahman, R.; Alam, M.d.G.R.; Reza, M.d.T.; Huq, A.; Jeon, G.; Uddin, M.d.Z.; Hassan, M.M. Demystifying evidential Dempster Shafer-based CNN architecture for fetal plane detection from 2D ultrasound images leveraging fuzzy-contrast enhancement and explainable AI. Ultrasonics 2023, 132, 107017. [Google Scholar] [CrossRef]
- Carneiro, G.; Georgescu, B.; Good, S.; Comaniciu, D. Automatic Fetal Measurements in Ultrasound Using Constrained Probabilistic Boosting Tree. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2007, Proceedings of the 10th International Conference, Brisbane, Australia, 29 October–2 November 2007; Ayache, N., Ourselin, S., Maeder, A., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4792, pp. 571–579. Available online: http://link.springer.com/10.1007/978-3-540-75759-7_69 (accessed on 9 June 2023).
- Carneiro, G.; Georgescu, B.; Good, S.; Comaniciu, D. Detection and Measurement of Fetal Anatomies from Ultrasound Images using a Constrained Probabilistic Boosting Tree. IEEE Trans. Med. Imaging 2008, 27, 1342–1355. [Google Scholar] [CrossRef]
- Luo, D.; Wen, H.; Peng, G.; Lin, Y.; Liang, M.; Liao, Y.; Qin, Y.; Zeng, Q.; Dang, J.; Li, S. A Prenatal Ultrasound Scanning Approach: One-Touch Technique in Second and Third Trimesters. Ultrasound Med. Biol. 2021, 47, 2258–2265. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Tsui, P.-H.; Wu, W.; Zhou, Z.; Wu, S. Fetal Ultrasound Image Segmentation for Automatic Head Circumference Biometry Using Deeply Supervised Attention-Gated V-Net. J. Digit. Imaging 2021, 34, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Heuvel, T.L.v.D.; Petros, H.; Santini, S.; de Korte, C.L.; van Ginneken, B. Automated Fetal Head Detection and Circumference Estimation from Free-Hand Ultrasound Sweeps Using Deep Learning in Resource-Limited Countries. Ultrasound Med. Biol. 2019, 45, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Lei, B.; Cheng, J.-Z.; Qin, J.; Wang, T.; Li, S.; Ni, D. Automatic Fetal Head Circumference Measurement in Ultrasound Using Random Forest and Fast Ellipse Fitting. IEEE J. Biomed. Health Inform. 2018, 22, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yang, Z.; Liao, S.; Guo, J.; Yin, S.; Liu, C.; Kang, Y. A new approach to automatic measure fetal head circumference in ultrasound images using convolutional neural networks. Comput. Biol. Med. 2022, 147, 105801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Petitjean, C.; Ainouz, S. Segmentation-Based vs. Regression-Based Biomarker Estimation: A Case Study of Fetus Head Circumference Assessment from Ultrasound Images. J. Imaging 2022, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Marini, T.J.; Saavedra, A.C.; Toscano, M.; Baran, T.M.; Drennan, K.; Dozier, A.; Zhao, Y.T.; Egoavil, M.; Tamayo, L.; et al. No sonographer, no radiologist: New system for automatic prenatal detection of fetal biometry, fetal presentation, and placental location. PLoS ONE 2022, 17, e0262107. [Google Scholar] [CrossRef]
- Li, P.; Zhao, H.; Liu, P.; Cao, F. Automated measurement network for accurate segmentation and parameter modification in fetal head ultrasound images. Med. Biol. Eng. Comput. 2020, 58, 2879–2892. [Google Scholar] [CrossRef]
- Kim, B.; Kim, K.C.; Park, Y.; Kwon, J.-Y.; Jang, J.; Seo, J.K. Machine-learning-based automatic identification of fetal abdominal circumference from ultrasound images. Physiol. Meas. 2018, 39, 105007. [Google Scholar] [CrossRef]
- Ni, D.; Yang, X.; Chen, X.; Chin, C.-T.; Chen, S.; Heng, P.A.; Li, S.; Qin, J.; Wang, T. Standard Plane Localization in Ultrasound by Radial Component Model and Selective Search. Ultrasound Med. Biol. 2014, 40, 2728–2742. [Google Scholar] [CrossRef]
- Chen, H.; Ni, D.; Qin, J.; Li, S.; Yang, X.; Wang, T.; Heng, P.A. Standard Plane Localization in Fetal Ultrasound via Domain Transferred Deep Neural Networks. IEEE J. Biomed. Health Inform. 2015, 19, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Liu, M.; Wang, F.; Qiu, D.; Li, R.; Dai, C. Automatic measurement of fetal femur length in ultrasound images: A comparison of random forest regression model and SegNet. Math. Biosci. Eng. 2021, 18, 7790–7805. [Google Scholar] [CrossRef] [PubMed]
- Van Den Heuvel, T.L.A.; De Bruijn, D.; De Korte, C.L.; Ginneken, B.V. Automated measurement of fetal head circumference using 2D ultrasound images. PLoS ONE 2018, 13, e0200412. [Google Scholar] [CrossRef] [PubMed]
- Donofrio, M.T.; Moon-Grady, A.J.; Hornberger, L.K.; Copel, J.A.; Sklansky, M.S.; Abuhamad, A.; Cuneao, B.F.; Huhta, J.C.; Jonas, R.A.; Krishnan, A.; et al. Diagnosis and Treatment of Fetal Cardiac Disease: A Scientific Statement from the American Heart Association. Circulation 2014, 129, 2183–2242. [Google Scholar] [CrossRef] [PubMed]
- Gembruch, U. Prenatal diagnosis of congenital heart disease. Prenat. Diagn. 1997, 17, 1283–1298. [Google Scholar] [CrossRef]
- Carvalho, J.; Allan, L.; Chaoui, R.; Copel, J.; DeVore, G.; Hecher, K.; Lee, W.; Munoz, H.; Paladini, D.; Tutschek, B.; et al. ISUOG Practice Guidelines (updated): Sonographic screening examination of the fetal heart. Ultrasound Obstet. Gynecol. 2013, 41, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Bensemlali, M.; Bajolle, F.; Laux, D.; Parisot, P.; Ladouceur, M.; Fermont, L.; Levy, M.; Le Bidois, J.; Raimondi, F.; Ville, Y.; et al. Neonatal management and outcomes of prenatally diagnosed CHDs. Cardiol. Young 2017, 27, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, R.; Curran, L.; Zhao, Y.; Levine, J.C.; Chinn, E.; Moon-Grady, A.J. An ensemble of neural networks provides expert-level prenatal detection of complex congenital heart disease. Nat. Med. 2021, 27, 882–891. [Google Scholar] [CrossRef]
- Yeo, L.; Romero, R. Fetal Intelligent Navigation Echocardiography (FINE): A novel method for rapid, simple, and automatic examination of the fetal heart: Fetal intelligent navigation echocardiography (FINE). Ultrasound Obstet. Gynecol. 2013, 42, 268–284. [Google Scholar] [CrossRef]
- Gembicki, M.; Hartge, D.R.; Dracopoulos, C.; Weichert, J. Semiautomatic Fetal Intelligent Navigation Echocardiography Has the Potential to Aid Cardiac Evaluations Even in Less Experienced Hands. J. Ultrasound Med. 2020, 39, 301–309. [Google Scholar] [CrossRef]
- Ma, M.; Li, Y.; Chen, R.; Huang, C.; Mao, Y.; Zhao, B. Diagnostic performance of fetal intelligent navigation echocardiography (FINE) in fetuses with double-outlet right ventricle (DORV). Int. J. Cardiovasc. Imaging 2020, 36, 2165–2172. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Guariento, A.; Cattapan, C.; Fedrigo, M.; Gervasi, M.T.; Angelini, A.; Riva, A.; Vida, V. Prenatal Diagnosis and Fetopsy Validation of Complete Atrioventricular Septal Defects Using the Fetal Intelligent Navigation Echocardiography Method. Diagnostics 2023, 13, 456. [Google Scholar] [CrossRef] [PubMed]
- Dozen, A.; Komatsu, M.; Sakai, A.; Komatsu, R.; Shozu, K.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules 2020, 10, 1526. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Jin, T.; Zhang, L.; Guo, C.; Gui, H.; Na, R.; Wang, X.; Bai, H. Adoption of Compound Echocardiography under Artificial Intelligence Algorithm in Fetal Congenial Heart Disease Screening during Gestation. Appl. Bionics Biomech. 2022, 2022, 6410103. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, M.; Shen, Z.; Wang, H.; Liu, X.; Wang, X.; Wang, S.; Li, T.; Yu, S.; Hou, M.; et al. DW-Net: A cascaded convolutional neural network for apical four-chamber view segmentation in fetal echocardiography. Comput. Med. Imaging Graph. 2020, 80, 101690. [Google Scholar] [CrossRef]
