From Vibrations to Visions: Raman Spectroscopy’s Impact on Skin Cancer Diagnostics
Abstract
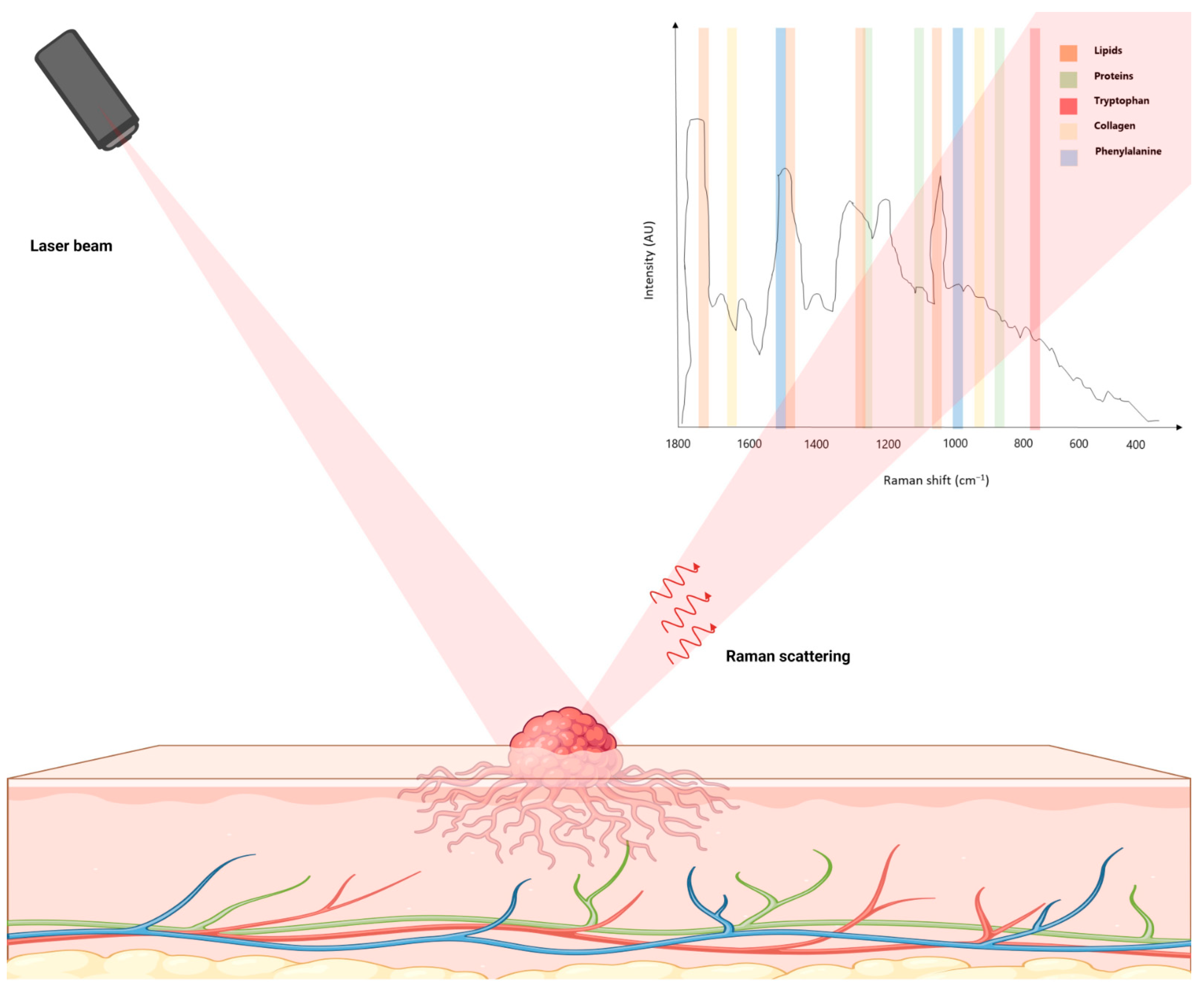
:1. Introduction
2. The Science behind Raman Spectroscopy
3. Raman Device Instrumentation
4. Raman Spectroscopy in Skin Cancer Diagnostics
4.1. Differentiation of Skin Cancer from Normal Tissue and between Skin Tumor Types
4.1.1. Cell Lines
4.1.2. Clinical Studies
4.2. Combination with Imaging Techniques
4.3. Multimodal Approaches
4.3.1. Multi-Wavelength Raman/Photoluminescence Microspectroscopy
4.3.2. Raman Spectroscopy and Machine Learning
4.3.3. Raman Spectroscopy and Optical Coherence Tomography
4.3.4. Raman and Autofluorescence Spectroscopy
4.3.5. Raman and Laser-Induced Breakdown Spectroscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman Techniques: Fundamentals and Frontiers. Nanoscale Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Canetta, E. Current and Future Advancements of Raman Spectroscopy Techniques in Cancer Nanomedicine. Int. J. Mol. Sci. 2021, 22, 13141. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Alvarez-Puebla, R.A. Surface-Enhanced Raman Spectroscopy in Cancer Diagnosis, Prognosis and Monitoring. Cancers 2019, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-Enhanced Raman Spectroscopy for in Vivo Biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar] [CrossRef]
- Huang, P.-J.; Lee, C.-K.; Lee, L.-H.; Huang, H.-F.; Huang, Y.-H.; Lan, J.-C.; Lee, C.-H. Surface-Enhanced Raman Scattering (SERS) by Gold Nanoparticle Characterizes Dermal Thickening by Collagen in Bleomycin-Treated Skin Ex Vivo. Ski. Res. Technol. 2023, 29, e13334. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Bonyár, A.; Veres, M.; Himics, L.; Balázs, L.; Juhász, L.; Csarnovics, I. A Generalized Exponential Relationship between the Surface-Enhanced Raman Scattering (SERS) Efficiency of Gold/Silver Nanoisland Arrangements and Their Non-Dimensional Interparticle Distance/Particle Diameter Ratio. Sens. Actuators A Phys. 2020, 314, 112225. [Google Scholar] [CrossRef]
- Fox, S.A.; Shanblatt, A.A.; Beckman, H.; Strasswimmer, J.; Terentis, A.C. Raman Spectroscopy Differentiates Squamous Cell Carcinoma (SCC) from Normal Skin Following Treatment with a High-Powered CO2 Laser. Lasers Surg. Med. 2014, 46, 757–772. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H. Near-Infrared Raman Spectroscopy for Optical Diagnosis of Lung Cancer. Int. J. Cancer 2003, 107, 1047–1052. [Google Scholar] [CrossRef]
- Michalska, M.; Chodorowska, G.; Krasowska, D. SIAscopy--a New Non-Invasive Technique of Melanoma Diagnosis. Ann. Univ. Mariae Curie Sklodowska Med. 2004, 59, 421–431. [Google Scholar]
- Leslie, D.G.; Kast, R.E.; Poulik, J.M.; Rabah, R.; Sood, S.; Auner, G.W.; Klein, M.D. Identification of Pediatric Brain Neoplasms Using Raman Spectroscopy. Pediatr. Neurosurg. 2012, 48, 109–117. [Google Scholar] [CrossRef]
- Kandurova, K.; Dremin, V.; Zherebtsov, E.; Potapova, E.; Alyanov, A.; Mamoshin, A.; Ivanov, Y.; Borsukov, A.; Dunaev, A. Fiber-Optic System for Intraoperative Study of Abdominal Organs during Minimally Invasive Surgical Interventions. Appl. Sci. 2019, 9, 217. [Google Scholar] [CrossRef]
- Khristoforova, Y.A.; Bratchenko, I.A.; Myakinin, O.O.; Artemyev, D.N.; Moryatov, A.A.; Orlov, A.E.; Kozlov, S.V.; Zakharov, V.P. Portable Spectroscopic System for in Vivo Skin Neoplasms Diagnostics by Raman and Autofluorescence Analysis. J. Biophotonics 2019, 12, e201800400. [Google Scholar] [CrossRef] [PubMed]
- Bersani, D.; Lottici, P.P. Applications of Raman Spectroscopy to Gemology. Anal. Bioanal. Chem. 2010, 397, 2631–2646. [Google Scholar] [CrossRef] [PubMed]
- Jehlička, J.; Vítek, P.; Edwards, H.G.M. Raman Spectra of Organic Acids Obtained Using a Portable Instrument at −5 °C in a Mountain Area at 2000 m above Sea Level. J. Raman Spectrosc. 2010, 41, 440–444. [Google Scholar] [CrossRef]
- Jehlička, J.; Vítek, P.; Edwards, H.G.M.; Hargreaves, M.D.; Čapoun, T. Fast Detection of Sulphate Minerals (Gypsum, Anglesite, Baryte) by a Portable Raman Spectrometer. J. Raman Spectrosc. 2009, 40, 1082–1086. [Google Scholar] [CrossRef]
- Cullum, B.M.; Mobley, J.; Chi, Z.; Stokes, D.L.; Miller, G.H.; Vo-Dinh, T. Development of a Compact, Handheld Raman Instrument with No Moving Parts for Use in Field Analysis. Rev. Sci. Instrum. 2000, 71, 1602–1607. [Google Scholar] [CrossRef]
- Vandenabeele, P.; Edwards, H.G.M.; Jehlička, J. The Role of Mobile Instrumentation in Novel Applications of Raman Spectroscopy: Archaeometry, Geosciences, and Forensics. Chem. Soc. Rev. 2014, 43, 2628–2649. [Google Scholar] [CrossRef] [PubMed]
