Technical Advances in the Measurement of Residual Disease in Acute Myeloid Leukemia
Abstract
:1. Introduction
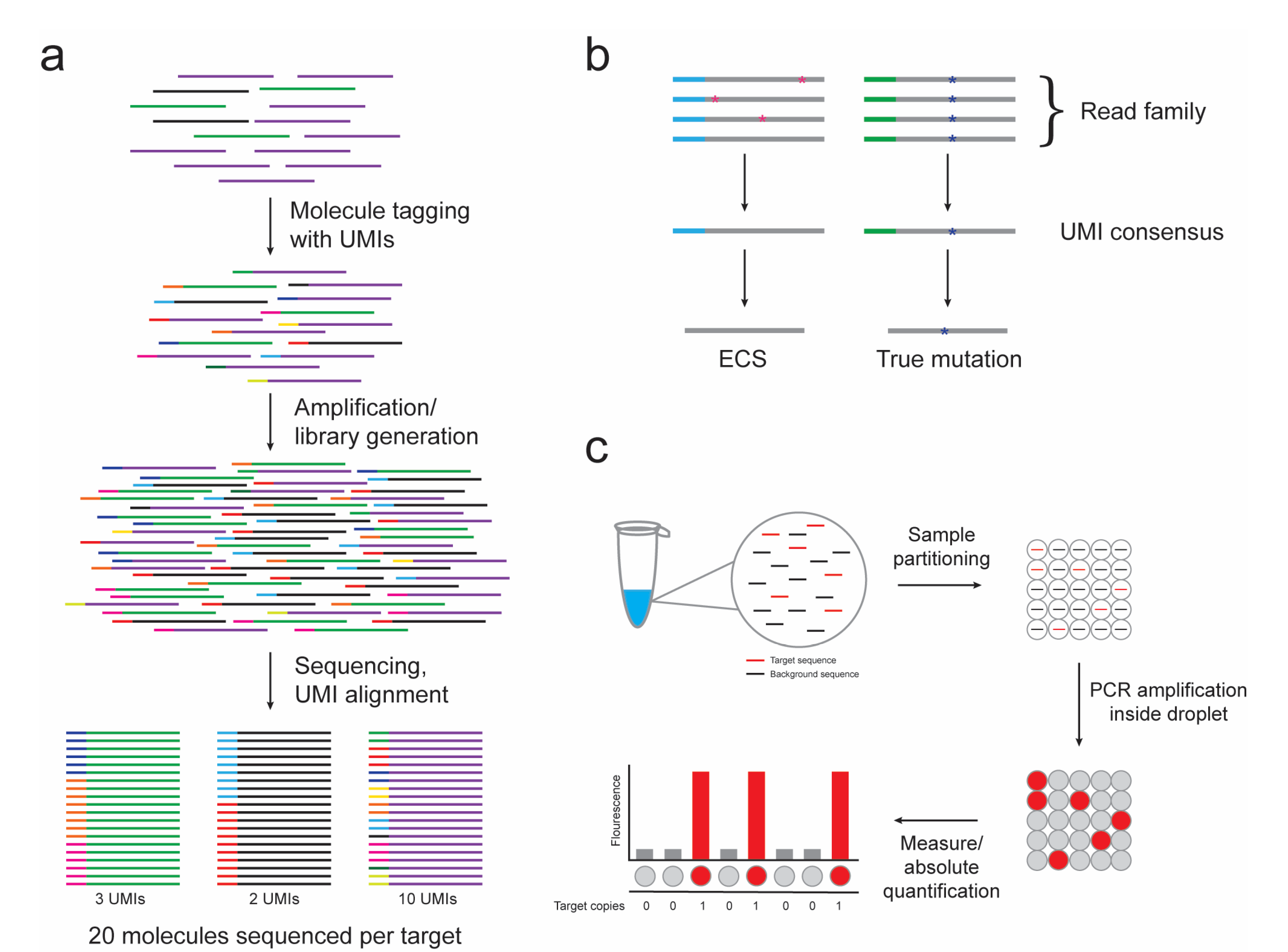
2. Next-Generation Sequencing
2.1. DNA Sequencing
2.2. Error-Corrected Sequencing
2.3. RNA Sequencing
3. Digital PCR
4. Novel Imaging Technologies
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014, 506, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.R.; Holland, J.F.; Glidewell, O.J.; Weinberg, V.; Brunner, K.; Obrecht, J.P.; Preisler, H.D.; Nawabi, I.W.; Prager, D.; Carey, R.W.; et al. Treatment of acute myelocytic leukemia: A study by cancer and leukemia group B. Blood 1981, 58, 1203–1212. [Google Scholar] [PubMed]
- Fernandez, H.F.; Sun, Z.; Yao, X.; Litzow, M.R.; Luger, S.M.; Paietta, E.M.; Racevskis, J.; Dewald, G.W.; Ketterling, R.P.; Bennett, J.M.; et al. Anthracycline dose intensification in acute myeloid leukemia. N. Engl. J. Med. 2009, 361, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Petersdorf, S.H.; Kopecky, K.J.; Slovak, M.; Willman, C.; Nevill, T.; Brandwein, J.; Larson, R.A.; Erba, H.P.; Stiff, P.J.; Stuart, R.K.; et al. A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood 2013, 121, 4854–4860. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.H. Acute myeloid leukemia: 2014 Update on risk-stratification and management. Am. J. Hematol. 2014, 89, 1063–1081. [Google Scholar] [CrossRef] [PubMed]
- Cheson, B.D.; Bennett, J.M.; Kopecky, K.J.; Buchner, T.; Willman, C.L.; Estey, E.H.; Schiffer, C.A.; Doehner, H.; Tallman, M.S.; Lister, T.A.; et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J. Clin. Oncol. 2003, 21, 4642–4649. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.S.; Shirakawa, S.; Trujillo, J.; Frei, E. The mechanism of induction of complete remission in acute myeloblastic leukemia in man. Cancer Res. 1969, 29, 2300–2307. [Google Scholar] [PubMed]
- Hourigan, C.S.; Karp, J.E. Minimal residual disease in acute myeloid leukaemia. Nat. Rev. Clin. Oncol. 2013, 10, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Renneville, A.; Hermitte, F.; Hills, R.K.; Daly, S.; Jovanovic, J.V.; Gottardi, E.; Fava, M.; Schnittger, S.; Weiss, T.; et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: A European LeukemiaNet study. J. Clin. Oncol. 2009, 27, 5195–5201. [Google Scholar] [CrossRef] [PubMed]
- Mule, M.P.; Mannis, G.N.; Wood, B.L.; Radich, J.P.; Hwang, J.; Ramos, N.R.; Andreadis, C.; Damon, L.; Logan, A.C.; Martin, T.G.; et al. Multigene Measurable Residual Disease Assessment Improves Acute Myeloid Leukemia Relapse Risk Stratification in Autologous Hematopoietic Cell Transplantation. Biol. Blood Marrow Transplant. 2016, 22, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Jovanovic, J.V.; Hills, R.K.; Nugent, E.A.; Patel, Y.; Flora, R.; Diverio, D.; Jones, K.; Aslett, H.; Batson, E.; et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J. Clin. Oncol. 2009, 27, 3650–3658. