Moonlighting with WDR5: A Cellular Multitasker
Abstract
:1. Introduction
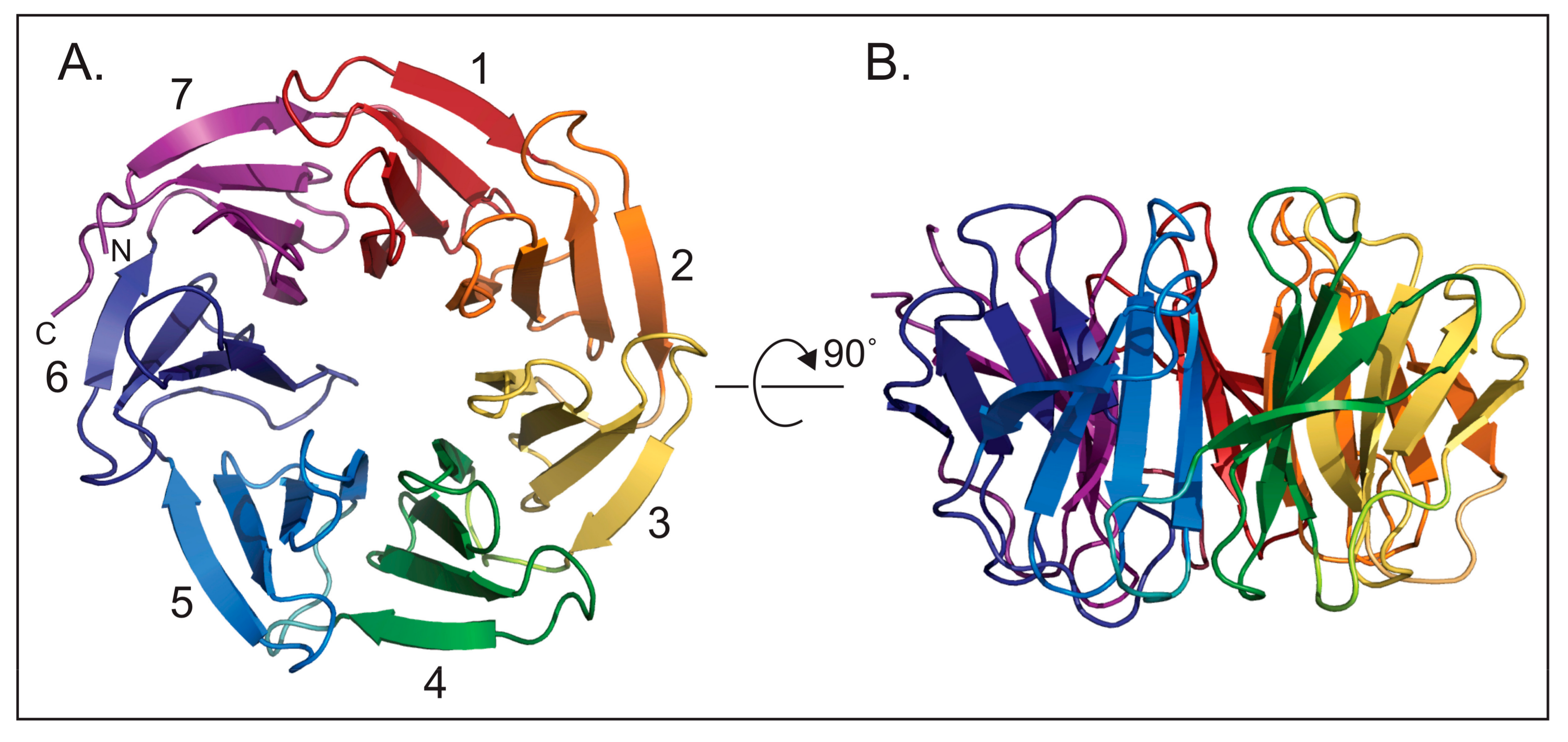
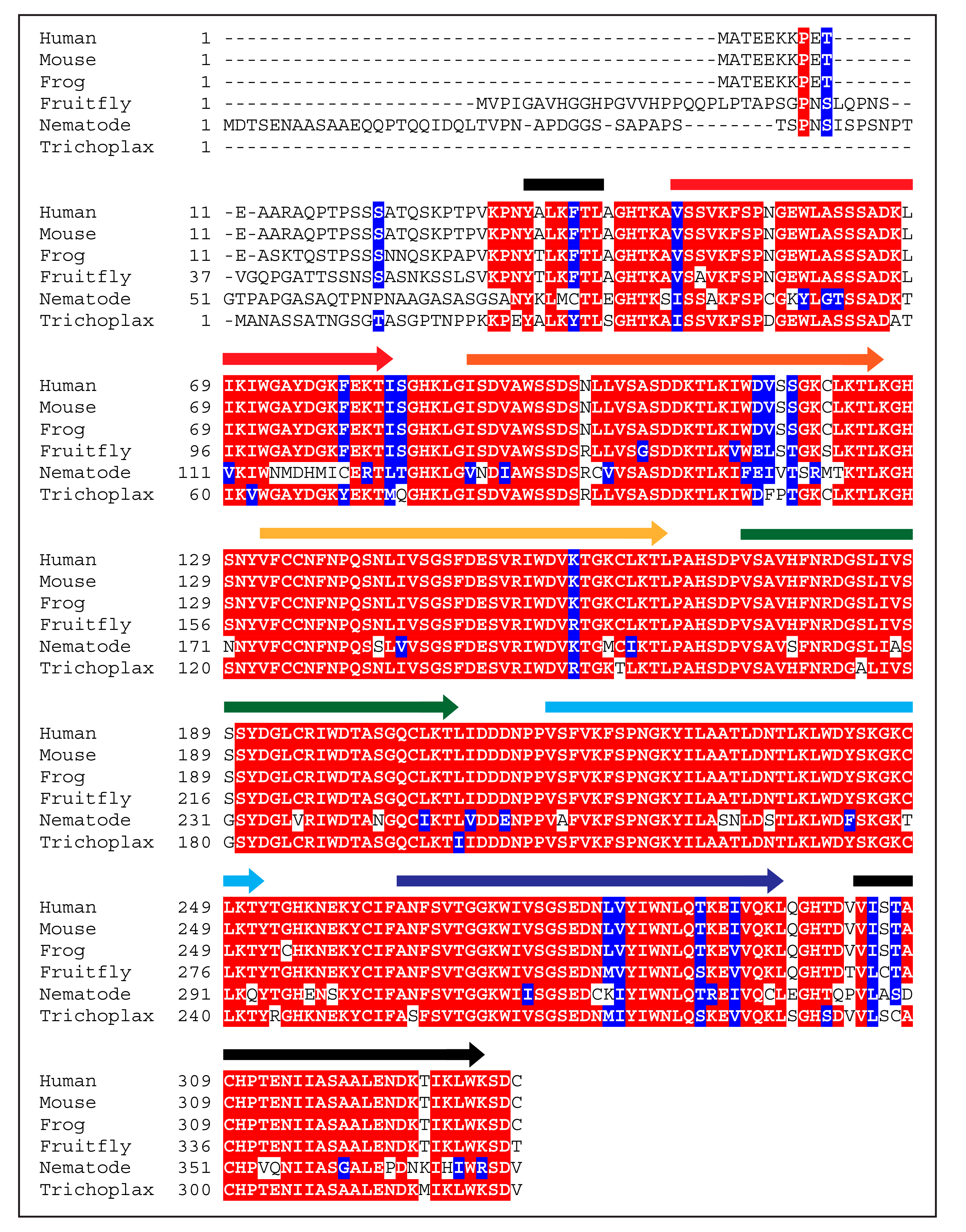
2. Beginnings and Basics
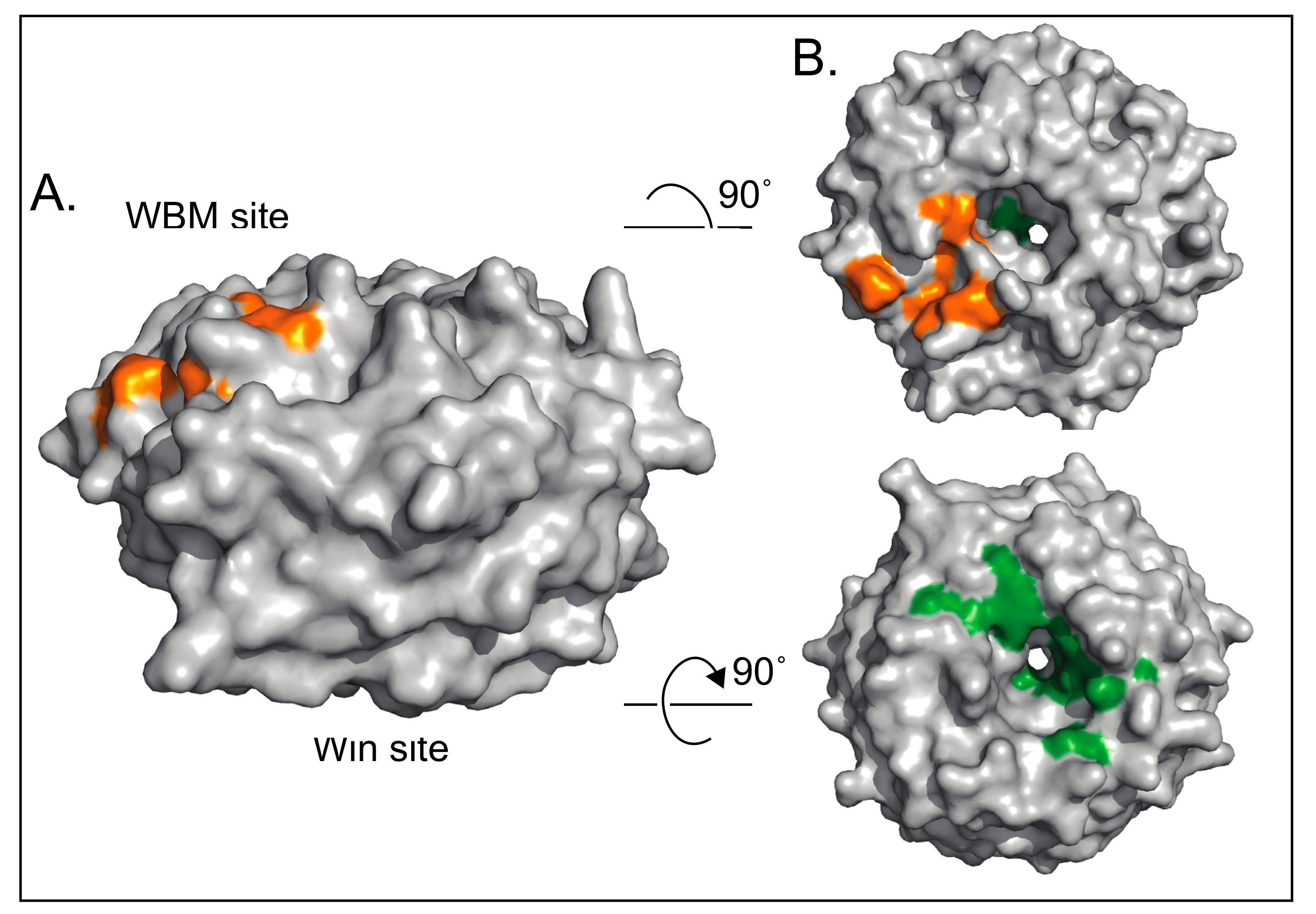
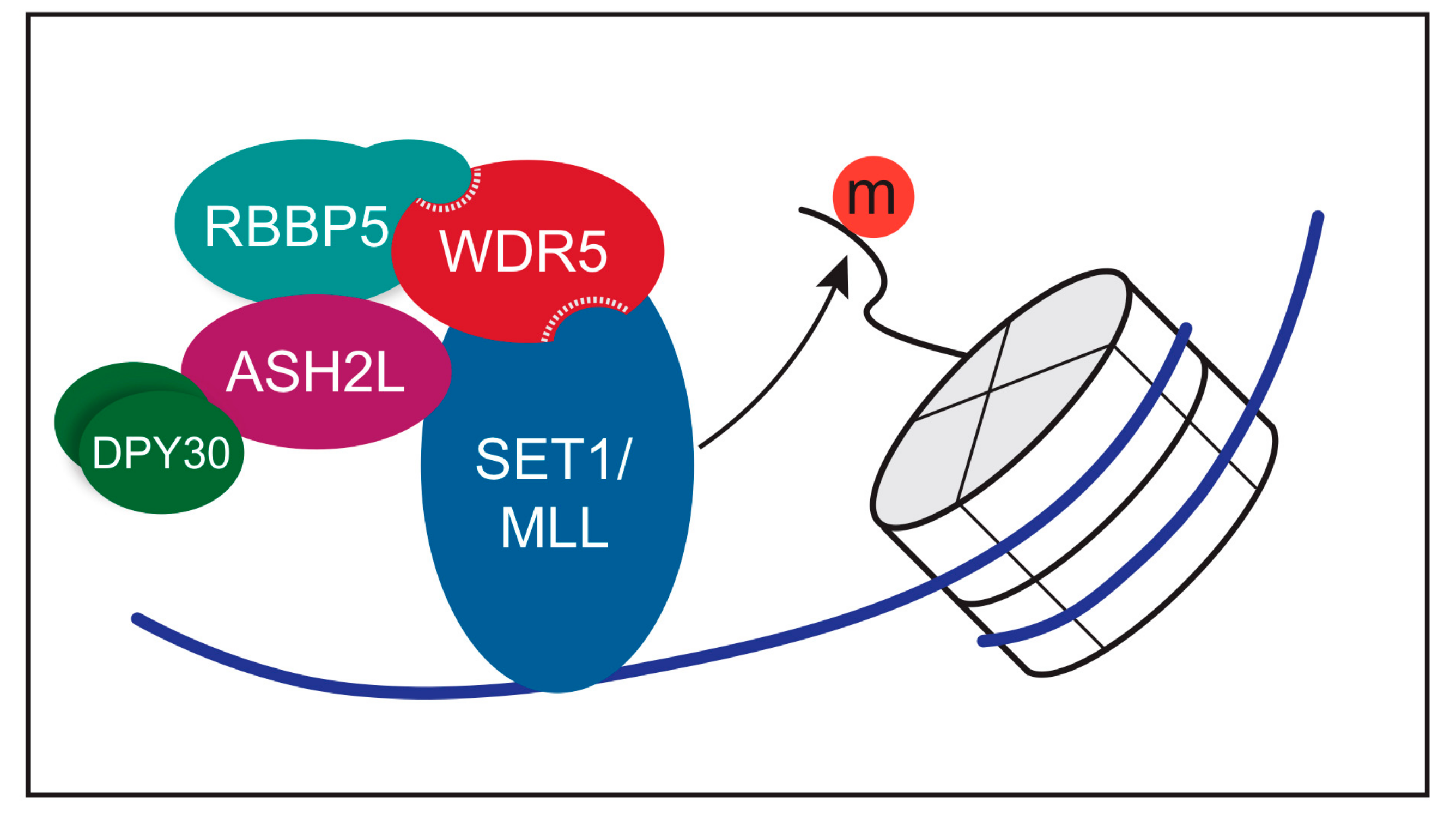
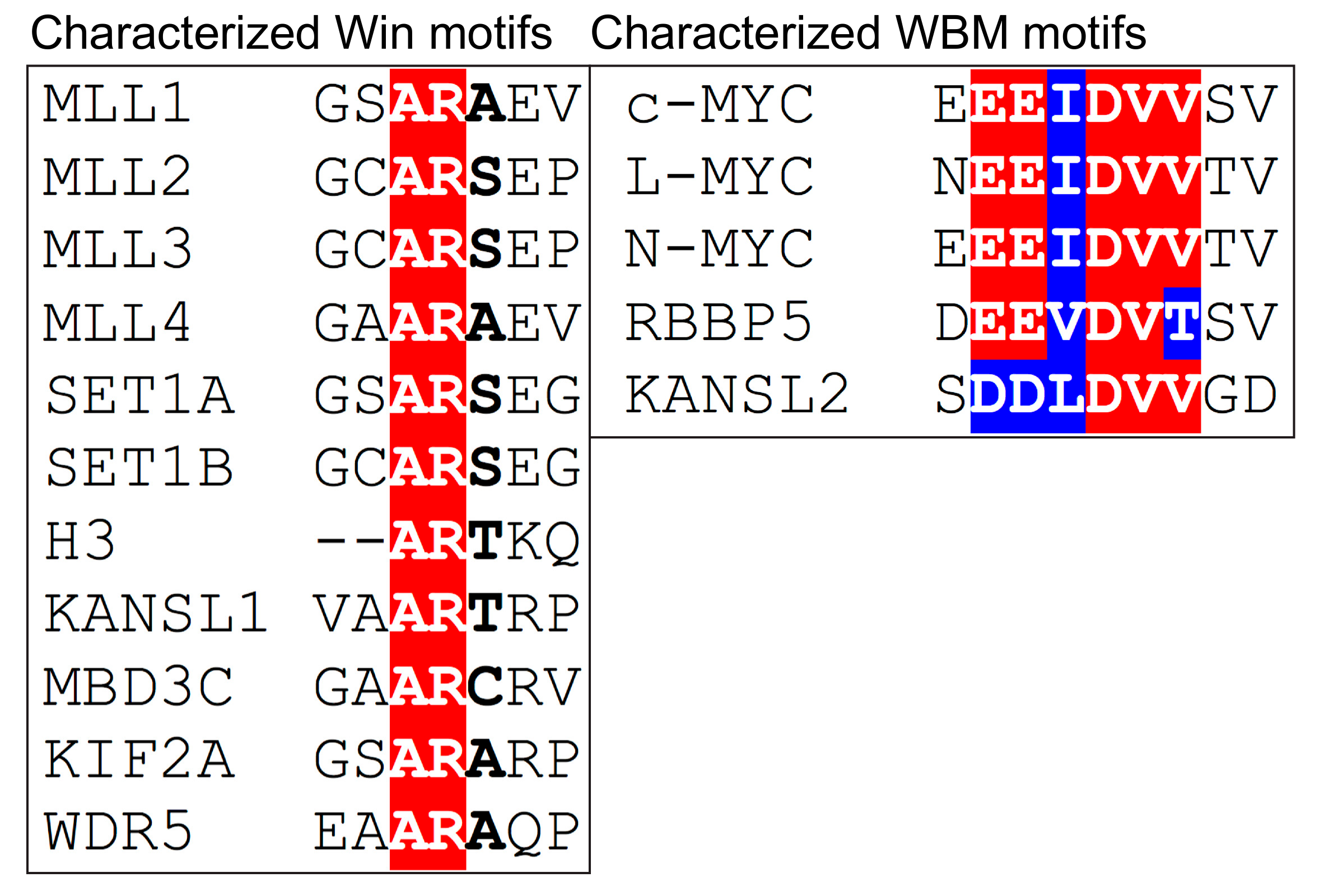
3. Function of WDR5 as a Core Member of Histone H3 Lysine 4 Methyltransferases
4. Moonlighting in the Nucleus
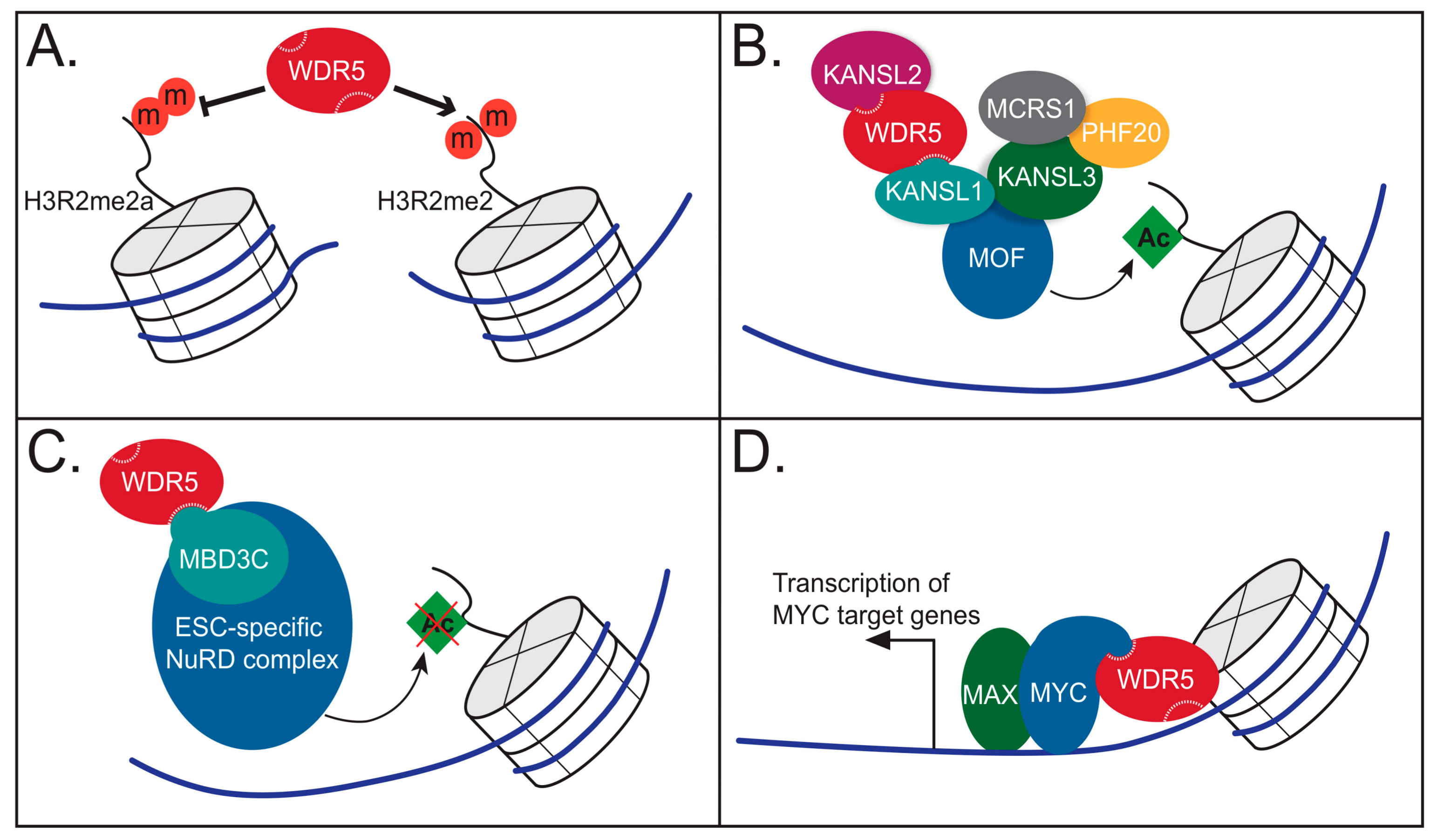
4.1. WDR5 as a Histone Tail Reader
4.2. WDR5 as Part of the NSL Complex
4.3. WDR5 as Part of the NuRD Complex
4.4. WDR5 Works with Sequence Specific Transcriptional Regulators
4.5. The Ever-Expanding Nuclear WDR5 Interactome
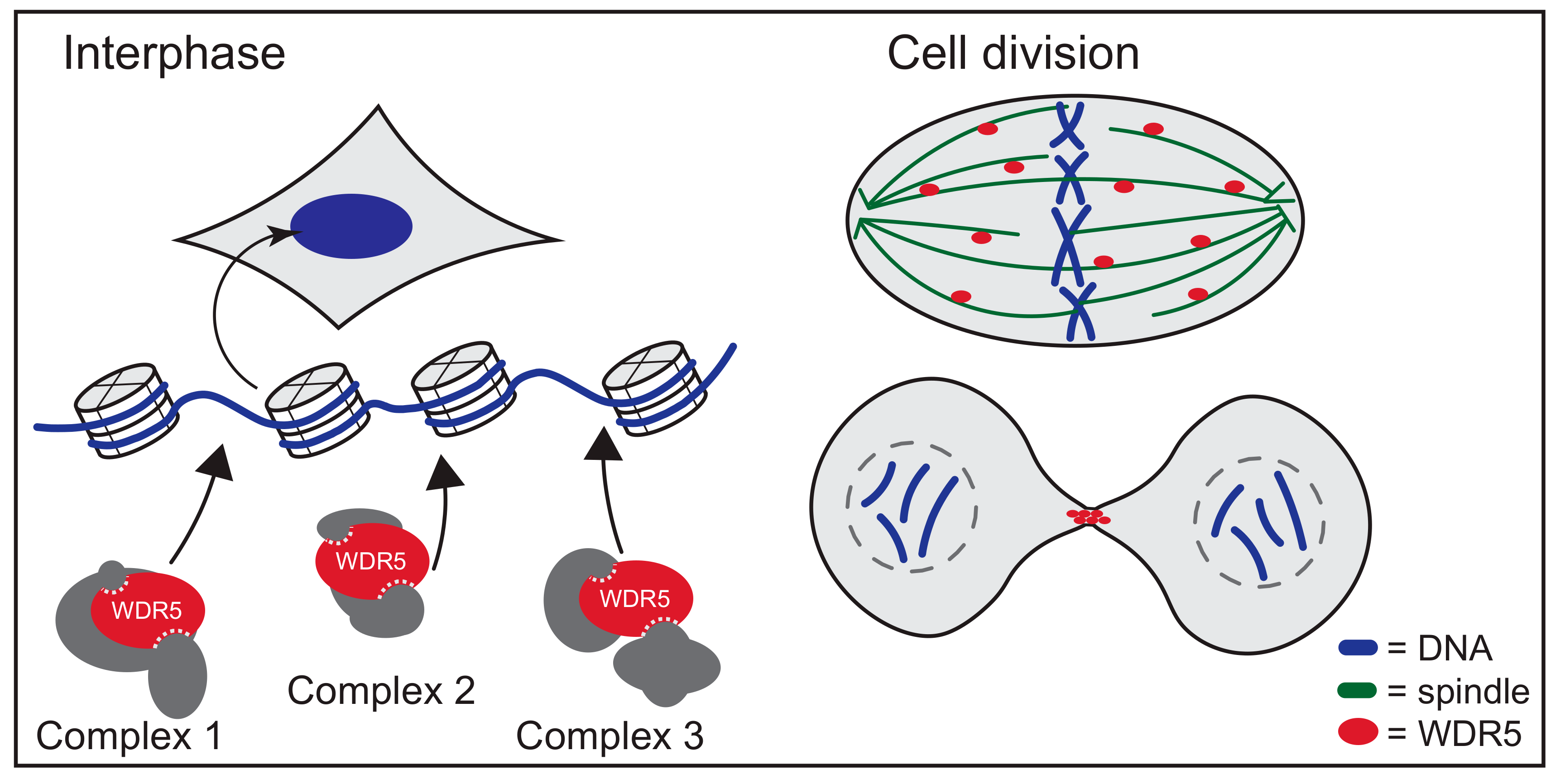
5. WDR5 Moonlights off Chromatin
6. Networking with WDR5
7. WDR5 and Drug Discovery
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Gori, F.; Divieti, P.; Demay, M.B. Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. J. Biol. Chem. 2001, 276, 46515–46522. [Google Scholar] [CrossRef] [PubMed]