- Brauchle, E.; Noor, S.; Holtorf, E.; Garbe, C.; Schenke-Layland, K.; Busch, C. Raman Spectroscopy as an Analytical Tool for Melanoma Research. Clin. Exp. Dermatol. 2014, 39, 636–645. [Google Scholar] [CrossRef]
- Piredda, P.; Berning, M.; Boukamp, P.; Volkmer, A. Subcellular Raman Microspectroscopy Imaging of Nucleic Acids and Tryptophan for Distinction of Normal Human Skin Cells and Tumorigenic Keratinocytes. Anal. Chem. 2015, 87, 6778–6785. [Google Scholar] [CrossRef]
- Wang, H.; Tsai, T.-H.; Zhao, J.; Lee, A.M.D.; Lo, B.K.K.; Yu, M.; Lui, H.; McLean, D.I.; Zeng, H. Differentiation of HaCaT Cell and Melanocyte from Their Malignant Counterparts Using Micro-Raman Spectroscopy Guided by Confocal Imaging. Photodermatol. Photoimmunol. Photomed. 2012, 28, 147–152. [Google Scholar] [CrossRef]
- Qiu, X.; He, T.; Wu, X.; Wang, P.; Wang, X.; Fu, Q.; Fang, X.; Li, S.; Li, Y. Combining Fiber Optical Tweezers and Raman Spectroscopy for Rapid Identification of Melanoma. J. Biophotonics 2022, 15, e202200158. [Google Scholar] [CrossRef] [PubMed]
- Bodanese, B.; Silveira, F.L.; Zângaro, R.A.; Pacheco, M.T.T.; Pasqualucci, C.A.; Silveira, L. Discrimination of Basal Cell Carcinoma and Melanoma from Normal Skin Biopsies In Vitro through Raman Spectroscopy and Principal Component Analysis. Photomed. Laser Surg. 2012, 30, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Gniadecka, M.; Philipsen, P.A.; Sigurdsson, S.; Wessel, S.; Nielsen, O.F.; Christensen, D.H.; Hercogova, J.; Rossen, K.; Thomsen, H.K.; Gniadecki, R.; et al. Melanoma Diagnosis by Raman Spectroscopy and Neural Networks: Structure Alterations in Proteins and Lipids in Intact Cancer Tissue. J. Investig. Dermatol. 2004, 122, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of Tumors during Tissue-Conserving Surgery with Integrated Autofluorescence and Raman Scattering Microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar] [CrossRef] [PubMed]
- Legesse, F.B.; Medyukhina, A.; Heuke, S.; Popp, J. Texture Analysis and Classification in Coherent Anti-Stokes Raman Scattering (CARS) Microscopy Images for Automated Detection of Skin Cancer. Comput. Med. Imaging Graph. 2015, 43, 36–43. [Google Scholar] [CrossRef]
- Lieber, C.A.; Majumder, S.K.; Billheimer, D.; Ellis, D.L.; Mahadevan-Jansen, A. Raman Microspectroscopy for Skin Cancer Detection in Vitro. J. Biomed. Opt. 2008, 13, 024013. [Google Scholar] [CrossRef]
- Lieber, C.A.; Majumder, S.K.; Ellis, D.L.; Billheimer, D.D.; Mahadevan-Jansen, A. In Vivo Nonmelanoma Skin Cancer Diagnosis Using Raman Microspectroscopy. Lasers Surg. Med. 2008, 40, 461–467. [Google Scholar] [CrossRef]
- Nijssen, A.; Maquelin, K.; Santos, L.F.; Caspers, P.J.; Bakker Schut, T.C.; den Hollander, J.C.; Neumann, M.H.A.; Puppels, G.J. Discriminating Basal Cell Carcinoma from Perilesional Skin Using High Wave-Number Raman Spectroscopy. J. Biomed. Opt. 2007, 12, 034004. [Google Scholar] [CrossRef]
- Nunes, L.D.O.; Martin, A.A.; Silveira, L., Jr.; Zampieri, M. FT-Raman Spectroscopy Study for Skin Cancer Diagnosis. J. Spectrosc. 2003, 17, 104696. [Google Scholar] [CrossRef]
- Philipsen, P.A.; Knudsen, L.; Gniadecka, M.; Ravnbak, M.H.; Wulf, H.C. Diagnosis of Malignant Melanoma and Basal Cell Carcinoma by in Vivo NIR-FT Raman Spectroscopy Is Independent of Skin Pigmentation. Photochem. Photobiol. Sci. 2013, 12, 770–776. [Google Scholar] [CrossRef]
- Schleusener, J.; Gluszczynska, P.; Reble, C.; Gersonde, I.; Helfmann, J.; Fluhr, J.W.; Lademann, J.; Röwert-Huber, J.; Patzelt, A.; Meinke, M.C. In Vivo Study for the Discrimination of Cancerous and Normal Skin Using Fibre Probe-Based Raman Spectroscopy. Exp. Dermatol. 2015, 24, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Silveira, F.L.; Pacheco, M.T.T.; Bodanese, B.; Pasqualucci, C.A.; Zângaro, R.A.; Silveira, L. Discrimination of Non-Melanoma Skin Lesions from Non-Tumor Human Skin Tissues in Vivo Using Raman Spectroscopy and Multivariate Statistics. Lasers Surg. Med. 2015, 47, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fan, Y.; Song, Y.; Xu, J. Accuracy of Raman Spectroscopy for Differentiating Skin Cancer from Normal Tissue. Medicine 2018, 97, e12022. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, G.; Youssef, K.K.; Vokaer, B.; Achouri, Y.; Michaux, C.; Sotiropoulou, P.A.; Blanpain, C. Identifying the Cellular Origin of Squamous Skin Tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 7431–7436. [Google Scholar] [CrossRef] [PubMed]
- Regad, T. Molecular and Cellular Pathogenesis of Melanoma Initiation and Progression. Cell. Mol. Life Sci. 2013, 70, 4055–4065. [Google Scholar] [CrossRef] [PubMed]