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Arif, M.; Eguchi, M.; Kumaravel, T.S.; Ueda, R.; Ohno, R.; Iwato, K.; Kyo, T.; Dohy, H.; Kamada, N. Application of fluorescence in situ hybridization to detect residual leukemic cells with 9;22 and 15;17 translocations. Leukemia 1997, 11, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.B.; Gooley, T.A.; Wood, B.L.; Milano, F.; Fang, M.; Sorror, M.L.; Estey, E.H.; Salter, A.I.; Lansverk, E.; Chien, J.W.; et al. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J. Clin. Oncol. 2011, 29, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.A.; Wood, B.L.; Othus, M.; Hourigan, C.S.; Ustun, C.; Linden, M.A.; DeFor, T.E.; Malagola, M.; Anthias, C.; Valkova, V.; et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: A meta-analysis. Haematologica 2017, 102, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Araki, D.; Wood, B.L.; Othus, M.; Radich, J.P.; Halpern, A.B.; Zhou, Y.; Mielcarek, M.; Estey, E.H.; Appelbaum, F.R.; Walter, R.B. Allogeneic Hematopoietic Cell Transplantation for Acute Myeloid Leukemia: Time to Move Toward a Minimal Residual Disease-Based Definition of Complete Remission? J. Clin. Oncol. 2016, 34, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Hourigan, C.S.; Goswami, M.; Battiwalla, M.; Barrett, A.J.; Sheela, S.; Karp, J.E.; Lai, C. When the Minimal Becomes Measurable. J. Clin. Oncol. 2016, 34, 2557–2558. [Google Scholar] [CrossRef] [PubMed]
- Gabert, J.; Beillard, E.; van der Velden, V.H.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cave, H.; et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—A Europe Against Cancer program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef] [PubMed]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dongen, J.J.; Macintyre, E.A.; Gabert, J.A.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.A.; O’Brien, M.A.; Hills, R.K.; Daly, S.B.; Wheatley, K.; Burnett, A.K. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: Results of the United Kingdom MRC AML-15 trial. Blood 2012, 120, 2826–2835. [Google Scholar] [CrossRef] [PubMed]
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S.; Ferrara, F.; Fazi, P.; Cicconi, L.; Di Bona, E.; et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013, 369, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Mardis, E.R.; Ding, L.; Fulton, B.; McLellan, M.D.; Chen, K.; Dooling, D.; Dunford-Shore, B.H.; McGrath, S.; Hickenbotham, M.; et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Thol, F.; Kolking, B.; Damm, F.; Reinhardt, K.; Klusmann, J.H.; Reinhardt, D.; von Neuhoff, N.; Brugman, M.H.; Schlegelberger, B.; Suerbaum, S.; et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosom. Cancer 2012, 51, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Kohlmann, A.; Nadarajah, N.; Alpermann, T.; Grossmann, V.; Schindela, S.; Dicker, F.; Roller, A.; Kern, W.; Haferlach, C.; Schnittger, S.; et al. Monitoring of residual disease by next-generation deep-sequencing of RUNX1 mutations can identify acute myeloid leukemia patients with resistant disease. Leukemia 2014, 28, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Getta, B.M.; Devlin, S.M.; Levine, R.L.; Arcila, M.E.; Mohanty, A.S.; Zehir, A.; Tallman, M.S.; Giralt, S.A.; Roshal, M. Multicolor Flow Cytometry and Multigene Next-Generation Sequencing Are Complementary and Highly Predictive for Relapse in Acute Myeloid Leukemia after Allogeneic Transplantation. Biol. Blood Marrow Transplant. 