- Wu, H.; Wu, B.; Lai, F.; Liu, P.; Lyu, G.; He, S.; Dai, J. Application of Artificial Intelligence in Anatomical Structure Recognition of Standard Section of Fetal Heart. Comput. Math. Methods Med. 2023, 2023, 5650378. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, B.; Wu, H.; Xu, W.; Lyu, G.; Liu, P.; He, S. Classification of normal and abnormal fetal heart ultrasound images and identification of ventricular septal defects based on deep learning. J. Perinat. Med. 2023, 51, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Nurmaini, S.; Rachmatullah, M.N.; Sapitri, A.I.; Darmawahyuni, A.; Tutuko, B.; Firdaus, F.; Partan, R.U.; Bernolian, N. Deep Learning-Based Computer-Aided Fetal Echocardiography: Application to Heart Standard View Segmentation for Congenital Heart Defects Detection. Sensors 2021, 21, 8007. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, Y.; Zhu, H.; Lv, J.; Cheng, Q.; Zhang, H.; He, Y.; Wang, S. Fetal Congenital Heart Disease Echocardiogram Screening Based on DGACNN: Adversarial One-Class Classification Combined with Video Transfer Learning. IEEE Trans. Med. Imaging 2020, 39, 1206–1222. [Google Scholar] [CrossRef]
- Nurmaini, S.; Partan, R.U.; Bernolian, N.; Sapitri, A.I.; Tutuko, B.; Rachmatullah, M.N.; Darmawahyuni, A.; Firdaus, F.; Mose, J.C. Deep Learning for Improving the Effectiveness of Routine Prenatal Screening for Major Congenital Heart Diseases. J. Clin. Med. 2022, 11, 6454. [Google Scholar] [CrossRef]
- Wang, X.; Yang, T.; Zhang, Y.; Liu, X.; Zhang, Y.; Sun, L.; Gu, X.; Chen, Z.; Guo, Y.; Xue, C.; et al. Diagnosis of fetal total anomalous pulmonary venous connection based on the post-left atrium space ratio using artificial intelligence. Prenat. Diagn. 2022, 42, 1323–1331. [Google Scholar] [CrossRef]
- Yu, L.; Guo, Y.; Wang, Y.; Yu, J.; Chen, P. Determination of Fetal Left Ventricular Volume Based on Two-Dimensional Echocardiography. J. Healthc. Eng. 2017, 2017, 4797315. [Google Scholar] [CrossRef] [PubMed]
- Herling, L.; Johnson, J.; Ferm-Widlund, K.; Zamprakou, A.; Westgren, M.; Acharya, G. Automated quantitative evaluation of fetal atrioventricular annular plane systolic excursion. Ultrasound Obstet. Gynecol. 2021, 58, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Scharf, J.L.; Dracopoulos, C.; Gembicki, M.; Welp, A.; Weichert, J. How Automated Techniques Ease Functional Assessment of the Fetal Heart: Applicability of MPI+TM for Direct Quantification of the Modified Myocardial Performance Index. Diagnostics 2023, 13, 1705. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Pan, S.; Luo, G.; Pang, S.; Chen, T.; Singh, A.K.; Lv, Z. A Pseudo-Siamese Feature Fusion Generative Adversarial Network for Synthesizing High-Quality Fetal Four-Chamber Views. IEEE J. Biomed. Health Informatics 2023, 27, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Noble, J.A. Hierarchical Class Incremental Learning of Anatomical Structures in Fetal Echocardiography Videos. IEEE J. Biomed. Health Informatics 2020, 24, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Emery, S.P.; Kreutzer, J.; Sherman, F.R.; Fujimoto, K.L.; Jaramaz, B.; Nikou, C.; Tobita, K.; Keller, B.B. Computer-assisted navigation applied to fetal cardiac intervention. Int. J. Med. Robot. Comput. Assist. Surg. 2007, 3, 187–198. [Google Scholar] [CrossRef]
- Dong, J.; Liu, S.; Liao, Y.; Wen, H.; Lei, B.; Li, S.; Wang, T. A Generic Quality Control Framework for Fetal Ultrasound Cardiac Four-Chamber Planes. IEEE J. Biomed. Health Inform. 2020, 24, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Pietrolucci, M.E.; Maqina, P.; Mappa, I.; Marra, M.C.; D’ Antonio, F.; Rizzo, G. Evaluation of an artificial intelligent algorithm (HeartassistTM) to automatically assess the quality of second trimester cardiac views: A prospective study. J. Perinat. Med. 2023, 51, 920–924. [Google Scholar] [CrossRef]
- Vijayalakshmi, S.; Sriraam, N.; Suresh, S.; Muttan, S. Automated region mask for four-chamber fetal heart biometry. J. Clin. Monit. Comput. 2013, 27, 205–209. [Google Scholar] [CrossRef]
- Sriraam, N.; Punyaprabha, V.; Sushma Tv Suresh, S. Performance evaluation of computer-aided automated master frame selection techniques for fetal echocardiography. Med. Biol. Eng. Comput. 2023, 61, 1723–1744. [Google Scholar] [CrossRef] [PubMed]
- Graupner, O.; Enzensberger, C.; Wieg, L.; Willruth, A.; Steinhard, J.; Gembruch, U.; Doelle, A.; Bahlmann, F.; Kawecki, A.; Degenhardt, J.; et al. Evaluation of right ventricular function in fetal hypoplastic left heart syndrome by color tissue Doppler imaging: Right ventricular function in fetal HLHS. Ultrasound Obstet. Gynecol. 2016, 47, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, J.; Su, X.; Chen, X.; Zhou, Y.; Zhang, X.; Lu, H.; Niu, J.; Yu, L.; Sun, C.; et al. Reference ranges of fetal heart function using a Modified Myocardial Performance Index: A prospective multicentre, cross-sectional study. BMJ Open 2021, 11, e049640. [Google Scholar] [CrossRef] [PubMed]
- Lane, E.S.; Jevsikov, J.; Shun-Shin, M.J.; Dhutia, N.; Matoorian, N.; Cole, G.D.; Francis, D.P.; Zolgharni, M. Automated multi-beat tissue Doppler echocardiography analysis using deep neural networks. Med. Biol. Eng. Comput. 2023, 61, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Sonographic examination of the fetal central nervous system: Guidelines for performing the ‘basic examination’ and the ‘fetal neurosonogram’. Ultrasound Obstet. Gynecol. 2007, 29, 109–116. [CrossRef] [PubMed]
- Yeung, P.-H.; Aliasi, M.; Papageorghiou, A.T.; Haak, M.; Xie, W.; Namburete, A.I. Learning to map 2D ultrasound images into 3D space with minimal human annotation. Med. Image Anal. 2021, 70, 101998. [Google Scholar] [CrossRef] [PubMed]
- Hesse, L.S.; Aliasi, M.; Moser, F.; INTERGROWTH-21(st) Consortium; Haak, M.C.; Xie, W.; Jenkinson, M.; Namburete, A.I. Subcortical segmentation of the fetal brain in 3D ultrasound using deep learning. NeuroImage 2022, 254, 119117. [Google Scholar] [CrossRef] [PubMed]
- Namburete, A.I.; Xie, W.; Yaqub, M.; Zisserman, A.; Noble, J.A. Fully-automated alignment of 3D fetal brain ultrasound to a canonical reference space using multi-task learning. Med. Image Anal. 2018, 46, 1–14. [Google Scholar] [CrossRef]
- Papageorghiou, A.T.; Ohuma, E.O.; Altman, D.G.; Todros, T.; Ismail, L.C.; Lambert, A.; Jaffer, Y.A.; Bertino, E.; Gravett, M.G.; Purwar, M.; et al. International standards for fetal growth based on serial ultrasound measurements: The Fetal Growth Longitudinal Study of the INTERGROWTH-21st Project. Lancet 2014, 384, 869–879. [Google Scholar] [CrossRef]
- Di Vece, C.; Dromey, B.; Vasconcelos, F.; David, A.L.; Peebles, D.; Stoyanov, D. Deep learning-based plane pose regression in obstetric ultrasound. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 833–839. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, Z.; Zhuang, Y.; Yi, H.; Han, L.; Chen, K.; Lin, J. A guiding approach of Ultrasound scan for accurately obtaining standard diagnostic planes of fetal brain malformation. J. X-ray Sci. Technol. 2022, 30, 1243–1260. [Google Scholar] [CrossRef]
- Xu, Y.; Lee, L.H.; Drukker, L.; Yaqub, M.; Papageorghiou, A.T.; Noble, A.J. Simulating realistic fetal neurosonography images with appearance and growth change using cycle-consistent adversarial networks and an evaluation. J. Med. Imaging 2020, 7, 057001. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhou, Y.; Shi, S.; Zhang, Y.; Yin, S.; Liu, X.; Peng, Q.; Huang, S.; Jiang, Y.; Cui, C.; et al. How much can AI see in early pregnancy: A multi-center study of fetus head characterization in week 10–14 in ultrasound using deep learning. Comput. Methods Programs Biomed. 2022, 226, 107170. [Google Scholar] [CrossRef]