- Youssef, K.K.; Van Keymeulen, A.; Lapouge, G.; Beck, B.; Michaux, C.; Achouri, Y.; Sotiropoulou, P.A.; Blanpain, C. Identification of the Cell Lineage at the Origin of Basal Cell Carcinoma. Nat. Cell Biol. 2010, 12, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.H.; From, L.; Bernardino, E.A.; Mihm, M.C. The Histogenesis and Biologic Behavior of Primary Human Malignant Melanomas of the Skin. Cancer Res. 1969, 29, 705–727. [Google Scholar] [PubMed]
- Feng, X.; Moy, A.J.; Nguyen, H.T.M.; Zhang, J.; Fox, M.C.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Raman Active Components of Skin Cancer. Biomed. Opt. Express 2017, 8, 2835–2850. [Google Scholar] [CrossRef]
- Santos, I.P.; Caspers, P.J.; Bakker Schut, T.C.; van Doorn, R.; Noordhoek Hegt, V.; Koljenović, S.; Puppels, G.J. Raman Spectroscopic Characterization of Melanoma and Benign Melanocytic Lesions Suspected of Melanoma Using High-Wavenumber Raman Spectroscopy. Anal. Chem. 2016, 88, 7683–7688. [Google Scholar] [CrossRef]
- Bodanese, B.; Silveira, L.; Albertini, R.; Zângaro, R.A.; Pacheco, M.T.T. Differentiating Normal and Basal Cell Carcinoma Human Skin Tissues in Vitro Using Dispersive Raman Spectroscopy: A Comparison between Principal Components Analysis and Simplified Biochemical Models. Photomed. Laser Surg. 2010, 28 (Suppl. S1), S119–S127. [Google Scholar] [CrossRef]
- Larraona-Puy, M.; Ghita, A.; Zoladek, A.; Perkins, W.; Varma, S.; Leach, I.H.; Koloydenko, A.A.; Williams, H.; Notingher, I. Development of Raman Microspectroscopy for Automated Detection and Imaging of Basal Cell Carcinoma. J. Biomed. Opt. 2009, 14, 054031. [Google Scholar] [CrossRef] [PubMed]
- Ly, E.; Durlach, A.; Antonicelli, F.; Bernard, P.; Manfait, M.; Piot, O. Probing Tumor and Peritumoral Tissues in Superficial and Nodular Basal Cell Carcinoma Using Polarized Raman Microspectroscopy. Exp. Dermatol. 2010, 19, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ly, E.; Piot, O.; Durlach, A.; Bernard, P.; Manfait, M. Polarized Raman Microspectroscopy Can Reveal Structural Changes of Peritumoral Dermis in Basal Cell Carcinoma. Appl. Spectrosc. 2008, 62, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Short, M.A.; Lui, H.; McLean, D.; Zeng, H.; Alajlan, A.; Chen, X.K. Changes in Nuclei and Peritumoral Collagen within Nodular Basal Cell Carcinomas via Confocal Micro-Raman Spectroscopy. J. Biomed. Opt. 2006, 11, 34004. [Google Scholar] [CrossRef]
- Choi, J.; Choo, J.; Chung, H.; Gweon, D.-G.; Park, J.; Kim, H.J.; Park, S.; Oh, C.-H. Direct Observation of Spectral Differences between Normal and Basal Cell Carcinoma (BCC) Tissues Using Confocal Raman Microscopy. Biopolymers 2005, 77, 264–272. [Google Scholar] [CrossRef]
- Nijssen, A.; Bakker Schut, T.C.; Heule, F.; Caspers, P.J.; Hayes, D.P.; Neumann, M.H.A.; Puppels, G.J. Discriminating Basal Cell Carcinoma from Its Surrounding Tissue by Raman Spectroscopy. J. Investig. Dermatol. 2002, 119, 64–69. [Google Scholar] [CrossRef]
- Kiss, N.; Krolopp, Á.; Lőrincz, K.; Bánvölgyi, A.; Szipőcs, R.; Wikonkál, N. Stain-Free Histopathology of Basal Cell Carcinoma by Dual Vibration Resonance Frequency CARS Microscopy. Pathol. Oncol. Res. 2018, 24, 927–930. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, F.; Li, J.; Song, D.; Li, H.; Wang, K.; He, Q.; Wang, S. Investigation on the Cancer Invasion and Metastasis of Skin Squamous Cell Carcinoma by Raman Spectroscopy. Molecules 2019, 24, 2059. [Google Scholar] [CrossRef]
- Vardaki, M.Z.; Pavlou, E.; Simantiris, N.; Lampri, E.; Seretis, K.; Kourkoumelis, N. Towards Non-Invasive Monitoring of Non-Melanoma Skin Cancer Using Spatially Offset Raman Spectroscopy. Analyst 2023, 148, 4386–4395. [Google Scholar] [CrossRef]
- Silveira, L.; Pasqualucci, C.A.; Bodanese, B.; Pacheco, M.T.T.; Zângaro, R.A. Normal-Subtracted Preprocessing of Raman Spectra Aiming to Discriminate Skin Actinic Keratosis and Neoplasias from Benign Lesions and Normal Skin Tissues. Lasers Med. Sci. 2020, 35, 1141–1151. [Google Scholar] [CrossRef]
- Zhao, J.; Zeng, H.; Kalia, S.; Lui, H. Wavenumber Selection Based Analysis in Raman Spectroscopy Improves Skin Cancer Diagnostic Specificity. Analyst 2016, 141, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, I.; Bratchenko, I.; Khristoforova, Y.; Bratchenko, L.; Moryatov, A.; Kozlov, S.; Kaganov, O.; Zakharov, V. Multivariate Curve Resolution Alternating Least Squares Analysis of In Vivo Skin Raman Spectra. Sensors 2022, 22, 9588. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.J.; Marro, M.; Galván, I.; Bernabeu-Wittel, J.; Conejo-Mir, J.; Zulueta-Dorado, T.; Guisado-Gil, A.B.; Loza-Álvarez, P. Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis. Cancers 2022, 14, 1056. [Google Scholar] [CrossRef] [PubMed]