2017, 23, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Klco, J.M.; Miller, C.A.; Griffith, M.; Petti, A.; Spencer, D.H.; Ketkar-Kulkarni, S.; Wartman, L.D.; Christopher, M.; Lamprecht, T.L.; Helton, N.M.; et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA 2015, 314, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.; Li, Z.; Fung, H.L.; Young, J.E.; Agarwal, S.; Antosiewicz-Bourget, J.; Canto, I.; Giorgetti, A.; Israel, M.A.; Kiskinis, E.; et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 2011, 471, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Dohm, J.C.; Lottaz, C.; Borodina, T.; Himmelbauer, H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008, 36, e105. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.H.; Mo, T.; Dong, W.; Lee, G.Q.; Swenson, L.C.; McCloskey, R.M.; Woods, C.K.; Brumme, C.J.; Ho, C.K.; Schinkel, J.; et al. Theoretical and experimental assessment of degenerate primer tagging in ultra-deep applications of next-generation sequencing. Nucleic Acids Res. 2014, 42, e98. [Google Scholar] [CrossRef] [PubMed]
- Jabara, C.B.; Jones, C.D.; Roach, J.; Anderson, J.A.; Swanstrom, R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. USA 2011, 108, 20166–20171. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.K.; Hu, J.; Wang, P.H.; Fodor, S.P. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc. Natl. Acad. Sci. USA 2011, 108, 9026–9031. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Minkah, N.; Leipzig, J.; Wang, G.; Arens, M.Q.; Tebas, P.; Bushman, F.D. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007, 35, e91. [Google Scholar] [CrossRef] [PubMed]
- Kivioja, T.; Vaharautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2011, 9, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Shiroguchi, K.; Jia, T.Z.; Sims, P.A.; Xie, X.S. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl. Acad. Sci. USA 2012, 109, 1347–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Young, A.L.; Wong, T.N.; Hughes, A.E.; Heath, S.E.; Ley, T.J.; Link, D.C.; Druley, T.E. Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia 2015, 29, 1608–1611. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.W.; Kennedy, S.R.; Salk, J.J.; Fox, E.J.; Hiatt, J.B.; Loeb, L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 14508–14513. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, D.; Bader, P.; Willasch, A.; Bartholomae, S.; Debatin, K.M.; Zimmermann, M.; Creutzig, U.; Reinhardt, D.; Gruhn, B. Prospective validation of a new method of monitoring minimal residual disease in childhood acute myelogenous leukemia. Clin. Cancer Res. 2015, 21, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; McGowan, K.S.; Lu, K.; Jain, N.; Candia, J.; Hensel, N.F.; Tang, J.; Calvo, K.R.; Battiwalla, M.; Barrett, A.J.; et al. A multigene array for measurable residual disease detection in AML patients undergoing SCT. Bone Marrow Transplant. 2015, 50, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Masetti, R.; Pigazzi, M.; Togni, M.; Astolfi, A.; Indio, V.; Manara, E.; Casadio, R.; Pession, A.; Basso, G.; Locatelli, F. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood 2013, 121, 3469–3472. [Google Scholar] [CrossRef] [PubMed]