- Lin, M.; He, X.; Guo, H.; He, M.; Zhang, L.; Xian, J.; Lei, T.; Xu, Q.; Zheng, J.; Feng, J.; et al. Use of real-time artificial intelligence in detection of abnormal image patterns in standard sonographic reference planes in screening for fetal intracranial malformations. Ultrasound Obstet. Gynecol. 2022, 59, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Alansary, A.; Oktay, O.; Li, Y.; Le Folgoc, L.; Hou, B.; Vaillant, G.; Kamnitsas, K.; Vlontzos, A.; Glocker, B.; Kainz, B.; et al. Evaluating reinforcement learning agents for anatomical landmark detection. Med. Image Anal. 2019, 53, 156–164. [Google Scholar] [CrossRef]
- Gofer, S.; Haik, O.; Bardin, R.; Gilboa, Y.; Perlman, S. Machine Learning Algorithms for Classification of First-Trimester Fetal Brain Ultrasound Images. J. Ultrasound Med. 2022, 41, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xie, W.; Noble, J.A. VP-Nets: Efficient automatic localization of key brain structures in 3D fetal neurosonography. Med. Image Anal. 2018, 47, 127–139. [Google Scholar] [CrossRef]
- Lin, Z.; Li, S.; Ni, D.; Liao, Y.; Wen, H.; Du, J.; Chen, S.; Wang, T.; Lei, B. Multi-task learning for quality assessment of fetal head ultrasound images. Med. Image Anal. 2019, 58, 101548. [Google Scholar] [CrossRef]
- Yaqub, M.; Kelly, B.; Papageorghiou, A.T.; Noble, J.A. A Deep Learning Solution for Automatic Fetal Neurosonographic Diagnostic Plane Verification Using Clinical Standard Constraints. Ultrasound Med. Biol. 2017, 43, 2925–2933. [Google Scholar] [CrossRef]
- Skelton, E.; Matthew, J.; Li, Y.; Khanal, B.; Martinez, J.C.; Toussaint, N.; Gupta, C.; Knight, C.; Kainz, B.; Hajnal, J.; et al. Towards automated extraction of 2D standard fetal head planes from 3D ultrasound acquisitions: A clinical evaluation and quality assessment comparison. Radiography 2021, 27, 519–526. [Google Scholar] [CrossRef]
- Xie, B.; Lei, T.; Wang, N.; Cai, H.; Xian, J.; He, M.; Zhang, L.; Xie, H. Computer-aided diagnosis for fetal brain ultrasound images using deep convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Sahli, H.; Mouelhi, A.; Ben Slama, A.; Sayadi, M.; Rachdi, R. Supervised classification approach of biometric measures for automatic fetal defect screening in head ultrasound images. J. Med. Eng. Technol. 2019, 43, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Vellvé, K.; Eixarch, E.; Crispi, F.; Bonet-Carne, E.; Bennasar, M.; Gratacos, E. Analysis of maturation features in fetal brain ultrasound via artificial intelligence for the estimation of gestational age. Am. J. Obstet. Gynecol. MFM 2021, 3, 100462. [Google Scholar] [CrossRef] [PubMed]
- Sreelakshmy, R.; Titus, A.; Sasirekha, N.; Logashanmugam, E.; Begam, R.B.; Ramkumar, G.; Raju, R. An Automated Deep Learning Model for the Cerebellum Segmentation from Fetal Brain Images. BioMed Res. Int. 2022, 2022, 8342767. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Z.; Du, Y.; Diao, Y.; Liu, P.; Lv, G.; Zhang, H. Recognition of Fetal Facial Ultrasound Standard Plane Based on Texture Feature Fusion. Comput. Math. Methods Med. 2021, 2021, 656942. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Tan, E.-L.; Ni, D.; Qin, J.; Chen, S.; Li, S.; Lei, B.; Wang, T. A Deep Convolutional Neural Network-Based Framework for Automatic Fetal Facial Standard Plane Recognition. IEEE J. Biomed. Health Inform. 2018, 22, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Han, J.; Xie, B.; Xue, J.; Zhou, H.; Jiang, Y.; Hu, L.; Chen, C.; Zhang, K.; Zhu, F.; et al. The Two-Stage Ensemble Learning Model Based on Aggregated Facial Features in Screening for Fetal Genetic Diseases. Int. J. Environ. Res. Public Health 2023, 20, 2377. [Google Scholar] [CrossRef]
- Hata, T. Current status of fetal neurodevelopmental assessment: Four-dimensional ultrasound study: Fetal neurodevelopmental assessment. J. Obstet. Gynaecol. Res. 2016, 42, 1211–1221. [Google Scholar] [CrossRef]
- AboEllail, M.A.M.; Hata, T. Fetal face as important indicator of fetal brain function. J. Perinat. Med. 2017, 45, 729–736. [Google Scholar] [CrossRef]
- Miyagi, Y.; Hata, T.; Bouno, S.; Koyanagi, A.; Miyake, T. Artificial intelligence to understand fluctuation of fetal brain activity by recognizing facial expressions. Int. J. Gynecol. Obstet. 2023, 161, 877–885. [Google Scholar] [CrossRef]
- Miyagi, Y.; Hata, T.; Bouno, S.; Koyanagi, A.; Miyake, T. Recognition of facial expression of fetuses by artificial intelligence (AI). J. Perinat. Med. 2021, 49, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, Y.; Hata, T.; Miyake, T. Fetal brain activity and the free energy principle. J. Perinat. Med. 2023, 51, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Groom, K.M.; Oyston, C.; Chamley, L.W.; Clark, A.R.; James, J.L. The placenta in fetal growth restriction: What is going wrong? Placenta 2020, 96, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Maltepe, E.; Fisher, S.J. Placenta: The Forgotten Organ. Annu. Rev. Cell Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Böckenhoff, P.; Geipel, A.; Gembruch, U.; Engels, A.C. Early sonographic evaluation of the placenta in cases with IUGR: A pilot study. Arch. Gynecol. Obstet. 2020, 302, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Singla, R.; Yan, R.; Mayer, C.; Rohling, R.N. Automated Placenta Segmentation with a Convolutional Neural Network Weighted by Acoustic Shadow Detection. In Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: New York, NY, USA, 2019; pp. 6718–6723. Available online: https://ieeexplore.ieee.org/document/8857448/ (accessed on 9 July 2023).
- Andreasen, L.A.; Feragen, A.; Christensen, A.N.; Thybo, J.K.; Svendsen, M.B.S.; Zepf, K.; Lekadir, K.; Tolsgaard, M.G. Multi-centre deep learning for placenta segmentation in obstetric ultrasound with multi-observer and cross-country generalization. Sci. Rep. 2023, 13, 2221. [Google Scholar] [CrossRef] [PubMed]
- Schilpzand, M.; Neff, C.; van Dillen, J.; van Ginneken, B.; Heskes, T.; de Korte, C.; Heuvel, T.v.D. Automatic Placenta Localization from Ultrasound Imaging in a Resource-Limited Setting Using a Predefined Ultrasound Acquisition Protocol and Deep Learning. Ultrasound Med. Biol. 2022, 48, 663–674. [Google Scholar] [CrossRef]
- Plasencia, W.; Akolekar, R.; Dagklis, T.; Veduta, A.; Nicolaides, K.H. Placental Volume at 11–13 Weeks’ Gestation in the Prediction of Birth Weight Percentile. Fetal Diagn. Ther. 2011, 30, 23–28. [Google Scholar] [CrossRef]
- Schwartz, N.; Oguz, I.; Wang, J.; Pouch, A.; Yushkevich, N.; Parameshwaran, S.; Gee, J.; Yushkevich, P.; Oguz, B. Fully Automated Placental Volume Quantification From 3D Ultrasound for Prediction of Small-for-Gestational-Age Infants. J. Ultrasound Med. 2022, 41, 1509–1524. [Google Scholar] [CrossRef]
- Looney, P.; Yin, Y.; Collins, S.L.; Nicolaides, K.H.; Plasencia, W.; Molloholli, M.; Natsis, S.; Stevenson, G.N. Fully Automated 3-D Ultrasound Segmentation of the Placenta, Amniotic Fluid, and Fetus for Early Pregnancy Assessment. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 2038–2047. [Google Scholar] [CrossRef]
- Qi, H.; Collins, S.; Noble, J.A. Automatic Lacunae Localization in Placental Ultrasound Images via Layer Aggregation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Proceedings of the 1st International Conference, Granada, Spain, 16–20 September 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11071, pp. 921–929. Available online: http://link.springer.com/10.1007/978-3-030-00934-2_102 (accessed on 9 June 2023).