- Mussi, V.; Ledda, M.; Polese, D.; Maiolo, L.; Paria, D.; Barman, I.; Lolli, M.G.; Lisi, A.; Convertino, A. Silver-Coated Silicon Nanowire Platform Discriminates Genomic DNA from Normal and Malignant Human Epithelial Cells Using Label-Free Raman Spectroscopy. Mater. Sci. Eng. C 2021, 122, 111951. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Balu, M.; Krasieva, T.; Potma, E.O.; Elkeeb, L.; Zachary, C.B.; Wilder-Smith, P. Evaluation of Stimulated Raman Scattering Microscopy for Identifying Squamous Cell Carcinoma in Human Skin. Lasers Surg. Med. 2013, 45, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.; Nichols, B.; Migden, M.R.; Rajaram, N.; Reichenberg, J.S.; Markey, M.K.; Ross, M.I.; Tunnell, J.W. Clinical Study of Noninvasive in Vivo Melanoma and Nonmelanoma Skin Cancers Using Multimodal Spectral Diagnosis. J. Biomed. Opt. 2014, 19, 117003. [Google Scholar] [CrossRef]
- Zhao, J.; Zeng, H.; Kalia, S.; Lui, H. Using Raman Spectroscopy to Detect and Diagnose Skin Cancer In Vivo. Dermatol. Clin. 2017, 35, 495–504. [Google Scholar] [CrossRef]
- Nguyen, H.T.M.; Zhang, Y.; Moy, A.J.; Feng, X.; Sebastian, K.R.; Reichenberg, J.S.; Fox, M.C.; Markey, M.K.; Tunnell, J.W. Characterization of Ex Vivo Nonmelanoma Skin Tissue Using Raman Spectroscopy. Photonics 2021, 8, 282. [Google Scholar] [CrossRef]
- Kourkoumelis, N.; Balatsoukas, I.; Moulia, V.; Elka, A.; Gaitanis, G.; Bassukas, I. Advances in the in Vivo Raman Spectroscopy of Malignant Skin Tumors Using Portable Instrumentation. Int. J. Mol. Sci. 2015, 16, 14554–14570. [Google Scholar] [CrossRef]
- Meksiarun, P.; Andriana, B.B.; Matsuyoshi, H.; Sato, H. Non-Invasive Quantitative Analysis of Specific Fat Accumulation in Subcutaneous Adipose Tissues Using Raman Spectroscopy. Sci. Rep. 2016, 6, 37068. [Google Scholar] [CrossRef]
- Edwards, H.G.M.; Williams, A.C.; Barry, B.W. Potential Applications of FT-Raman Spectroscopy for Dermatological Diagnostics. J. Mol. Struct. 1995, 347, 379–387. [Google Scholar] [CrossRef]
- Choi, M.J.; Maibach, H.I. Role of Ceramides in Barrier Function of Healthy and Diseased Skin. Am. J. Clin. Dermatol. 2005, 6, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Fox, M.C.; Reichenberg, J.S.; Lopes, F.C.P.S.; Sebastian, K.R.; Markey, M.K.; Tunnell, J.W. Biophysical Basis of Skin Cancer Margin Assessment Using Raman Spectroscopy. Biomed. Opt. Express 2019, 10, 104–118. [Google Scholar] [CrossRef]
- Silver, F.; Kelkar, N.; Deshmukh, T.; Shah, R. Biomechanical Relationship between Cells and Collagen in Skin and Skin Lesions. J. Dermatol. Surg. Res. Ther. 2019, 2, 70–76. [Google Scholar]
- Feng, X.; Moy, A.J.; Nguyen, H.T.M.; Zhang, Y.; Zhang, J.; Fox, M.C.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Raman Biophysical Markers in Skin Cancer Diagnosis. J. Biomed. Opt. 2018, 23, 057002. [Google Scholar] [CrossRef] [PubMed]
- Franzen, L.; Windbergs, M. Applications of Raman Spectroscopy in Skin Research—From Skin Physiology and Diagnosis up to Risk Assessment and Dermal Drug Delivery. Adv. Drug Deliv. Rev. 2015, 89, 91–104. [Google Scholar] [CrossRef]
- Heuke, S.; Vogler, N.; Meyer, T.; Akimov, D.; Kluschke, F.; Röwert-Huber, H.-J.; Lademann, J.; Dietzek, B.; Popp, J. Detection and Discrimination of Non-Melanoma Skin Cancer by Multimodal Imaging. Healthcare 2013, 1, 64–83. [Google Scholar] [CrossRef]
- Bratchenko, I.A.; Artemyev, D.N.; Myakinin, O.O.; Khristoforova, Y.A.; Moryatov, A.A.; Kozlov, S.V.; Zakharov, V.P. Combined Raman and Autofluorescence Ex Vivo Diagnostics of Skin Cancer in Near-Infrared and Visible Regions. J. Biomed. Opt. 2017, 22, 27005. [Google Scholar] [CrossRef]
- Lui, H.; Zhao, J.; McLean, D.; Zeng, H. Real-Time Raman Spectroscopy for In Vivo Skin Cancer Diagnosis. Cancer Res. 2012, 72, 2491–2500. [Google Scholar] [CrossRef]
- Wu, M.; Gao, B.; Wei, X. Recent Advances in Raman Spectroscopy for Skin Diagnosis. J. Innov. Opt. Health Sci. 2023, 16, 2330003. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, S.; Yue, S. Raman Spectroscopy and Imaging for Cancer Diagnosis. J. Healthc. Eng. 2018, 2018, 8619342. [Google Scholar] [CrossRef] [PubMed]
- Calin, M.A.; Parasca, S.V.; Savastru, D.; Manea, D. Hyperspectral Imaging in the Medical Field: Present and Future. Appl. Spectrosc. Rev. 2014, 49, 435–447. [Google Scholar] [CrossRef]
- Himmelsbach, D. Pittcon 2004 NIR Awards. NIR News 2004, 15, 3. [Google Scholar] [CrossRef]
- Tang, Y.; Song, S.; Gui, S.; Chao, W.; Cheng, C.; Qin, R. Active and Low-Cost Hyperspectral Imaging for the Spectral Analysis of a Low-Light Environment. Sensors 2023, 23, 1437. [Google Scholar] [CrossRef] [PubMed]