- Togni, M.; Masetti, R.; Pigazzi, M.; Astolfi, A.; Zama, D.; Indio, V.; Serravalle, S.; Manara, E.; Bisio, V.; Rizzari, C.; et al. Identification of the NUP98-PHF23 fusion gene in pediatric cytogenetically normal acute myeloid leukemia by whole-transcriptome sequencing. J. Hematol. Oncol. 2015, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Li, Y.; Malek, S.N.; Kim, Y.C.; Xu, J.; Chen, P.; Xiao, F.; Huang, X.; Zhou, X.; Xuan, Z.; et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS ONE 2012, 7, e51203. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.; Qin, J.; Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 2008, 3, e2876. [Google Scholar] [CrossRef] [PubMed]
- Kinz, E.; Leiherer, A.; Lang, A.H.; Drexel, H.; Muendlein, A. Accurate quantitation of JAK2 V617F allele burden by array-based digital PCR. Int J. Lab. Hematol. 2015, 37, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Olmedillas-Lopez, S.; Garcia-Arranz, M.; Garcia-Olmo, D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol. Diagn. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Livak, K.J.; Flood, S.J.; Marmaro, J.; Giusti, W.; Deetz, K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995, 4, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Brambati, C.; Galbiati, S.; Xue, E.; Toffalori, C.; Crucitti, L.; Greco, R.; Sala, E.; Crippa, A.; Chiesa, L.; Soriani, N.; et al. Droplet digital polymerase chain reaction for DNMT3A and IDH1/2 mutations to improve early detection of acute myeloid leukemia relapse after allogeneic hematopoietic stem cell transplantation. Haematologica 2016, 101, e157–e161. [Google Scholar] [CrossRef] [PubMed]
- Mencia-Trinchant, N.; Hu, Y.; Alas, M.A.; Ali, F.; Wouters, B.J.; Lee, S.; Ritchie, E.K.; Desai, P.; Guzman, M.L.; Roboz, G.J.; et al. Minimal Residual Disease Monitoring of Acute Myeloid Leukemia by Massively Multiplex Digital PCR in Patients with NPM1 Mutations. J. Mol. Diagn. 2017, 19, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Percival, M.E.; Lai, C.; Estey, E.; Hourigan, C.S. Bone marrow evaluation for diagnosis and monitoring of acute myeloid leukemia. Blood Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ganzel, C.; Manola, J.; Douer, D.; Rowe, J.M.; Fernandez, H.F.; Paietta, E.M.; Litzow, M.R.; Lee, J.W.; Luger, S.M.; Lazarus, H.M.; et al. Extramedullary Disease in Adult Acute Myeloid Leukemia Is Common but Lacks Independent Significance: Analysis of Patients in ECOG-ACRIN Cancer Research Group Trials, 1980–2008. J. Clin. Oncol. 2016, 34, 3544–3553. [Google Scholar] [CrossRef] [PubMed]
- Stolzel, F.; Rollig, C.; Radke, J.; Mohr, B.; Platzbecker, U.; Bornhauser, M.; Paulus, T.; Ehninger, G.; Zophel, K.; Schaich, M. (1)(8)F-FDG-PET/CT for detection of extramedullary acute myeloid leukemia. Haematologica 2011, 96, 1552–1556. [Google Scholar] [CrossRef] [PubMed]
