Diagnostic Accuracy of FebriDx: A Rapid Test to Detect Immune Responses to Viral and Bacterial Upper Respiratory Infections
Abstract
:1. Introduction
2. Experimental Section
2.1. Design and URI Study Population
2.2. FebriDx Measurements
2.3. Outcome
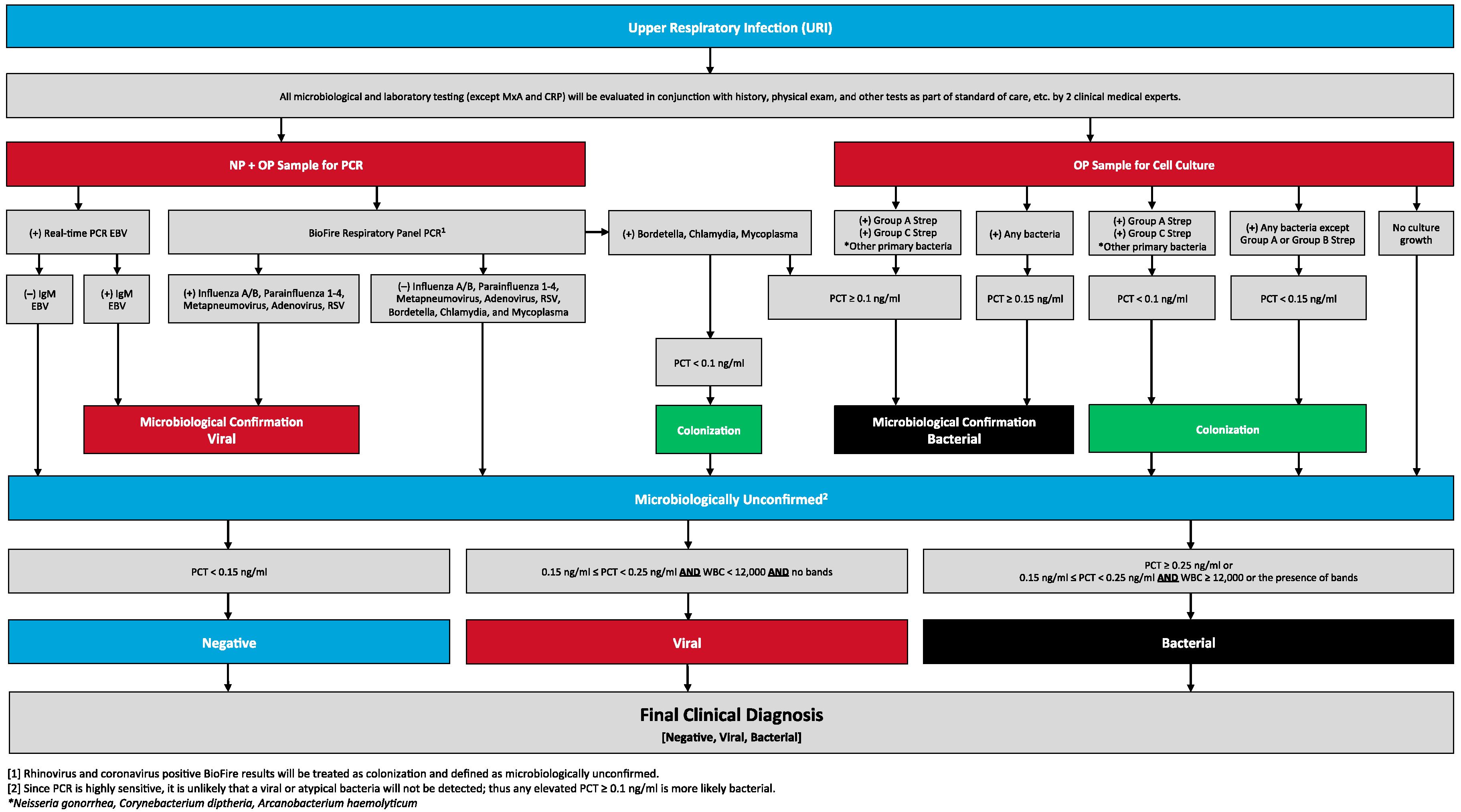
2.4. Reference Testing Algorithm
2.5. Physician Panel Over-Read
2.6. Statistical Analysis
2.7. Asymptomatic Control Population
3. Results
3.1. URI Study Population
3.1.1. Reference Standard Results
3.1.2. Diagnostic Accuracy of FebriDx
3.1.3. Asymptomatic Control Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shapiro, D.J.; Hick, L.A.; Pavia, A.T.; Hersh, A.L. Antibiotic prescribing for adults in ambulatory care in the USA, 2007–2009. J. Antimicrob. Chemother. 2014, 69, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Fleming-Dutra, K.E.; Hersh, A.L.; Shapiro, D.J. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010–2011. JAMA 2016, 315, 1864–1873. [Google Scholar] [CrossRef] [PubMed]
- Dellit, T.H.; Owens, R.C.; McGowan, J.E., Jr.; Gerding, D.N.; Weinstein, R.A.; Burke, J.P.; Huskins, W.C.; Paterson, D.L.; Fishman, N.O.; Carpenter, C.F.; et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin. Infect. Dis. 2007, 44, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Get Smart: Know when Antibiotics Work. Available online: http://www.cdc.gov/getsmart/community/index.html (accessed on 5 May 2016).