- Withagen, P.J.; Den Breejen, E.; Franken, E.M.; De Jong, A.N.; Winkel, H. Band Selection from a Hyperspectral Data-Cube for a Real-Time Multispectral 3CCD Camera. In Algorithms for Multispectral, Hyperspectral, and Ultraspectral Imagery VII, Proceedings of the Aerospace/Defense Sensing, Simulation, and Controls, Orlando, FL, USA, 16–20 April 2001; Shen, S.S., Descour, M.R., Eds.; SPIE: Bellingham, WT, USA, 2001; pp. 84–93. [Google Scholar]
- Gómez-Sanchis, J.; Lorente, D.; Soria-Olivas, E.; Aleixos, N.; Cubero, S.; Blasco, J. Development of a Hyperspectral Computer Vision System Based on Two Liquid Crystal Tuneable Filters for Fruit Inspection. Application to Detect Citrus Fruits Decay. Food Bioprocess Technol. 2014, 7, 1047–1056. [Google Scholar] [CrossRef]
- Li, Y.; Shen, F.; Hu, L.; Lang, Z.; Liu, Q.; Cai, F.; Fu, L. A Stare-Down Video-Rate High-Throughput Hyperspectral Imaging System and Its Applications in Biological Sample Sensing. IEEE Sens. J. 2023, 23, 23629–23637. [Google Scholar] [CrossRef]
- Anastassopoulou, J.; Kyriakidou, M.; Malesiou, E.; Rallis, M.; Theophanides, T. Infrared and Raman Spectroscopic Studies of Molecular Disorders in Skin Cancer. In Vivo 2019, 33, 567–572. [Google Scholar] [CrossRef]
- Rimskaya, E.; Shelygina, S.; Timurzieva, A.; Saraeva, I.; Perevedentseva, E.; Melnik, N.; Kudrin, K.; Reshetov, D.; Kudryashov, S. Multispectral Raman Differentiation of Malignant Skin Neoplasms In Vitro: Search for Specific Biomarkers and Optimal Wavelengths. Int. J. Mol. Sci. 2023, 24, 14748. [Google Scholar] [CrossRef]
- Wu, M.; Wang, S.; Pan, S.; Terentis, A.C.; Strasswimmer, J.; Zhu, X. Deep Learning Data Augmentation for Raman Spectroscopy Cancer Tissue Classification. Sci. Rep. 2021, 11, 23842. [Google Scholar] [CrossRef]
- Kalatzis, D.; Spyratou, E.; Karnachoriti, M.; Kouri, M.A.; Orfanoudakis, S.; Koufopoulos, N.; Pouliakis, A.; Danias, N.; Seimenis, I.; Kontos, A.G.; et al. Advanced Raman Spectroscopy Based on Transfer Learning by Using a Convolutional Neural Network for Personalized Colorectal Cancer Diagnosis. Optics 2023, 4, 310–320. [Google Scholar] [CrossRef]
- Sigurdsson, S.; Philipsen, P.A.; Hansen, L.K.; Larsen, J.; Gniadecka, M.; Wulf, H.C. Detection of Skin Cancer by Classification of Raman Spectra. IEEE Trans. Biomed. Eng. 2004, 51, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Hofmann-Wellenhof, R.; Wurm, E.M.T.; Ahlgrimm-Siess, V.; Richtig, E.; Koller, S.; Smolle, J.; Gerger, A. Reflectance Confocal Microscopy--State-of-Art and Research Overview. Semin. Cutan. Med. Surg. 2009, 28, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Rajadhyaksha, M.; Marghoob, A.; Rossi, A.; Halpern, A.C.; Nehal, K.S. Reflectance Confocal Microscopy of Skin in Vivo: From Bench to Bedside. Lasers Surg. Med. 2017, 49, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Feng, X.; Fox, M.C.; Reichenberg, J.S.; Lopes, F.C.P.S.; Sebastian, K.R.; Markey, M.K.; Tunnell, J.W. Deep Learning on Reflectance Confocal Microscopy Improves Raman Spectral Diagnosis of Basal Cell Carcinoma. J. Biomed. Opt. 2022, 27, 065004. [Google Scholar] [CrossRef] [PubMed]
- Bratchenko, I.A.; Bratchenko, L.A.; Khristoforova, Y.A.; Moryatov, A.A.; Kozlov, S.V.; Zakharov, V.P. Classification of Skin Cancer Using Convolutional Neural Networks Analysis of Raman Spectra. Comput. Methods Programs Biomed. 2022, 219, 106755. [Google Scholar] [CrossRef]
- Araújo, D.C.; Veloso, A.A.; de Oliveira Filho, R.S.; Giraud, M.-N.; Raniero, L.J.; Ferreira, L.M.; Bitar, R.A. Finding Reduced Raman Spectroscopy Fingerprint of Skin Samples for Melanoma Diagnosis through Machine Learning. Artif. Intell. Med. 2021, 120, 102161. [Google Scholar] [CrossRef]
- Chang, M.; He, C.; Du, Y.; Qiu, Y.; Wang, L.; Chen, H. RaT: Raman Transformer for Highly Accurate Melanoma Detection with Critical Features Visualization. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 305, 123475. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Li, C. Signal Pathways of Melanoma and Targeted Therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef]
- Shin, S.-S.; Jeong, B.-S.; Wall, B.A.; Li, J.; Shan, N.L.; Wen, Y.; Goydos, J.S.; Chen, S. Participation of xCT in Melanoma Cell Proliferation in Vitro and Tumorigenesis in Vivo. Oncogenesis 2018, 7, 86. [Google Scholar] [CrossRef]
- Böhme, I.; Schönherr, R.; Eberle, J.; Bosserhoff, A.K. Membrane Transporters and Channels in Melanoma. Rev. Physiol. Biochem. Pharmacol. 2021, 181, 269–374. [Google Scholar] [CrossRef]
- McArdle, L.; Bergin, O.; Fallowfield, M.E.; Dervan, P.A.; Easty, D.J. Tyrosine Phosphate in Melanoma Progression. Br. J. Dermatol. 2003, 149, 289–295. [Google Scholar] [CrossRef] [PubMed]