- Qi, H.; Collins, S.; Noble, A. Weakly Supervised Learning of Placental Ultrasound Images with Residual Networks. In Medical Image Understanding and Analysis; Valdés Hernández, M., González-Castro, V., Eds.; Communications in Computer and Information Science; Springer International Publishing: Cham, Switzerland, 2017; Volume 723, pp. 98–108. Available online: http://link.springer.com/10.1007/978-3-319-60964-5_9 (accessed on 9 June 2023).
- Lei, B.; Yao, Y.; Chen, S.; Li, S.; Li, W.; Ni, D.; Wang, T. Discriminative Learning for Automatic Staging of Placental Maturity via Multi-layer Fisher Vector. Sci. Rep. 2015, 5, 12818. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yao, Y.; Ni, D.; Chen, S.; Li, S.; Lei, B.; Wang, T. Automatic staging of placental maturity based on dense descriptor. Bio-Med. Mater. Eng. 2014, 24, 2821–2829. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Balyan, K.; Lamba, B.; Puri, M.; Sengupta, D.; Kumar, M. Ultrasound placental image texture analysis using artificial intelligence to predict hypertension in pregnancy. J. Matern. Neonatal Med. 2022, 35, 5587–5594. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jiao, J.; Ren, Y.; Guo, Y.; Wang, Y. Multimodal fusion model for classifying placenta ultrasound imaging in pregnancies with hypertension disorders. Pregnancy Hypertens. 2023, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jiao, J.; Ren, Y.; Guo, Y.; Wang, Y. Model application to quantitatively evaluate placental features from ultrasound images with gestational diabetes. J. Clin. Ultrasound 2022, 50, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, Z.; Jia, X. Deep Learning Algorithm-Based Ultrasound Image Information in Diagnosis and Treatment of Pernicious Placenta Previa. Comput. Math. Methods Med. 2022, 2022, 3452176. [Google Scholar] [CrossRef] [PubMed]
- Asadpour, V.; Puttock, E.J.; Getahun, D.; Fassett, M.J.; Xie, F. Automated placental abruption identification using semantic segmentation, quantitative features, SVM, ensemble and multi-path CNN. Heliyon 2023, 9, e13577. [Google Scholar] [CrossRef]
- Pradipta, G.A.; Wardoyo, R.; Musdholifah, A.; Sanjaya, I.N.H. Machine learning model for umbilical cord classification using combination coiling index and texture feature based on 2-D Doppler ultrasound images. Health Inform. J. 2022, 28, 146045822210842. [Google Scholar] [CrossRef]
- Beksaç, M.; Egemen, A.; Izzetoǵlu, K.; Ergün, G.; Erkmen, A.M. An automated intelligent diagnostic system for the interpretation of umbilical artery Doppler velocimetry. Eur. J. Radiol. 1996, 23, 162–167. [Google Scholar] [CrossRef]
- Beksaç, M.; Başaran, F.; Eskiizmirliler, S.; Erkmen, A.M.; Yörükan, S. A computerized diagnostic system for the interpretation of umbilical artery blood flow velocity waveforms. Eur. J. Obstet. Gynecol. Reprod. Biol. 1996, 64, 37–42. [Google Scholar] [CrossRef]
- Baykal, N.; A Reggia, J.; Yalabik, N.; Erkmen, A.; Beksac, M.S. Interpretation of Doppler blood flow velocity waveforms using neural networks. Proc. Annu. Symp. Comput. Appl. Med. Care 1994, 865–869. [Google Scholar]
- Torrents-Barrena, J.; Monill, N.; Piella, G.; Gratacós, E.; Eixarch, E.; Ceresa, M.; Ballester, M.A.G. Assessment of Radiomics and Deep Learning for the Segmentation of Fetal and Maternal Anatomy in Magnetic Resonance Imaging and Ultrasound. Acad. Radiol. 2021, 28, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Torrents-Barrena, J.; López-Velazco, R.; Piella, G.; Masoller, N.; Valenzuela-Alcaraz, B.; Gratacós, E.; Eixarch, E.; Ceresa, M.; Ballester, M.G. TTTS-GPS: Patient-specific preoperative planning and simulation platform for twin-to-twin transfusion syndrome fetal surgery. Comput. Methods Programs Biomed. 2019, 179, 104993. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, K.H. The 11–13+6 Weeks Scan; Fetal Medicine Foundation: London, UK, 2004. [Google Scholar]
- Salomon, L.J.; Alfirevic, Z.; Bilardo, C.M.; Chalouhi, G.E.; Ghi, T.; Kagan, K.O.; Lau, T.K.; Papageorghiou, A.T.; Raine-Fenning, N.J.; Stirnemann, J.; et al. ISUOG Practice Guidelines: Performance of first-trimester fetal ultrasound scan. Ultrasound Obstet. Gynecol. 2013, 41, 102–113. [Google Scholar] [PubMed]
- Snijders, R.; Noble, P.; Sebire, N.; Souka, A.; Nicolaides, K. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10–14 weeks of gestation. Lancet 1998, 352, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.C.; Willner, I.; Miguel, O.X.; Murphy, M.S.Q.; El-Chaâr, D.; Moretti, F.; Harvey, A.L.J.D.; White, R.R.; Muldoon, K.A.; Carrington, A.M.; et al. Using deep-learning in fetal ultrasound analysis for diagnosis of cystic hygroma in the first trimester. PLoS ONE 2022, 17, e0269323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dong, D.; Sun, Y.; Hu, C.; Sun, C.; Wu, Q.; Tian, J. Development and Validation of a Deep Learning Model to Screen for Trisomy 21 During the First Trimester from Nuchal Ultrasonographic Images. JAMA Netw. Open 2022, 5, e2217854. [Google Scholar] [CrossRef]
- Sciortino, G.; Tegolo, D.; Valenti, C. Automatic detection and measurement of nuchal translucency. Comput. Biol. Med. 2017, 82, 12–20. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, Y.; Chen, P.; Yu, J. A hierarchical model for automatic nuchal translucency detection from ultrasound images. Comput. Biol. Med. 2012, 42, 706–713. [Google Scholar] [CrossRef]
- Tsai, P.-Y.; Hung, C.-H.; Chen, C.-Y.; Sun, Y.-N. Automatic Fetal Middle Sagittal Plane Detection in Ultrasound Using Generative Adversarial Network. Diagnostics 2020, 11, 21. [Google Scholar] [CrossRef]
- Ryou, H.; Yaqub, M.; Cavallaro, A.; Papageorghiou, A.T.; Noble, J.A. Automated 3D ultrasound image analysis for first trimester assessment of fetal health. Phys. Med. Biol. 2019, 64, 185010. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, L.; Li, S.; Wen, H.; Luo, D.; Bian, C.; Qin, J.; Ni, D.; Heng, P.-A. Towards Automated Semantic Segmentation in Prenatal Volumetric Ultrasound. IEEE Trans. Med Imaging 2019, 38, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Salomon, L.J.; Alfirevic, Z.; Berghella, V.; Bilardo, C.; Hernandez-Andrade, E.; Johnsen, S.L.; Kalache, K.; Leung, K.; Malinger, G.; Munoz, H.; et al. Practice guidelines for performance of the routine mid-trimester fetal ultrasound scan. Ultrasound Obstet. Gynecol. 2010, 37, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Matthew, J.; Skelton, E.; Day, T.G.; Zimmer, V.A.; Gomez, A.; Wheeler, G.; Toussaint, N.; Liu, T.; Budd, S.; Lloyd, K.; et al. Exploring a new paradigm for the fetal anomaly ultrasound scan: Artificial intelligence in real time. Prenat. Diagn. 2022, 42, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Cengizler, Ç.; Ün, M.K.; Büyükkurt, S. A Nature-Inspired Search Space Reduction Technique for Spine Identification on Ultrasound Samples of Spina Bifida Cases. Sci. Rep. 2020, 10, 9280. [Google Scholar] [CrossRef] [PubMed]