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A.; et al. Better tests, better care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013, 57, S139–S170. [Google Scholar] [CrossRef] [PubMed]
- FebriDx. RPS Diagnostics. Available online: https://www.rpsdetectors.com/in/products/febridx/ (accessed on 29 April 2016).
- Davidson, M. FebriDx Point-of-Care Testing to Guide Antibiotic Therapy for Acute Respiratory Tract Infection in UK Primary Care: A Retrospective Outcome Analysis. J. Infect. Dis. Prev. Med. 2017, 5, 165. [Google Scholar]
- Engelmann, I.; Dubos, F.; Lobert, P.E.; Houssin, C.; Degas, V.; Sardet, A.; Decoster, A.; Dewilde, A.; Martinot, A.; Hober, D. Diagnosis of viral infections using myxovirus resistance protein A (MxA). Pediatrics 2015, 135, e985–e993. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Gauvin, F.; Amre, D.K.; Saint-Louis, P.; Lacroix, J. Serum procalcitonin and c-reactive protein levels as markers of bacterial infection: A systematic review and meta-analysis. Clin. Infect. Dis. 2004, 39, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, M.; Kusano, A.; Furuya, A.; Hanai, N.; Tanigaki, H.; Tomita, A.; Horiguchi, A.; Nagata, K.; Itazawa, T.; Adachi, Y.; et al. New sandwich-type enzyme-linked immunosorbent assay for human MxA protein in a whole blood using monoclonal antibodies against GTP-binding domain for recognition of viral infection. J. Clin. Lab. Anal. 2012, 26, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Briel, M.; Mueller, B. Clinical outcomes associated with procalcitonin algorithms to guide antibiotic therapy in respiratory tract infections. JAMA 2013, 309, 717–718. [Google Scholar] [CrossRef] [PubMed]
- Musher, D.M.; Bebko, S.P.; Roig, I.L. Serum procalcitonin level, viral polymerase chain reaction analysis, and lower respiratory tract infection. J. Infect. Dis. 2014, 209, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Self, W.H.; Williams, D.J.; Zhu, Y.W.; Ampofo, K.; Pavia, A.T.; Chappell, J.D.; Hymas, W.C.; Stockmann, C.; Bramley, A.M.; Schneider, E.; et al. Respiratory viral detection in children and adults: Comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016, 213, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.R.; Wieringa, J.; Koekkoek, S.M.; Visser, C.E.; Pajkrt, D.; Molenkamp, R.; de Jong, M.D.; Schinkel, J. Frequent detection of respiratory viruses without symptoms: Toward defining clinically relevant cutoff values. J. Clin. Microbiol. 2011, 49, 2631–2636. [Google Scholar] [CrossRef] [PubMed]
- Jartti, T.; Jartti, L.; Peltola, V.; Maris, M.; Ruuskanen, O. Identification of respiratory viruses in asymptomatic subjects: Asymptomatic respiratory viral infections. Pediatr. Infect. Dis. J. 2008, 27, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- FilmArray Respiratory Panel. BioMerieux. Available online: http://www.biomerieux-diagnostics.com/filmarrayr-respiratory-panel (accessed on 29 April 2016).