- Cribe, A.S.; Steenhof, M.; Marcher, C.W.; Petersen, H.; Frederiksen, H.; Friis, L.S. Extramedullary disease in patients with acute myeloid leukemia assessed by 18F-FDG PET. Eur. J. Haematol. 2013, 90, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.L.; Wu, H.B.; Wang, L.J.; Tian, Y.; Dong, Y.; Wang, Q.S. Usefulness and pitfalls of F-18-FDG PET/CT for diagnosing extramedullary acute leukemia. Eur J. Radiol. 2016, 85, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, I.; Kohno, B. 18 FDG-PET/CT: 21st Century approach to leukemic tumors in 124 cases. Am. J. Hematol. 2016, 91, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Shields, A.F.; Grierson, J.R.; Dohmen, B.M.; Machulla, H.J.; Stayanoff, J.C.; Lawhorn-Crews, J.M.; Obradovich, J.E.; Muzik, O.; Mangner, T.J. Imaging proliferation in vivo with [F-18]FLT and positron emission tomography. Nat. Med. 1998, 4, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.K.; Bommer, M.; Juweid, M.E.; Glatting, G.; Stilgenbauer, S.; Mottaghy, F.M.; Schulz, M.; Kull, T.; Bunjes, D.; Moller, P.; et al. First demonstration of leukemia imaging with the proliferation marker 18F-fluorodeoxythymidine. J. Nucl. Med. 2008, 49, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Vanderhoek, M.; Juckett, M.B.; Perlman, S.B.; Nickles, R.J.; Jeraj, R. Early assessment of treatment response in patients with AML using [(18)F]FLT PET imaging. Leuk. Res. 2011, 35, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Hourigan, C.S.; Gale, R.P.; Gormley, N.J.; Ossenkoppele, G.J.; Walter, R.B. Measurable residual disease testing in acute myeloid leukaemia. Leukemia 2017, 31, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Ommen, H.B. Monitoring minimal residual disease in acute myeloid leukaemia: A review of the current evolving strategies. Ther. Adv. Hematol. 2016, 7, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Jovanovic, J.V.; Hills, R.K. Can we say farewell to monitoring minimal residual disease in acute promyelocytic leukaemia? Best Pract. Res. Clin. Haematol. 2014, 27, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kuderer, N.M.; Burton, K.A.; Blau, S.; Rose, A.L.; Parker, S.; Lyman, G.H.; Blau, C.A. Comparison of 2 Commercially Available Next-Generation Sequencing Platforms in Oncology. JAMA Oncol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Hokland, P.; Ommen, H.B.; Mule, M.P.; Hourigan, C.S. Advancing the Minimal Residual Disease Concept in Acute Myeloid Leukemia. Semin. Hematol. 2015, 52, 184–192. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roloff, G.W.; Lai, C.; Hourigan, C.S.; Dillon, L.W. Technical Advances in the Measurement of Residual Disease in Acute Myeloid Leukemia. J. Clin. Med. 2017, 6, 87. https://doi.org/10.3390/jcm6090087
Roloff GW, Lai C, Hourigan CS, Dillon LW. Technical Advances in the Measurement of Residual Disease in Acute Myeloid Leukemia. Journal of Clinical Medicine. 2017; 6(9):87. https://doi.org/10.3390/jcm6090087
Chicago/Turabian StyleRoloff, Gregory W., Catherine Lai, Christopher S. Hourigan, and Laura W. Dillon. 2017. "Technical Advances in the Measurement of Residual Disease in Acute Myeloid Leukemia" Journal of Clinical Medicine 6, no. 9: 87. https://doi.org/10.3390/jcm6090087