- Meenakshi, S.; Suganthi, M.; Sureshkumar, P. Segmentation and Boundary Detection of Fetal Kidney Images in Second and Third Trimesters Using Kernel-Based Fuzzy Clustering. J. Med Syst. 2019, 43, 203. [Google Scholar] [CrossRef] [PubMed]
- Shozu, K.; Komatsu, M.; Sakai, A.; Komatsu, R.; Dozen, A.; Machino, H.; Yasutomi, S.; Arakaki, T.; Asada, K.; Kaneko, S.; et al. Model-Agnostic Method for Thoracic Wall Segmentation in Fetal Ultrasound Videos. Biomolecules 2020, 10, 1691. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Willis, A.; Chen, C.; Sieniek, M.; Watters, A.; Stetson, B.; Uddin, A.; Wong, J.; Pilgrim, R.; Chou, K.; et al. Development of a Machine Learning Model for Sonographic Assessment of Gestational Age. JAMA Netw. Open 2023, 6, e2248685. [Google Scholar] [CrossRef]
- Gomes, R.G.; Vwalika, B.; Lee, C.; Willis, A.; Sieniek, M.; Price, J.T.; Chen, C.; Kasaro, M.P.; Taylor, J.A.; Stringer, E.M.; et al. A mobile-optimized artificial intelligence system for gestational age and fetal malpresentation assessment. Commun. Med. 2022, 2, 128. [Google Scholar] [CrossRef]
- Beksaç, M.S.; Odçikin, Z.; Egemen, A.; Karakaş, U. An intelligent diagnostic system for the assessment of gestational age based on ultrasonic fetal head measurements. Technol. Health Care 1996, 4, 223–231. [Google Scholar] [CrossRef]
- Namburete, A.I.; Stebbing, R.V.; Kemp, B.; Yaqub, M.; Papageorghiou, A.T.; Noble, J.A. Learning-based prediction of gestational age from ultrasound images of the fetal brain. Med. Image Anal. 2015, 21, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Alzubaidi, M.; Agus, M.; Shah, U.; Makhlouf, M.; Alyafei, K.; Househ, M. Ensemble Transfer Learning for Fetal Head Analysis: From Segmentation to Gestational Age and Weight Prediction. Diagnostics 2022, 12, 2229. [Google Scholar] [CrossRef] [PubMed]
- Dan, T.; Chen, X.; He, M.; Guo, H.; He, X.; Chen, J.; Xian, J.; Hu, Y.; Zhang, B.; Wang, N.; et al. DeepGA for automatically estimating fetal gestational age through ultrasound imaging. Artif. Intell. Med. 2023, 135, 102453. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.H.; Bradburn, E.; Craik, R.; Yaqub, M.; Norris, S.A.; Ismail, L.C.; Ohuma, E.O.; Barros, F.C.; Lambert, A.; Carvalho, M.; et al. Machine learning for accurate estimation of fetal gestational age based on ultrasound images. NPJ Digit. Med. 2023, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Maraci, M.A.; Yaqub, M.; Craik, R.; Beriwal, S.; Self, A.; Von Dadelszen, P.; Papageorghiou, A.; Noble, J.A. Toward point-of-care ultrasound estimation of fetal gestational age from the trans-cerebellar diameter using CNN-based ultrasound image analysis. J. Med. Imaging 2020, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Pokaprakarn, T.; Prieto, J.C.; Price, J.T.; Kasaro, M.P.; Sindano, N.; Shah, H.R.; Peterson, M.; Akapelwa, M.M.; Kapilya, F.M.; Sebastião, Y.V.; et al. AI Estimation of Gestational Age from Blind Ultrasound Sweeps in Low-Resource Settings. NEJM Évid. 2022, 1, EVIDoa2100058. [Google Scholar] [CrossRef] [PubMed]
- Prieto, J.C.; Shah, H.; Rosenbaum, A.; Jiang, X.; Musonda, P.; Price, J.; Stringer, E.M.; Vwalika, B.; Stamilio, D.M.; Stringer, J.S.A. An automated framework for image classification and segmentation of fetal ultrasound images for gestational age estimation. In Medical Imaging 2021: Image Processing; Landman, B.A., Išgum, I., Eds.; SPIE: Bellingham, WA, USA, 2021; p. 55. Available online: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11596/2582243/An-automated-framework-for-image-classification-and-segmentation-of-fetal/10.1117/12.2582243.full (accessed on 16 October 2023).
- Drukker, L.; Sharma, H.; Droste, R.; Alsharid, M.; Chatelain, P.; Noble, J.A.; Papageorghiou, A.T. Transforming obstetric ultrasound into data science using eye tracking, voice recording, transducer motion and ultrasound video. Sci. Rep. 2021, 11, 14109. [Google Scholar] [CrossRef]
- Sharma, H.; Drukker, L.; Chatelain, P.; Droste, R.; Papageorghiou, A.T.; Noble, J.A. Knowledge representation and learning of operator clinical workflow from full-length routine fetal ultrasound scan videos. Med. Image Anal. 2021, 69, 101973. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Q.; Drukker, L.; Papageorghiou, A.; Hu, Y.; Noble, J.A. Task model-specific operator skill assessment in routine fetal ultrasound scanning. Int. J. CARS 2022, 17, 1437–1444. [Google Scholar] [CrossRef]
- Zhao, C.; Droste, R.; Drukker, L.; Papageorghiou, A.T.; Noble, J.A. Visual-Assisted Probe Movement Guidance for Obstetric Ultrasound Scanning Using Landmark Retrieval. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021, Proceedings of the 24th International Conference, Strasbourg, France, 27 September–1 October 2021; De Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12908, pp. 670–679. Available online: https://link.springer.com/10.1007/978-3-030-87237-3_64 (accessed on 9 June 2023).
- Drukker, L.; Sharma, H.; Karim, J.N.; Droste, R.; Noble, J.A.; Papageorghiou, A.T. Clinical workflow of sonographers performing fetal anomaly ultrasound scans: Deep-learning-based analysis. Ultrasound Obstet. Gynecol. 2022, 60, 759–765. [Google Scholar] [CrossRef]
- Alsharid, M.; Cai, Y.; Sharma, H.; Drukker, L.; Papageorghiou, A.T.; Noble, J.A. Gaze-assisted automatic captioning of fetal ultrasound videos using three-way multi-modal deep neural networks. Med. Image Anal. 2022, 82, 102630. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Drukker, L.; Papageorghiou, A.T.; Noble, J.A. Multi-Modal Learning from Video, Eye Tracking, and Pupillometry for Operator Skill Characterization in Clinical Fetal Ultrasound. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1646–1649. Available online: https://ieeexplore.ieee.org/document/9433863/ (accessed on 9 June 2023).