- EBV-VCA IgM Enzyme Immunoassay Test Kit. Diamedix. Available online: http://diamedix.com/wp-content/uploads/2015/10/PI-EBVVCAIgM-720610-Rev7-June-15.pdf (accessed on 29 April 2016).
- BRAHMS PCT Sensitive KRYPTOR. Thermo Fisher Scientific. Available online: http://www.procalcitonin.com/ (accessed on 29 April 2016).
- Korppi, M.; Kroger, L.; Laitinen, M. White blood cell and differential counts in acute respiratory viral and bacterial infections in children. Scand. J. Infect. Dis. 1993, 25, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.R.; Marsocci, S.M.; Murphy, M.L.; Francis, A.B.; Pichichero, M.E. White blood cell count can aid judicious antibiotic prescribing in acute upper respiratory infections in children. Clin. Pediatr. 2003, 42, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, O.; Ewig, S.; Haagen, U.; Giersdorf, S.; Hartmann, O.; Wegscheider, K.; Hummers-Pradier, E.; Welte, T. Procalcitonin guidance and reduction of antibiotic use in acute respiratory tract infection. Eur. Respir. J. 2010, 36, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Briel, M.; Schuetz, P.; Mueller, B.; Young, J.; Schildm, U.; Nusbaumer, C.; Périat, P.; Bucher, H.C.; Christ-Crain, M. Procalcitonin-guided antibiotic use vs a standard approach for acute respiratory tract infections in primary care. Arch. Intern. Med. 2008, 168, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Dandona, P.; Nix, D.; Wilson, M.F.; Aljada, A.; Love, J.; Assicot, M. Procalcitonin increase after endotoxin injection in normal subjects. J. Clin. Endocrinol. Metab. 1994, 79, 1605–1608. [Google Scholar] [PubMed]
- Stolz, D.; Christ-Crain, M.; Gencay, M.M.; Bingisser, R.; Huber, P.R.; Müller, B.; Tamm, M. Diagnostic value of signs, symptoms and laboratory values in lower respiratory tract infection. Swiss Med. Wkly. 2006, 136, 434–440. [Google Scholar] [PubMed]
- Elsammak, M.; Hanna, H.; Ghazal, A.; Edeen, F.B.; Kandil, M. Diagnostic value of serum procalcitonin and C-reactive protein in Egyptian children with streptococcal tonsillopharyngitis. Pediatr. Infect. Dis. J. 2006, 25, 174–176. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.M.; Thomsen, M.K.; Ovesen, T.; Klug, T.E. Are procalcitonin or other infection markers useful in the detection of group A streptococcal acute tonsillitis? Scand. J. Infect. Dis. 2014, 46, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Reichenheim, M.E. Confidence intervals for the kappa statistic. Stata J. 2004, 4, 421–428. [Google Scholar]
- Stewart, E.H.; Davis, B.; Clemans-Taylor, B.L.; Littenberg, B.; Estrada, C.A.; Centor, R.M. Rapid antigen Group A Streptococcus Test to diagnose pharyngitis: A systematic review and meta-analysis. PLoS ONE 2014, 9, e111727. [Google Scholar] [CrossRef] [PubMed]
- Dingle, T.C.; Abbott, A.N.; Fang, F.C. Reflexive culture in adolescents and adults with Group A Streptococcal pharyngitis. Clin. Infect. Dis. 2014, 59, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Inverness Medical: BinaxNow Influenza A&B CLIA Waived (Package Insert Training Packet). Available online: http://www.amms.net/images/healthcare_industry_clia_binax_now_influenza_test_2009.pdf (accessed on 02 June 2016).