- McArdle, L.; Rafferty, M.; Maelandsmo, G.M.; Bergin, O.; Farr, C.J.; Dervan, P.A.; O’Loughlin, S.; Herlyn, M.; Easty, D.J. Protein Tyrosine Phosphatase Genes Downregulated in Melanoma. J. Investig. Dermatol. 2001, 117, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Hubková, B.; Valko-Rokytovská, M.; Čižmárová, B.; Zábavníková, M.; Mareková, M.; Birková, A. Tryptophan: Its Metabolism along the Kynurenine, Serotonin, and Indole Pathway in Malignant Melanoma. Int. J. Mol. Sci. 2022, 23, 9160. [Google Scholar] [CrossRef] [PubMed]
- Oscilowska, I.; Rolkowski, K.; Baszanowska, W.; Huynh, T.Y.L.; Lewoniewska, S.; Nizioł, M.; Sawicka, M.; Bielawska, K.; Szoka, P.; Miltyk, W.; et al. Proline Dehydrogenase/Proline Oxidase (PRODH/POX) Is Involved in the Mechanism of Metformin-Induced Apoptosis in C32 Melanoma Cell Line. Int. J. Mol. Sci. 2022, 23, 2354. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Müller, M.; Rios, A.; Sennrich, R. Why Self-Attention? A Targeted Evaluation of Neural Machine Translation Architectures. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018; Association for Computational Linguistics: Brussels, Belgium, 2018; pp. 4263–4272. [Google Scholar]
- Li, Z.; Li, Z.; Chen, Q.; Ramos, A.; Zhang, J.; Boudreaux, J.P.; Thiagarajan, R.; Bren-Mattison, Y.; Dunham, M.E.; McWhorter, A.J.; et al. Detection of Pancreatic Cancer by Convolutional-Neural-Network-Assisted Spontaneous Raman Spectroscopy with Critical Feature Visualization. Neural Netw. 2021, 144, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, W.; Zhang, J.; Xu, Z. An Improved K-Nearest Neighbour Method to Diagnose Breast Cancer. Analyst 2018, 143, 2807–2811. [Google Scholar] [CrossRef]
- Baria, E.; Cicchi, R.; Malentacchi, F.; Mancini, I.; Pinzani, P.; Pazzagli, M.; Pavone, F.S. Supervised Learning Methods for the Recognition of Melanoma Cell Lines through the Analysis of Their Raman Spectra. J. Biophotonics 2021, 14, e202000365. [Google Scholar] [CrossRef]
- Qiu, X.; Wu, X.; Fang, X.; Fu, Q.; Wang, P.; Wang, X.; Li, S.; Li, Y. Raman Spectroscopy Combined with Deep Learning for Rapid Detection of Melanoma at the Single Cell Level. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 286, 122029. [Google Scholar] [CrossRef]
- Qiu, S.; Li, M.; Liu, J.; Chen, X.; Lin, T.; Xu, Y.; Chen, Y.; Weng, Y.; Pan, Y.; Feng, S.; et al. Study on the Chemodrug-Induced Effect in Nasopharyngeal Carcinoma Cells Using Laser Tweezer Raman Spectroscopy. Biomed. Opt. Express 2020, 11, 1819. [Google Scholar] [CrossRef]
- Zhang, L.; Li, C.; Peng, D.; Yi, X.; He, S.; Liu, F.; Zheng, X.; Huang, W.E.; Zhao, L.; Huang, X. Raman Spectroscopy and Machine Learning for the Classification of Breast Cancers. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120300. [Google Scholar] [CrossRef]
- Hanlon, E.B.; Manoharan, R.; Koo, T.-W.; Shafer, K.E.; Motz, J.T.; Fitzmaurice, M.; Kramer, J.R.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Prospects for in Vivo Raman Spectroscopy. Phys. Med. Biol. 2000, 45, R1–R59. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Kendall, C.; Shepherd, N.; Crow, P.; Barr, H. Near-infrared Raman Spectroscopy for the Classification of Epithelial Pre-cancers and Cancers. J. Raman Spectrosc. 2002, 33, 564–573. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, L.; Xu, J.; Yu, Y.; Shen, L.; Gao, J.; Ye, A. Dynamic Characterization of Drug Resistance and Heterogeneity of the Gastric Cancer Cell BGC823 Using Single-Cell Raman Spectroscopy. Analyst 2017, 143, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Popp, J.; Bocklitz, T. Deep Learning for Raman Spectroscopy: A Review. Analytica 2022, 3, 287–301. [Google Scholar] [CrossRef]
- Lunter, D.; Klang, V.; Kocsis, D.; Varga-Medveczky, Z.; Berkó, S.; Erdő, F. Novel Aspects of Raman Spectroscopy in Skin Research. Exp. Dermatol. 2022, 31, 1311–1329. [Google Scholar] [CrossRef]
- Mazurenka, M.; Behrendt, L.; Meinhardt-Wollweber, M.; Morgner, U.; Roth, B. Development of a Combined OCT-Raman Probe for the Prospective in Vivo Clinical Melanoma Skin Cancer Screening. Rev. Sci. Instrum. 2017, 88, 105103. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Yi, J.-Y.; Hsu, T.-W.; Huang, S.-L. Integration of Cellular-Resolution Optical Coherence Tomography and Raman Spectroscopy for Discrimination of Skin Cancer Cells with Machine Learning. J. Biomed. Opt. 2023, 28, 096005. [Google Scholar] [CrossRef]
- Zakharov, V.P.; Bratchenko, I.A.; Artemyev, D.N.; Myakinin, O.O.; Kornilin, D.V.; Kozlov, S.V.; Moryatov, A.A. Comparative Analysis of Combined Spectral and Optical Tomography Methods for Detection of Skin and Lung Cancers. J. Biomed. Opt. 2015, 20, 25003. [Google Scholar] [CrossRef]
- Patil, C.A.; Kirshnamoorthi, H.; Ellis, D.L.; van Leeuwen, T.G.; Mahadevan-Jansen, A. A Clinical Instrument for Combined Raman Spectroscopy-Optical Coherence Tomography of Skin Cancers. Lasers Surg. Med. 2011, 43, 143–151. [Google Scholar] [CrossRef]