- Xia, T.-H.; Tan, M.; Li, J.-H.; Wang, J.-J.; Wu, Q.-Q.; Kong, D.-X. Establish a normal fetal lung gestational age grading model and explore the potential value of deep learning algorithms in fetal lung maturity evaluation. Chin. Med. J. 2021, 134, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Fang, Z.; Jiao, J.; Xi, G.; Zhu, C.; Ren, Y.; Guo, Y.; Wang, Y. Application of ultrasound-based radiomics technology in fetal-lung-texture analysis in pregnancies complicated by gestational diabetes and/or pre-eclampsia. Ultrasound Obstet. Gynecol. 2021, 57, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, Y.; Deng, Y.; Wang, Y.; He, P.; Lv, X.; Yu, J. A preliminary study to quantitatively evaluate the development of maturation degree for fetal lung based on transfer learning deep model from ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Jiao, J.; Ji, C.; Li, M.; Guo, Y.; Wang, Y.; Zhou, J.; Ren, Y. Ultrasound-based radiomics technology in fetal lung texture analysis prediction of neonatal respiratory morbidity. Sci. Rep. 2022, 12, 12747. [Google Scholar] [CrossRef] [PubMed]
- Bonet-Carne, E.; Palacio, M.; Cobo, T.; Perez-Moreno, A.; Lopez, M.; Piraquive, J.P.; Ramirez, J.C.; Botet, F.; Marques, F.; Gratacos, E. Quantitative ultrasound texture analysis of fetal lungs to predict neonatal respiratory morbidity. Ultrasound Obstet. Gynecol. 2015, 45, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Fraser, R.F.; Chen, C.W. A Novel Algorithm for Computer-Assisted Measurement of Cervical Length from Transvaginal Ultrasound Images. IEEE Trans. Inform. Technol. Biomed. 2004, 8, 333–342. [Google Scholar] [CrossRef]
- He, H.; Liu, R.; Zhou, X.; Zhang, Y.; Yu, B.; Xu, Z.; Huang, H. B-Ultrasound Image Analysis of Intrauterine Pregnancy Residues after Mid-Term Pregnancy Based on Smart Medical Big Data. J. Healthc. Eng. 2022, 2022, 9937051. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, D.; Liu, G. Value of Ultrasonic Image Features in Diagnosis of Perinatal Outcomes of Severe Preeclampsia on account of Deep Learning Algorithm. Comput. Math. Methods Med. 2022, 2022, 4010339. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Luo, N. Doppler Ultrasound Imaging Combined with Fetal Heart Detection in Predicting Fetal Distress in Pregnancy-Induced Hypertension under the Guidance of Artificial Intelligence Algorithm. J. Healthc. Eng. 2021, 2021, 4405189. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.; Yin, C.; Chen, L.; Yang, Z.; Jia, S.; Sun, X.; Bai, Y.; Han, F.; Yuan, Z. Automated prediction of early spontaneous miscarriage based on the analyzing ultrasonographic gestational sac imaging by the convolutional neural network: A case-control and cohort study. BMC Pregnancy Childbirth 2022, 22, 621. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.D.; Jayaprakasan, K.; Jones, N.W.; Clewes, J.; Winter, B.; Cash, N.; Campbell, B.; Raine-Fenning, N.J. A Novel Technique for the Semi-Automated Measurement of Embryo Volume: An Intraobserver Reliability Study. Ultrasound Med. Biol. 2010, 36, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Ghi, T.; Conversano, F.; Zegarra, R.R.; Pisani, P.; Dall’Asta, A.; Lanzone, A.; Lau, W.; Vimercati, A.; Iliescu, D.G.; Mappa, I.; et al. Novel artificial intelligence approach for automatic differentiation of fetal occiput anterior and non-occiput anterior positions during labor. Ultrasound Obstet. Gynecol. 2021, 59, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhi, D.; Zhou, M.; Lai, F.; Chen, G.; Ou, Z.; Zeng, R.; Long, S.; Qiu, R.; Zhou, M.; et al. Multitask Deep Neural Network for the Fully Automatic Measurement of the Angle of Progression. Comput. Math. Methods Med. 2022, 2022, 5192338. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Sun, Z.; Yu, S.; Lu, Y.; Long, S.; Wang, H.; Qiu, R.; Ou, Z.; Zhou, M.; Zhi, D.; et al. A framework for computing angle of progression from transperineal ultrasound images for evaluating fetal head descent using a novel double branch network. Front. Physiol. 2022, 13, 940150. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Cheng, J.-Z.; Li, S.; Lei, B.; Wang, T.; Ni, D. FUIQA: Fetal Ultrasound Image Quality Assessment with Deep Convolutional Networks. IEEE Trans. Cybern. 2017, 47, 1336–1349. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Housden, J.; Matthew, J.; Rueckert, D.; Schnabel, J.A.; Kainz, B.; Sinclair, M.; Zimmer, V.; Hou, B.; Rajchl, M.; et al. Weakly Supervised Estimation of Shadow Confidence Maps in Fetal Ultrasound Imaging. IEEE Trans. Med. Imaging 2019, 38, 2755–2767. [Google Scholar] [CrossRef]
- Gupta, L.; Sisodia, R.S.; Pallavi, V.; Firtion, C.; Ramachandran, G. Segmentation of 2D fetal ultrasound images by exploiting context information using conditional random fields. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; IEEE: New York, NY, USA, 2011; pp. 7219–7222. [Google Scholar]
- Yin, P.; Wang, H. Evaluation of Nursing Effect of Pelvic Floor Rehabilitation Training on Pelvic Organ Prolapse in Postpartum Pregnant Women under Ultrasound Imaging with Artificial Intelligence Algorithm. Comput. Math. Methods Med. 2022, 2022, 1786994. [Google Scholar] [CrossRef]
- Cho, H.C.; Sun, S.; Hyun, C.M.; Kwon, J.-Y.; Kim, B.; Park, Y.; Seo, J.K. Automated ultrasound assessment of amniotic fluid index using deep learning. Med. Image Anal. 2021, 69, 101951. [Google Scholar] [CrossRef]
- Compagnone, C.; Borrini, G.; Calabrese, A.; Taddei, M.; Bellini, V.; Bignami, E. Artificial intelligence enhanced ultrasound (AI-US) in a severe obese parturient: A case report. Ultrasound J. 2022, 14, 34. [Google Scholar] [CrossRef]
- Rueda, S.; Knight, C.L.; Papageorghiou, A.T.; Noble, J.A. Feature-based fuzzy connectedness segmentation of ultrasound images with an object completion step. Med. Image Anal. 2015, 26, 30–46. [Google Scholar] [CrossRef]
- Kaplan, E.; Ekinci, T.; Kaplan, S.; Barua, P.D.; Dogan, S.; Tuncer, T.; Tan, R.S.; Arunkumar, N.; Acharya, U.R. PFP-LHCINCA: Pyramidal Fixed-Size Patch-Based Feature Extraction and Chi-Square Iterative Neighborhood Component Analysis for Automated Fetal Sex Classification on Ultrasound Images. Contrast Media Mol. Imaging 2022, 2022, 6034971. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, D.; Testa, A.C.; Bourne, T.; Ameye, L.; Jurkovic, D.; Van Holsbeke, C.; Paladini, D.; Van Calster, B.; Vergote, I.; Van Huffel, S.; et al. Simple ultrasound-based rules for the diagnosis of ovarian cancer. Ultrasound Obstet. Gynecol. 2008, 31, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Amor, F.; Vaccaro, H.; Alcázar, J.L.; León, M.; Craig, J.M.; Martinez, J. Gynecologic Imaging Reporting and Data System: A New Proposal for Classifying Adnexal Masses on the Basis of Sonographic Findings. J. Ultrasound Med. 2009, 28, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-T.; Su, Y.-J.; Hung, C.-H.; Chen, M.-J.; Lu, C.-H.; Kuo, C.-E. Automatic ovarian tumors recognition system based on ensemble convolutional neural network with ultrasound imaging. BMC Med. Inform. Decis. Mak. 2022, 22, 298. [Google Scholar] [CrossRef] [PubMed]
- Al-Karawi, D.; Al-Assam, H.; Du, H.; Sayasneh, A.; Landolfo, C.; Timmerman, D.; Bourne, T.; Jassim, S. An Evaluation of the Effectiveness of Image-based Texture Features Extracted from Static B-mode Ultrasound Images in Distinguishing between Benign and Malignant Ovarian Masses. Ultrason. Imaging 2021, 43, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Aramendía-Vidaurreta, V.; Cabeza, R.; Villanueva, A.; Navallas, J.; Alcázar, J.L. Ultrasound Image Discrimination between Benign and Malignant Adnexal Masses Based on a Neural Network Approach. Ultrasound Med. Biol. 2016, 42, 742–752. [Google Scholar] [CrossRef]