- Uyeki, T.M.; Prasad, R.; Vukotich, C.; Stebbins, S.; Rinaldo, C.; Ferng, Y.H.; Morse, S.S.; Larson, E.L.; Aiello, A.E.; Davis, B.; et al. Low sensitivity of rapid diagnostics tests for influenza. Clin. Infect. Dis. 2009, 48, e89–e92. [Google Scholar] [CrossRef] [PubMed]
- Self, W.H.; McNaughton, C.D.; Grijalva, C.G.; Zhu, Y.; Chappell, J.D.; Williams, J.V.; Talbot, H.K.; Shay, D.K.; Griffin, M.R. Diagnostic performance of the BinaxNow Influenza A&B rapid antigen test in ED patients. Am. J. Emerg. Med. 2012, 30, 1955–1961. [Google Scholar] [PubMed]
- Andreola, B.; Bressan, S.; Callegaro, S.; Liverani, A.; Plebani, M.; Da Dalt, L. Procalcitonin and C-reactive protein as diagnostic markers of severe bacterial infections in febrile infants and children in the emergency department. Pediatr. Infect. Dis. J. 2007, 26, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Bafadhel, M.; Clark, T.W.; Reid, C.; Medina, M.J.; Batham, S.; Barer, M.R.; Nicholson, K.G.; Brightling, C.E. Procalcitonin and c-reactive protein in hospitalized adult patients with community-acquired pneumonia or exacerbation of asthma or COPD. Chest 2011, 139, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Sambursky, R.; Shapiro, N.I. Evaluation of a combined MxA and CRP point-of-care immunoassay to identify viral and/or bacterial immune response in patients with acute febrile respiratory infection. Eur. Clin. Respir. J. 2015, 2, 28245. [Google Scholar] [CrossRef] [PubMed]
- Putto, A.; Ruuskanen, O.; Meurman, O.; Ekblad, H.; Korvenranta, H.; Mertsola, J.; Peltola, H.; Sarkkinen, H.; Viljanen, M.K.; Halonen, P. C reactive protein in the evaluation of febrile illness. Arch. Dis. Child. 1986, 61, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Hatherill, M.; Tibby, S.M.; Sykes, K.; Turner, C.; Murdoch, I.A. Diagnostic markers of infection: Comparison of procalcitonin with c reactive protein and leukocyte count. Arch. Dis. Child. 1999, 81, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Roers, A.; Hochkeppel, H.K.; Horisberger, M.A.; Hovanessian, A.; Haller, O. MxA gene expression after live virus vaccination: A sensitive marker for endogenous type I interferon. J. Infect. Dis. 1994, 169, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Staeheli, P.; Pitossi, F.; Pa vlovic, J. Mx proteins: GTPases with antiviral activity. Trends Cell Biol. 1993, 3, 268–272. [Google Scholar] [CrossRef]
- Staeheli, P. Interferon induced proteins and the antiviral state. Adv. Virus Res. 1990, 38, 147–200. [Google Scholar] [PubMed]
- Chieux, V.; Hober, D.; Harvey, J.; Lion, G.; Lucidarme, D.; Forzy, G.; Duhamel, M.; Cousin, J.; Ducoulombier, H.; Wattré, P. The MxA protein levels in while blood lysates of patients with various viral infections. J. Virol. Methods 1998, 70, 183–191. [Google Scholar] [CrossRef]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balket, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, M.; et al. Community-acquired pneumonia requiring hospitalization among US adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Branche, A.R.; Walsh, E.E.; Vargas, R.; Hulbert, B.; Formica, M.A.; Baran, A.; Peterson, D.R.; Falsey, A.R. Serum procalcitonin measurement and viral testing to guide antibiotic use for respiratory infections in hospitalized adults: A randomized controlled trial. J. Infect. Dis. 2015, 212, 1692–1700. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.A.; Biesbroek, G.; Trzcinski, K.; Sanders, E.A.; Bogaert, D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013, 9, e1003057. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L.; Top, F.H., Jr.; Dudding, B.A.; Wannamaker, L.W. Diagnosis of streptococcal pharyngitis: Differentiation of active infection from the carrier state in the symptomatic child. J. Infect. Dis. 1971, 123, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L. The group A streptococcal upper respiratory tract carrier state: An enigma. J. Pediatr. 1980, 97, 337–345. [Google Scholar] [CrossRef]