- Varkentin, A.; Mazurenka, M.; Blumenröther, E.; Behrendt, L.; Emmert, S.; Morgner, U.; Meinhardt-Wollweber, M.; Rahlves, M.; Roth, B. Trimodal System for in Vivo Skin Cancer Screening with Combined Optical Coherence tomography-Raman and Colocalized Optoacoustic Measurements. J. Biophotonics 2018, 11, e201700288. [Google Scholar] [CrossRef]
- Patil, C.A.; Kalkman, J.; Faber, D.J.; Nyman, J.S.; Van Leeuwen, T.G.; Mahadevan-Jansen, A. Integrated System for Combined Raman Spectroscopy–Spectral Domain Optical Coherence Tomography. J. Biomed. Opt. 2011, 16, 011007. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, S.; Akhtar, J.; Schartner, E.; Ebendorff-Heidepriem, H.; Mahadevan-Jansen, A.; Li, J. Multimodal Raman Spectroscopy and Optical Coherence Tomography for Biomedical Analysis. J. Biophotonics 2023, 16, e202200231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Moy, A.J.; Feng, X.; Nguyen, H.T.M.; Sebastian, K.R.; Reichenberg, J.S.; Wilke, C.O.; Markey, M.K.; Tunnell, J.W. Assessment of Raman Spectroscopy for Reducing Unnecessary Biopsies for Melanoma Screening. Molecules 2020, 25, 2852. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.M.; Moy, A.J.; Zhang, Y.; Feng, X.; Reichenberg, J.S.; Fox, M.; Tunnell, J.W. Tumor Margin Assessment in Mohs Surgery Using Reflectance, Fluorescence and Raman Spectroscopy. In Advanced Biomedical and Clinical Diagnostic and Surgical Guidance Systems XV, Proceedings of the Proceedings Volume 10054, San Francisco, CA, USA, 28 January–2 February 2017; Mahadevan-Jansen, A., Vo-Dinh, T., Grundfest, W.S., Eds.; SPIE: San Francisco, CA, USA, 2017; p. 1005403. [Google Scholar]
- Santos, I.P.; van Doorn, R.; Caspers, P.J.; Bakker Schut, T.C.; Barroso, E.M.; Nijsten, T.E.C.; Noordhoek Hegt, V.; Koljenović, S.; Puppels, G.J. Improving Clinical Diagnosis of Early-Stage Cutaneous Melanoma Based on Raman Spectroscopy. Br. J. Cancer 2018, 119, 1339–1346. [Google Scholar] [CrossRef]
- Sinjab, F.; Kong, K.; Gibson, G.; Varma, S.; Williams, H.; Padgett, M.; Notingher, I. Tissue Diagnosis Using Power-Sharing Multifocal Raman Micro-Spectroscopy and Auto-Fluorescence Imaging. Biomed. Opt. Express 2016, 7, 2993–3006. [Google Scholar] [CrossRef]
- Bratchenko, I.A.; Bratchenko, L.A.; Moryatov, A.A.; Khristoforova, Y.A.; Artemyev, D.N.; Myakinin, O.O.; Orlov, A.E.; Kozlov, S.V.; Zakharov, V.P. In Vivo Diagnosis of Skin Cancer with a Portable Raman Spectroscopic Device. Exp. Dermatol. 2021, 30, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, J.; Lui, H.; He, Q.; Zeng, H. In Vivo Near-infrared Autofluorescence Imaging of Pigmented Skin Lesions: Methods, Technical Improvements and Preliminary Clinical Results. Ski. Res. Technol. 2013, 19, 20–26. [Google Scholar] [CrossRef]
- Mahadevan-Jansen, A.; Richards-Kortum, R.R. Raman Spectroscopy for the Detection of Cancers and Precancers. J. Biomed. Opt. 1996, 1, 31–70. [Google Scholar] [CrossRef]
- Khristoforova, Y.; Bratchenko, I.; Bratchenko, L.; Moryatov, A.; Kozlov, S.; Kaganov, O.; Zakharov, V. Combination of Optical Biopsy with Patient Data for Improvement of Skin Tumor Identification. Diagnostics 2022, 12, 2503. [Google Scholar] [CrossRef]
- Khan, M.N.; Wang, Q.; Idrees, B.S.; Teng, G.; Xiangli, W.; Cui, X.; Wei, K. Evaluation of Human Melanoma and Normal Formalin Paraffin-Fixed Samples Using Raman and LIBS Fused Data. Lasers Med. Sci. 2022, 37, 2489–2499. [Google Scholar] [CrossRef]
- Kallaway, C.; Almond, L.M.; Barr, H.; Wood, J.; Hutchings, J.; Kendall, C.; Stone, N. Advances in the Clinical Application of Raman Spectroscopy for Cancer Diagnostics. Photodiagnosis Photodyn. Ther. 2013, 10, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Bohorfoush, A.G. Tissue Spectroscopy for Gastrointestinal Diseases. Endoscopy 1996, 28, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Bakker Schut, T.C.; Witjes, M.J.; Sterenborg, H.J.; Speelman, O.C.; Roodenburg, J.L.; Marple, E.T.; Bruining, H.A.; Puppels, G.J. In Vivo Detection of Dysplastic Tissue by Raman Spectroscopy. Anal. Chem. 2000, 72, 6010–6018. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.P.S.; Shu, C.; Zheng, W.; Lin, K.; Huang, Z. Advances in Real-time Fiber-optic Raman Spectroscopy for Early Cancer Diagnosis: Pushing the Frontier into Clinical Endoscopic Applications. Transl. Biophotonics 2021, 3, e202000018. [Google Scholar] [CrossRef]
- Zeng, H.; Zhao, J.; Short, M.; Mclean, D.I.; Lam, S.; Mcwilliams, A.; Lui, H. Raman Spectroscopy for In Vivo Tissue Analysis and Diagnosis, from Instrument Development to Clinical Applications. J. Innov. Opt. Health Sci. 2008, 1, 95–106. [Google Scholar] [CrossRef]