- Christiansen, F.; Epstein, E.L.; Smedberg, E.; Åkerlund, M.; Smith, K. Ultrasound image analysis using deep neural networks for discriminating between benign and malignant ovarian tumors: Comparison with expert subjective assessment. Ultrasound Obstet. Gynecol. 2021, 57, 155–163. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, S.; Xu, X.; Li, H.; Yao, S.; Song, K.; Li, X.; Chen, L.; Tang, J.; Xing, H.; et al. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: A retrospective, multicentre, diagnostic study. Lancet Digit. Health 2022, 4, e179–e187. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, T.; Han, M.R.; Kim, S.; Kim, G.; Lee, S.; Choi, Y.J. Ovarian tumor diagnosis using deep convolutional neural networks and a denoising convolutional autoencoder. Sci Rep. 2022, 12, 17024. [Google Scholar] [CrossRef]
- Martínez-Más, J.; Bueno-Crespo, A.; Khazendar, S.; Remezal-Solano, M.; Martínez-Cendán, J.-P.; Jassim, S.; Du, H.; Al Assam, H.; Bourne, T.; Timmerman, D. Evaluation of machine learning methods with Fourier Transform features for classifying ovarian tumors based on ultrasound images. PLoS ONE 2019, 14, e0219388. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, B.-W.; Qian, L.; Meng, Y.-S.; Bai, X.-H.; Hong, X.-W.; He, X.; Jiang, M.-J.; Yuan, F.; Du, Q.-W.; et al. Deep Learning Prediction of Ovarian Malignancy at US Compared with O-RADS and Expert Assessment. Radiology 2022, 304, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Nero, C.; Ciccarone, F.; Boldrini, L.; Lenkowicz, J.; Paris, I.; Capoluongo, E.D.; Testa, A.C.; Fagotti, A.; Valentini, V.; Scambia, G. Germline BRCA 1-2 status prediction through ovarian ultrasound images radiogenomics: A hypothesis generating study (PROBE study). Sci. Rep. 2020, 10, 16511. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qiao, C.; Wu, M.; Cai, L.; Yin, C.; Yang, M.; Sang, X.; Bai, W. Improving the Segmentation Accuracy of Ovarian-Tumor Ultrasound Images Using Image Inpainting. Bioengineering 2023, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Chen, M.; Qiao, Y.; Zhao, F. Global guidelines for breast cancer screening: A systematic review. Breast 2022, 64, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Berg, W.A.; Aldrete, A.-L.L.; Jairaj, A.; Parea, J.C.L.; García, C.Y.; McClennan, R.C.; Cen, S.Y.; Larsen, L.H.; de Lara, M.T.S.; Love, S. Toward AI-supported US Triage of Women with Palpable Breast Lumps in a Low-Resource Setting. Radiology 2023, 307, e223351. [Google Scholar] [CrossRef]
- Huang, X.; Qiu, Y.; Bao, F.; Wang, J.; Lin, C.; Lin, Y.; Wu, J.; Yang, H. Artificial intelligence breast ultrasound and handheld ultrasound in the BI-RADS categorization of breast lesions: A pilot head to head comparison study in screening program. Front. Public Health 2023, 10, 1098639. [Google Scholar] [CrossRef]
- Browne, J.L.; Pascual, M.Á.; Perez, J.; Salazar, S.; Valero, B.; Rodriguez, I.; Cassina, D.; Alcazar, J.L.; Guerriero, S.; Graupera, B. AI: Can It Make a Difference to the Predictive Value of Ultrasound Breast Biopsy? Diagnostics 2023, 13, 811. [Google Scholar] [CrossRef]
- Pfob, A.; Sidey-Gibbons, C.; Barr, R.G.; Duda, V.; Alwafai, Z.; Balleyguier, C.; Clevert, D.-A.; Fastner, S.; Gomez, C.; Goncalo, M.; et al. Intelligent multi-modal shear wave elastography to reduce unnecessary biopsies in breast cancer diagnosis (INSPiRED 002): A retrospective, international, multicentre analysis. Eur. J. Cancer 2022, 177, 1–14. [Google Scholar] [CrossRef]
- Dong, F.; She, R.; Cui, C.; Shi, S.; Hu, X.; Zeng, J.; Wu, H.; Xu, J.; Zhang, Y. One step further into the blackbox: A pilot study of how to build more confidence around an AI-based decision system of breast nodule assessment in 2D ultrasound. Eur. Radiol. 2021, 31, 4991–5000. [Google Scholar] [CrossRef] [PubMed]
- Pfob, A.; Sidey-Gibbons, C.; Barr, R.G.; Duda, V.; Alwafai, Z.; Balleyguier, C.; Clevert, D.-A.; Fastner, S.; Gomez, C.; Goncalo, M.; et al. The importance of multi-modal imaging and clinical information for humans and AI-based algorithms to classify breast masses (INSPiRED 003): An international, multicenter analysis. Eur. Radiol. 2022, 32, 4101–4115. [Google Scholar] [CrossRef] [PubMed]
- Heremans, R.; Bosch, T.V.D.; Valentin, L.; Wynants, L.; Pascual, M.A.; Fruscio, R.; Testa, A.C.; Buonomo, F.; Guerriero, S.; Epstein, E.; et al. Ultrasound features of endometrial pathology in women without abnormal uterine bleeding: Results from the International Endometrial Tumor Analysis study (IETA3). Ultrasound Obstet. Gynecol. 2022, 60, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Vitale, S.G.; Riemma, G.; Haimovich, S.; Carugno, J.; Pacheco, L.A.; Perez-Medina, T.; Parry, J.P.; Török, P.; Tesarik, J.; Della Corte, L.; et al. Risk of endometrial cancer in asymptomatic postmenopausal women in relation to ultrasonographic endometrial thickness: Systematic review and diagnostic test accuracy meta-analysis. Am. J. Obstet. Gynecol. 2022, 228, 22–35.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, S.; Zhang, B.; Burjoo, A.; Yang, Y.; Xu, D. Artificial intelligence diagnosis of intrauterine adhesion by 3D ultrasound imaging: A prospective study. Quant. Imaging Med. Surg. 2023, 13, 2314–2327. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bao, N.; Xin, X.; Tan, J.; Li, H.; Zhou, S.; Liu, H. Automatic evaluation of endometrial receptivity in three-dimensional transvaginal ultrasound images based on 3D U-Net segmentation. Quant. Imaging Med. Surg. 2022, 12, 4095–4108. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, H.J.; Kim, H.G.; Ro, Y.M.; Shin, D.; Lee, S.R.; Kim, S.H.; Kong, M. Endometrium segmentation on transvaginal ultrasound image using key-point discriminator. Med. Phys. 2019, 46, 3974–3984. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Q.; Peng, B.; Jiang, J.; Fang, L.; Weng, W.; Wang, W.; Wang, S.; Zhu, X. Automatic Measurement of Endometrial Thickness from Transvaginal Ultrasound Images. Front. Bioeng. Biotechnol. 2022, 10, 853845. [Google Scholar] [CrossRef]
- Moro, F.; Albanese, M.; Boldrini, L.; Chiappa, V.; Lenkowicz, J.; Bertolina, F.; Mascilini, F.; Moroni, R.; Gambacorta, M.A.; Raspagliesi, F.; et al. Developing and validating ultrasound-based radiomics models for predicting high-risk endometrial cancer. Ultrasound Obstet. Gynecol. 2022, 60, 256–268. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, J.; Ji, Z.; Liu, W.; Li, M.; Xia, E.; Zhang, J.; Wang, J. Ultrasound Evaluation of Pelvic Floor Function after Transumbilical Laparoscopic Single-Site Total Hysterectomy Using Deep Learning Algorithm. Comput. Math. Methods Med. 2022, 2022, 1116332. [Google Scholar] [CrossRef]
- Williams, H.; Cattani, L.; Van Schoubroeck, D.; Yaqub, M.; Sudre, C.; Vercauteren, T.; D’Hooge, J.; Deprest, J. Automatic Extraction of Hiatal Dimensions in 3-D Transperineal Pelvic Ultrasound Recordings. Ultrasound Med. Biol. 2021, 47, 3470–3479. [Google Scholar] [CrossRef] [PubMed]
- Szentimrey, Z.; Ameri, G.; Hong, C.X.; Cheung, R.Y.K.; Ukwatta, E.; Eltahawi, A. Automated segmentation and measurement of the female pelvic floor from the mid-sagittal plane of 3D ultrasound volumes. Med. Phys. 2023, 50, 6215–6227. [Google Scholar] [CrossRef] [PubMed]