- Gerber, M.A.; Randolph, M.F.; Mayo, D.R. The group A streptococcal carrier state. A reexamination. Am. J. Dis. Child. 1988, 142, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L.; Pass, T.M.; Aronson, M.D.; Ervin, C.T.; Cretin, S.; Winickoff, R.N.; Branch, W.T., Jr. The prediction of streptococcal pharyngitis in adults. J. Gen. Intern. Med. 1986, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.A.; Randolph, M.F.; Chanatry, J.; Wright, L.L.; DeMeo, K.K.; Anderson, L.R. Antigen detection test for streptococcal pharyngitis: Evaluation of sensitivity with respect totrue infection. J. Pediatr. 1986, 108, 654–658. [Google Scholar] [CrossRef]
- Nussinovitch, M.; Finkelstein, Y.; Amir, J.; Varsano, I. Group A beta-hemolytic streptococcal pharyngitis in preschool children aged 3 months to 5 years. Clin. Pediatr. 1999, 38, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Ivaska, L.; Niemelä, J.; Lempainen, J.; Österback, R.; Warism, M.; Vuorinen, T.; Hytönen, J.; Rantakokko-Jalava, K.; Peltola, V. Aetiology of febrile pharyngitis in children: Potential of myxovirus resistance protein A (MxA) as a biomarker of viral infection. J. Infect. 2017, 74, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, M.J.; Puhakka, T.; Ruuskanen, O.; Leinonen, M.; Saikku, P.; Kimpimäki, M.; Blomqvist, S.; Hyypiä, T.; Arstila, P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998, 36, 539–542. [Google Scholar] [PubMed]
- Lingard, H.; Zehetmayer, S.; Maier, M. Bacterial superinfection in upper respiratory tract infections estimated by increases in CRP values: A diagnostic follow-up in primary care. Scand. J. Prim. Health Care 2008, 26, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Salonen, E.M.; Vaheri, A. C-Reactive protein in acute viral infections. J. Med. Virol. 1981, 8, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Sarov, I.; Shainkin-Kestenbaum, R.; Zimlichman, R.; Winikoff, Y.; Chaimovitz, C.; Pras, M. Serum amyloid A levels in patients with infections due to cytomegalovirus, varicella-zoster virus and Herpes simplex virus. J. Infect. Dis. 1982, 146, 443. [Google Scholar] [CrossRef] [PubMed]
- Whicher, J.T.; Chambers, R.E.; Higginson, J.; Nashef, L.; Higgins, P.G. Acute phase response of serum amyloid A protein and C reactive protein to the common cold and influenza. J. Clin. Pathol. 1985, 38, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Melbye, H.; Hvidsten, D.; Holm, A.; Nordbø, S.A.; Brox, J. The course of C-reactive protein response in untreated upper respiratory tract infection. Br. J. Gen. Pract. 2004, 54, 653–658. [Google Scholar] [PubMed]
- Holm, A.; Pedersen, S.S.; Nexoe, J.; Obel, N.; Nielsen, L.P.; Koldkjaer, O.; Pedersen, C. Procalcitonin versus C-reactive protein for predicting pneumonia in adults with lower respiratory tract infection in primary care. Br. J. Gen. Pract. 2007, 57, 555–560. [Google Scholar] [PubMed]
- Kutz, A.; Briel, M.; Christ-Crain, M.; Stolz, D.; Bouadma, L.; Wolff, M.; Kristoffersen, K.B.; Wei, L.; Burkhardt, O.; Welte, T.; et al. Prognostic value of procalcitonin in respiratory tract infections across clinical settings. Crit. Care 2015, 19, 74. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L.; Wannamaker, L.W. C-reactive protein in streptococcal pharyngitis. Pediatrics 1977, 60, 28–32. [Google Scholar] [PubMed]