- Zhao, J.; Lui, H.; Kalia, S.; Zeng, H. Real-Time Raman Spectroscopy for Automatic in Vivo Skin Cancer Detection: An Independent Validation. Anal. Bioanal. Chem. 2015, 407, 8373–8379. [Google Scholar] [CrossRef]
| Technique | Applications | Key Findings | Ref |
|---|---|---|---|
| Raman spectroscopy | Skin cancer detection | Higher precision in ex vivo detection compared to in vivo. Sensitivity and specificity values reported for BCC, MM, and SCC. Ex vivo imaging revealed spatial distribution of tissue structures, but was time-intensive. | [7,22,23,24,25,26,27,28,29,30,31,32] |
| Near-infrared Raman spectroscopy | Skin tissue biochemical analysis | Identified key biochemicals like actin, collagen, elastin, and triolein. Classification model differentiated normal tissues from BCC and MM tissues. | [32] |
| Raman spectroscopy | Skin cancer diagnosis | PCA and Euclidean distance distinguished Raman spectra of BCC and MM from normal tissues in vitro, achieving a high diagnostic accuracy. | [22] |
| Raman spectroscopy | BCC diagnosis | Identified spectral variances between normal skin tissues and BCC-affected tissues. Diagnostic algorithms based on PCA and Mahalanobis distance identified tissue types effectively. | [28,40,41,42,43,44,45,46] |
| Coherent anti-Stokes Raman scattering assessments | BCC imaging | Produced pseudo H&E stained ex vivo microscopic imagery of BCC human skin specimens. | [47] |
| Raman microspectroscopy | SCC molecular analysis | Identified decrease in collagen intensities and increase in DNA and lipid intensities in SCC lesions. Stimulated Raman scattering microscopy revealed pathological characteristics within SCC tissues. | [48] |
| Spatially offset Raman spectroscopy | NMSC diagnosis | Effective for diagnosing NMSCs prior to histopathological examination. Identified specific wavenumbers associated with SCC and BCC. | [49] |
| Raman spectroscopy | BCC and SCC lipid and collagen analysis | Identified spectral characteristics distinguishing BCC and SCC groups based on lipid and collagen content. | [49] |
| Raman spectroscopy | Skin condition differentiation | Enhanced diagnostic accuracy in distinguishing among skin conditions using normal-subtracted preprocessing on Raman spectra. | [50] |
| Raman spectroscopy | Skin lesion diagnosis | Utilizing LASSO-based wavenumber selection with PCA and GDA analysis enhanced diagnostic performance in a merged cohort of skin lesions. | [51] |
| Raman spectroscopy | Differentiation of BCC, MM, and other conditions | MCR-ALS technique applied to analyze in vivo Raman spectra for differentiating BCC, MM, and other conditions. ROC AUC values reported for various discrimination models. | [52] |
| Raman spectroscopy imaging with MCR-ALS algorithm | MM and dysplastic nevi analysis | MM, dysplastic nevi, and compound nevus tumors. High sensitivity and specificity in identifying dysplastic nevi lesions and differentiating dysplastic nevi and MM samples. | [53] |
| Raman spectroscopy with silver-coated silicon nanowires | Genomic DNA differentiation in MM | Facilitated differentiation between healthy and cancerous genomic DNA extracted from normal human skin cells and MM cells. Achieved discrimination with no false-negative detections and a minimal false-positive rate of less than 2%. | [54] |
| Advanced Raman techniques (SRS and CARS) | Ex vivo characterization of NMSCs | Employing a multimodal methodology that includes coherent anti-Stokes Raman scattering, second harmonic generation, and two-photon excited fluorescence, distinctive morphological attributes of BCC and SCC were identified through H&E staining. Differences in coherent anti-Stokes Raman scattering imagery between BCC and SCC were noted, providing insights into lipid metabolism, bioenergetics, and tumor–stroma interactions. The amalgamated information aids in distinguishing healthy skin from NMSCs and between BCC and SCC. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delrue, C.; Speeckaert, R.; Oyaert, M.; De Bruyne, S.; Speeckaert, M.M. From Vibrations to Visions: Raman Spectroscopy’s Impact on Skin Cancer Diagnostics. J. Clin. Med. 2023, 12, 7428. https://doi.org/10.3390/jcm12237428
Delrue C, Speeckaert R, Oyaert M, De Bruyne S, Speeckaert MM. From Vibrations to Visions: Raman Spectroscopy’s Impact on Skin Cancer Diagnostics. Journal of Clinical Medicine. 2023; 12(23):7428. https://doi.org/10.3390/jcm12237428
Chicago/Turabian StyleDelrue, Charlotte, Reinhart Speeckaert, Matthijs Oyaert, Sander De Bruyne, and Marijn M. Speeckaert. 2023. "From Vibrations to Visions: Raman Spectroscopy’s Impact on Skin Cancer Diagnostics" Journal of Clinical Medicine 12, no. 23: 7428. https://doi.org/10.3390/jcm12237428