- Van Den Noort, F.; Manzini, C.; Van Der Vaart, C.H.; Van Limbeek, M.A.J.; Slump, C.H.; Grob, A.T.M. Automatic identification and segmentation of slice of minimal hiatal dimensions in transperineal ultrasound volumes. Ultrasound Obstet. Gynecol. 2022, 60, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Van den Noort, F.; van der Vaart, C.H.; Grob, A.T.M.; van de Waarsenburg, M.K.; Slump, C.H.; van Stralen, M. Deep learning enables automatic quantitative assessment of puborectalis muscle and urogenital hiatus in plane of minimal hiatal dimensions. Ultrasound Obstet. Gynecol. 2019, 54, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Ren, Y.; Lin, X.; Huang, Z.; Zheng, Z.; Zhang, X. Development and validation of a composite AI model for the diagnosis of levator ani muscle avulsion. Eur. Radiol. 2022, 32, 5898–5906. [Google Scholar] [CrossRef] [PubMed]
- Maicas, G.; Leonardi, M.; Avery, J.; Panuccio, C.; Carneiro, G.; Hull, M.L.; Condous, G. Deep learning to diagnose pouch of Douglas obliteration with ultrasound sliding sign. Reprod. Fertil. 2021, 2, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, D.; Raffone, A.; Aru, A.C.; Giorgi, M.; Giaquinto, I.; Spagnolo, E.; Travaglino, A.; Galatolo, F.A.; Cimino, M.G.C.A.; Lenzi, J.; et al. Application of Deep Learning Model in the Sonographic Diagnosis of Uterine Adenomyosis. Int. J. Environ. Res. Public Health 2023, 20, 1724. [Google Scholar] [CrossRef]
- Zhang, Y.; Hou, J.; Wang, Q.; Hou, A.; Liu, Y. Application of Transfer Learning and Feature Fusion Algorithms to Improve the Identification and Prediction Efficiency of Premature Ovarian Failure. J. Healthc. Eng. 2022, 2022, 3269692. [Google Scholar] [CrossRef]
- Yu, L.; Qing, X. Diagnosis of Idiopathic Premature Ovarian Failure by Color Doppler Ultrasound under the Intelligent Segmentation Algorithm. Comput. Math. Methods Med. 2022, 2022, 2645607. [Google Scholar] [CrossRef]
- Huo, T.; Li, L.; Chen, X.; Wang, Z.; Zhang, X.; Liu, S.; Huang, J.; Zhang, J.; Yang, Q.; Wu, W.; et al. Artificial intelligence-aided method to detect uterine fibroids in ultrasound images: A retrospective study. Sci. Rep. 2023, 13, 3714. [Google Scholar] [CrossRef]
- Yang, T.; Yuan, L.; Li, P.; Liu, P. Real-Time Automatic Assisted Detection of Uterine Fibroid in Ultrasound Images Using a Deep Learning Detector. Ultrasound Med. Biol. 2023, 49, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Yousef Kalafi, E.; Cheah, E.; Wang, S.; Wang, J.; Ozturk, A.; Li, Q.; Eldar, Y.C.; Samir, A.E.; Kumar, V. HaTU-Net: Harmonic Attention Network for Automated Ovarian Ultrasound Quantification in Assisted Pregnancy. Diagnostics 2022, 12, 3213. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.; Vignarajan, C.; Malhotra, N.; Vanamail, P. Three-Dimensional Automated Volume Calculation (Sonography-Based Automated Volume Count) versus Two-Dimensional Manual Ultrasonography for Follicular Tracking and Oocyte Retrieval in Women Undergoing in vitro Fertilization-Embryo Transfer: A Randomized Controlled Trial. J. Hum. Reprod. Sci. 2020, 13, 296. [Google Scholar] [PubMed]
- Maurice, P.; Dhombres, F.; Blondiaux, E.; Friszer, S.; Guilbaud, L.; Lelong, N.; Khoshnood, B.; Charlet, J.; Perrot, N.; Jauniaux, E.; et al. Towards ontology-based decision support systems for complex ultrasound diagnosis in obstetrics and gynecology. J. Gynecol. Obstet. Hum. Reprod. 2017, 46, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Dhombres, F.; Maurice, P.; Friszer, S.; Guilbaud, L.; Lelong, N.; Khoshnood, B.; Charlet, J.; Perrot, N.; Jauniaux, E.; Jurkovic, D.; et al. Developing a knowledge base to support the annotation of ultrasound images of ectopic pregnancy. J. Biomed. Semant. 2017, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.; Khan, S.; Choi, S.; Shin, D.; Lee, J.E.; Lee, E.S.; Ye, J.C. Tunable image quality control of 3-D ultrasound using switchable CycleGAN. Med. Image Anal. 2023, 83, 102651. [Google Scholar] [CrossRef] [PubMed]
- Kalantaridou, S.N.; Nelson, L.M. Premature ovarian failure is not premature menopause. Ann. N. Y. Acad. Sci. 2006, 900, 393–402. [Google Scholar] [CrossRef]
- Watzenboeck, M.L.; Heidinger, B.H.; Rainer, J.; Schmidbauer, V.; Ulm, B.; Rubesova, E.; Prayer, D.; Kasprian, G.; Prayer, F. Reproducibility of 2D versus 3D radiomics for quantitative assessment of fetal lung development: A retrospective fetal MRI study. Insights Imaging 2023, 14, 31. [Google Scholar] [CrossRef]
- Prayer, F.; Watzenböck, M.L.; Heidinger, B.H.; Rainer, J.; Schmidbauer, V.; Prosch, H.; Ulm, B.; Rubesova, E.; Prayer, D.; Kasprian, G. Fetal MRI radiomics: Non-invasive and reproducible quantification of human lung maturity. Eur. Radiol. 2023, 33, 4205–4213. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Cheng, R.; Liu, S.; Qu, F.; Yin, X.; Wang, Q.; Xiao, B.; Ye, Z. Radiomics analysis of apparent diffusion coefficient in cervical cancer: A preliminary study on histological grade evaluation: Radiomic Features in Uterine Cervical Cancer. J. Magn. Reson. Imaging 2019, 49, 280–290. [Google Scholar] [CrossRef]
- Drukker, L.; Noble, J.A.; Papageorghiou, A.T. Introduction to Artificial Intelligence in Ultrasound Imaging in Obstetrics and Gynecology. Obstet. Gynecol. Surv. 2021, 76, 127–129. [Google Scholar] [CrossRef]
- Jani, J.; Peralta, C.F.A.; Benachi, A.; Deprest, J.; Nicolaides, K.H. Assessment of lung area in fetuses with congenital diaphragmatic hernia. Ultrasound Obstet. Gynecol. 2007, 30, 72–76. [Google Scholar] [CrossRef]
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance; Licence: CC BY-NC-SA 3.0 IGO; WHO: Switzerland, Geneva, 2021. [Google Scholar]
- Dhombres, F.; Bonnard, J.; Bailly, K.; Maurice, P.; Papageorghiou, A.T.; Jouannic, J.-M. Contributions of Artificial Intelligence Reported in Obstetrics and Gynecology Journals: Systematic Review. J. Med. Internet Res. 2022, 24, e35465. [Google Scholar] [CrossRef]

| PICOS Search Tool Headings for Literature Evaluation | |
|---|---|
| Participants | Examiner: Healthcare professionals in OB/GYN or radiology, AI specialists Patients: Healthy pregnant and non-pregnant women or women with any gynecological or obstetric disease/complication, OB/GYN training models |
| Intervention or Exposure | AI-assisted US applications |
| Comparison | Comparison of AI US algorithms to human US examiners or another AI algorithm |
| Outcome | Fields of AI applications in OB/GYN US imaging, benefits and limitations of AI usage, future aspects for emerging fields of applications |
| Study type | Published literature of any design, excluding trial protocols and reviews |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jost, E.; Kosian, P.; Jimenez Cruz, J.; Albarqouni, S.; Gembruch, U.; Strizek, B.; Recker, F. Evolving the Era of 5D Ultrasound? A Systematic Literature Review on the Applications for Artificial Intelligence Ultrasound Imaging in Obstetrics and Gynecology. J. Clin. Med. 2023, 12, 6833. https://doi.org/10.3390/jcm12216833
Jost E, Kosian P, Jimenez Cruz J, Albarqouni S, Gembruch U, Strizek B, Recker F. Evolving the Era of 5D Ultrasound? A Systematic Literature Review on the Applications for Artificial Intelligence Ultrasound Imaging in Obstetrics and Gynecology. Journal of Clinical Medicine. 2023; 12(21):6833. https://doi.org/10.3390/jcm12216833
Chicago/Turabian StyleJost, Elena, Philipp Kosian, Jorge Jimenez Cruz, Shadi Albarqouni, Ulrich Gembruch, Brigitte Strizek, and Florian Recker. 2023. "Evolving the Era of 5D Ultrasound? A Systematic Literature Review on the Applications for Artificial Intelligence Ultrasound Imaging in Obstetrics and Gynecology" Journal of Clinical Medicine 12, no. 21: 6833. https://doi.org/10.3390/jcm12216833
APA StyleJost, E., Kosian, P., Jimenez Cruz, J., Albarqouni, S., Gembruch, U., Strizek, B., & Recker, F. (2023). Evolving the Era of 5D Ultrasound? A Systematic Literature Review on the Applications for Artificial Intelligence Ultrasound Imaging in Obstetrics and Gynecology. Journal of Clinical Medicine, 12(21), 6833. https://doi.org/10.3390/jcm12216833