- Wannamaker, L.W.; Ayoub, E.M. Antibody titers in acute rheumatic fever. Circulation 1960, 21, 598–614. [Google Scholar] [CrossRef] [PubMed]
- Hanson, L.A.; Jodal, U.; Sabel, K.G.; Wadsworth, C. The diagnostic value of C-reactive protein. Pediatr. Infect. Dis. 1983, 2, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Steurer, J.; Held, U.; Spaar, A.; Bausch, B.; Zoller, M.; Hunziker, R.; Bachmann, L.M. A decision aid to rule out pneumonia and reduce unnecessary prescriptions of antibiotics in primary care patients with cough and fever. BMC Med. 2011, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valkenburg, H.A.; Haverkorn, M.J.; Goslings, W.R. Streptococcal pharyngitis in the general population. II. The attack rate of rheumatic fever and acute glomerulonephritis in patients. J. Infect. Dis. 1971, 124, 348–358. [Google Scholar] [CrossRef] [PubMed]
- National Clinical Guideline Centre (UK). Pneumonia: Diagnosis and management of community and hospital acquired pneumonia in Adults. Clin. Guidel. 2014. [Google Scholar] [PubMed]
- Huang, Y.; Chen, R.; Wu, T.; Wei, X.; Guo, A. Association between point-of-care CRP testing and antibiotic prescribing in respiratory tract infections: A systematic review and meta-analysis of primary care studies. Br. J. Gen. Pract. 2013, 63, e787–e794. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.; Butler, C.; Hopstaken, R.; Dryden, M.S.; McNulty, C.; Hurding, S.; Moore, M.; Livermore, D.M. Narrative review of primary care point-of-care testing (POCT) and antibacterial use in respiratory tract infection (RTI). BMJ Open Respir. Res. 2015, 2, e000086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cals, J.W.L.; Butler, C.C.; Hopstaken, R.M.; Hood, K.; Dinant, G.-J. Effect of point of care testing for C reactive protein and training in communication skills on antibiotic use in lower respiratory tract infections: Cluster randomised trial. BMJ 2009, 338, b1374. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Chiappa, V.; Briel, M.; Greenwald, J.L. Procalcitonin algorithms for antibiotic therapy decisions: A systematic review of randomized controlled trials and recommendations for clinical algorithms. Arch. Intern. Med. 2011, 171, 1322–1331. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | URI Population (n = 205) | Asymptomatic Control Population (n = 163) |
|---|---|---|
| Female Gender, n (%) | 123 (60.0) | 90 (55.2) |
| Median age, years (IQR) | 29 (15, 46) | 44 (29, 55) |
| Age Groups, n (%) | ||
| <18 years | 56 (27.3) | 5 (3.1) |
| 18–50 years | 112 (54.6) | 99 (60.7) |
| >50 years | 37 (18.1) | 59 (36.2) |
| Race, n (%) | ||
| White | 142 (69.3) | 134 (82.2) |
| Black | 46 (22.4) | 14 (8.6) |
| Other | 17 (8.3) | 15 (9.2) |
| Hispanic Ethnicity, n (%) | 88 (42.9) | 64 (39.3) |
| Median body temperature, °F (IQR) | 101.5 (101.0, 102.1) | n/a |
| Median white blood cell count, thousand cells/mcL (IQR) | 7.8 (5.9, 10.3) | n/a |
| Median serum procalcitonin, ng/mL (IQR) | <0.05 (<0.05, 0.07) | n/a |
| Median CRP, mg/L (IQR) | 12.1 (3.8, 45.2) | 2.0 (1.0, 4.0) |
| Median MxA, ng/mL (IQR) | 4.9 (0.65, 20.2) | <1.0 (0.03, 2.0) |
| Reference Standard Classification Criterion | n | Organisms Detected |
|---|---|---|
| Bacterial | 25 | |
| Throat culture positive for bacteria commonly causing pharyngitis and PCT ≥ 0.10 ng/mL | 9 | 6 Group A Streptococcus 3 Group C Streptococcus |
| Throat culture positive for other bacteria and PCT ≥ 0.15 ng/mL | 2 | 1 Haemophilus parainfluenza 1 Enterobacter cloacae |
| NP/OP PCR positive for atypical bacteria and PCT ≥ 0.10 ng/mL | 1 | 1 Chlamydophila pneumoniae |
| Throat culture negative or contaminant, PCR negative, and PCT PCT ≥ 0.25 ng/mL | 5 | 5 none |
| Throat culture negative or contaminant, PCR negative, and PCT between 0.15 ng/mL and 0.25 ng/mL, and WBC > 15 k or bands present | 6 | 6 none |
| Physician panel over-read classified as bacterial after algorithm suggested negative | 2 | 1 Group A Streptococcus 1 none |
| Viral | 53 | |
| NP/OP positive PCR for pathogenic virus | 47 | 33 influenza 9 parainfluenza 3 RSV 1 hMPV 1 hMPV and parainfluenza |
| NP/OP positive PCR and IgM positive for EBV | 3 | 3 EBV |
| Throat culture negative or contaminant, PCR negative or contaminant , and PCT between 0.15 ng/mL and 0.25 ng/mL, and WBC < 15 k, and no bands present | 3 | 1 none 1 Enterobacter gergoviae 1 Group B Streptococcus |
| Negative | 127 | |
| Throat culture negative or contaminant, P/OP PCR negative, and PCT < 0.15 ng/mL | 79 | 79 none |
| Throat culture positive for bacteria commonly causing pharyngitis and PCT < 0.10 ng/mL | 11 | 10 Group A Streptococcus 1 Group C Streptococcus |
| Throat culture positive for other bacteria and PCT < 0.15 ng/mL | 37 | 18 S. aureus 3 Group B Streptococcus 3 Klebsiella pneumoniae 3 Group B Strep and S. aureus 3 Enterobacter sp. 2 Streptococcus pneumoniae 2 Proteus mirabilis 1 Citrobacter freundi 1 M. catarrhalis and S. aureus 1 Anicetobacter sp and S. aureus |
| FebriDx Test | Reference Standard | Total | |||
|---|---|---|---|---|---|
| Test | FebriDx Result | Bacterial | Viral | Negative | |
| FebriDx | Bacterial | 20 | 1 | 11 | 32 |
| Viral | 1 | 46 | 25 | 72 | |
| Negative | 4 | 6 | 91 | 101 | |
| Total | 25 | 53 | 127 | 205 | |
| Population | Total n | Bacterial/Viral by Reference Standard, n (%) | FebriDx Test Characteristics | ||||
|---|---|---|---|---|---|---|---|
| Sens (%) (95% CI) | Spec (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | ||||
| (A) Bacterial | |||||||
| Full URI population | 205 | 25 (12%) | 80% (59–93%) | 93% (90–97%) | 63% (45–79%) | 97% (94–99%) | |
| Age < 18 | 56 | 5 (8.9%) | 60% (16–95%) | 100% (94–100%) * | 100% (30–100%) * | 96% (88–100%) | |
| Age 18–50 | 112 | 13 (12%) | 85% (56–98%) | 90% (84–96%) | 55% (33–77%) | 98% (93–100%) | |
| Age > 50 | 37 | 7 (19%) | 86% (43–100%) | 90% (75–98%) | 67% (31–93%) | 96% (83–100%) | |
| (B) Viral | |||||||
| Full URI population | 205 | 53 (26%) | 87% (75–95%) | 83% (77–89%) | 64% (53–75%) | 95% (90–98%) | |
| Age <18 | 56 | 25 (45%) | 96% (80–100%) | 55% (37–73%) | 63% (47–78%) | 94% (73–100%) | |
| Age 18–50 | 112 | 21 (19%) | 76% (53–92%) | 87% (80–93%) | 57% (38–76%) | 94% (87–98%) | |
| Age >50 | 37 | 7 (19%) | 86% (42–100%) | 100% (89–100%)* | 100% (54–100%) * | 97% (83–100%) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Self, W.H.; Rosen, J.; Sharp, S.C.; Filbin, M.R.; Hou, P.C.; Parekh, A.D.; Kurz, M.C.; Shapiro, N.I. Diagnostic Accuracy of FebriDx: A Rapid Test to Detect Immune Responses to Viral and Bacterial Upper Respiratory Infections. J. Clin. Med. 2017, 6, 94. https://doi.org/10.3390/jcm6100094
Self WH, Rosen J, Sharp SC, Filbin MR, Hou PC, Parekh AD, Kurz MC, Shapiro NI. Diagnostic Accuracy of FebriDx: A Rapid Test to Detect Immune Responses to Viral and Bacterial Upper Respiratory Infections. Journal of Clinical Medicine. 2017; 6(10):94. https://doi.org/10.3390/jcm6100094
Chicago/Turabian StyleSelf, Wesley H., Jeffrey Rosen, Stephan C. Sharp, Michael R. Filbin, Peter C. Hou, Amisha D. Parekh, Michael C. Kurz, and Nathan. I. Shapiro. 2017. "Diagnostic Accuracy of FebriDx: A Rapid Test to Detect Immune Responses to Viral and Bacterial Upper Respiratory Infections" Journal of Clinical Medicine 6, no. 10: 94. https://doi.org/10.3390/jcm6100094