- Gori, F.; Friedman, L.G.; Demay, M.B. Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol. 2006, 295, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Gori, F.; Demay, M.B. BIG-3, a novel WD-40 repeat protein, is expressed in the developing growth plate and accelerates chondrocyte differentiation in vitro. Endocrinology 2004, 145, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Gori, F.; Zhu, E.D.; Demay, M.B. Perichondrial expression of Wdr5 regulates chondrocyte proliferation and differentiation. Dev. Biol. 2009, 329, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhu, E.D.; Provot, S.; Gori, F. Wdr5 is required for chick skeletal development. J. Bone Miner. Res. 2010, 25, 2504–2514. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Krogan, N.J.; Dover, J.; Erdjument-Bromage, H.; Tempst, P.; Johnston, M.; Greenblatt, J.F.; Shilatifard, A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 2001, 98, 12902–12907. [Google Scholar] [CrossRef] [PubMed]
- Roguev, A.; Schaft, D.; Shevchenko, A.; Pijnappel, W.W.; Wilm, M.; Aasland, R.; Stewart, A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001, 20, 7137–7148. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Swigut, T.; Milne, T.A.; Dou, Y.; Zhang, X.; Burlingame, A.L.; Roeder, R.G.; Brivanlou, A.H.; Allis, C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Ang, Y.S.; Tsai, S.Y.; Lee, D.F.; Monk, J.; Su, J.; Ratnakumar, K.; Ding, J.; Ge, Y.; Darr, H.; Chang, B.; et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 2011, 145, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Kong, N.C.; Gu, X.; Li, Z.; He, Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.H.; Westfield, G.H.; Oleskie, A.N.; Trievel, R.C.; Shilatifard, A.; Skiniotis, G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. USA 2011, 108, 20526–20531. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Allali-Hassani, A.; Martin, F.; Loppnau, P.; Vedadi, M.; Bochkarev, A.; Plotnikov, A.N.; Arrowsmith, C.H.; Min, J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006, 25, 4245–4252. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.M.; Rozenblatt-Rosen, O.; Milne, T.A.; Copeland, T.D.; Levine, S.S.; Lee, J.C.; Hayes, D.N.; Shanmugam, K.S.; Bhattacharjee, A.; Biondi, C.A.; et al. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol. Cell 2004, 13, 587–597. [Google Scholar] [CrossRef]
- Yokoyama, A.; Wang, Z.; Wysocka, J.; Sanyal, M.; Aufiero, D.J.; Kitabayashi, I.; Herr, W.; Cleary, M.L. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell Biol. 2004, 24, 5639–5649. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Skalnik, D.G. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005, 280, 41725–41731. [Google Scholar] [CrossRef] [PubMed]
- Glaser, S.; Schaft, J.; Lubitz, S.; Vintersten, K.; van der Hoeven, F.; Tufteland, K.R.; Aasland, R.; Anastassiadis, K.; Ang, S.L.; Stewart, A.F. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 2006, 133, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Mo, R.; Rao, S.M.; Zhu, Y.J. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 2006, 281, 15714–15720. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Tate, C.M.; You, J.S.; Skalnik, D.G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007, 282, 13419–13428. [Google Scholar] [CrossRef] [PubMed]
- Ernst, P.; Vakoc, C.R. WRAD: Enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genom. 2012, 11, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Milne, T.A.; Ruthenburg, A.J.; Lee, S.; Lee, J.W.; Verdine, G.L.; Allis, C.D.; Roeder, R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006, 13, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Shinsky, S.A.; Monteith, K.E.; Viggiano, S.; Cosgrove, M.S. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J. Biol. Chem. 2015, 290, 6361–6375. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, J.; Zhang, Y.; Cao, F.; Liu, Z.; Li, S.; Wu, J.; Hu, C.; Wang, Y.; Shuai, J.; et al. Structural basis for activity regulation of MLL family methyltransferases. Nature 2016, 530, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Odho, Z.; Southall, S.M.; Wilson, J.R. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J. Biol. Chem. 2010, 285, 32967–32976. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Dharmarajan, V.; Cosgrove, M.S. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 2008, 283, 32158–32161. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Vought, V.E.; Dharmarajan, V.; Cosgrove, M.S. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008, 283, 32162–32175. [Google Scholar] [CrossRef] [PubMed]
- Dharmarajan, V.; Lee, J.H.; Patel, A.; Skalnik, D.G.; Cosgrove, M.S. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J. Biol. Chem. 2012, 287, 27275–27289. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Kingston, R.E. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008, 283, 35258–35264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lee, H.; Brunzelle, J.S.; Couture, J.F. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012, 40, 4237–4246. [Google Scholar] [CrossRef] [PubMed]
- Karatas, H.; Townsend, E.C.; Bernard, D.; Dou, Y.; Wang, S. Analysis of the binding of mixed lineage leukemia 1 (MLL1) and histone 3 peptides to WD repeat domain 5 (WDR5) for the design of inhibitors of the MLL1-WDR5 interaction. J. Med. Chem. 2010, 53, 5179–5185. [Google Scholar] [CrossRef] [PubMed]
- Alicea-Velazquez, N.L.; Shinsky, S.A.; Loh, D.M.; Lee, J.H.; Skalnik, D.G.; Cosgrove, M.S. Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes. J. Biol. Chem. 2016, 291, 22357–22372. [Google Scholar] [CrossRef] [PubMed]
- Van Nuland, R.; Smits, A.H.; Pallaki, P.; Jansen, P.W.; Vermeulen, M.; Timmers, H.T. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 2013, 33, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.F.; Collazo, E.; Trievel, R.C. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006, 13, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Guo, L.; Wang, H.; Shen, Y.; Deng, X.W.; Chai, J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell 2006, 22, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Ruthenburg, A.J.; Wang, W.; Graybosch, D.M.; Li, H.; Allis, C.D.; Patel, D.J.; Verdine, G.L. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006, 13, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Iberg, A.N.; Espejo, A.; Cheng, D.; Kim, D.; Michaud-Levesque, J.; Richard, S.; Bedford, M.T. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 2008, 283, 3006–3010. [Google Scholar] [CrossRef] [PubMed]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Luscher, B.; Amati, B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Hyllus, D.; Stein, C.; Schnabel, K.; Schiltz, E.; Imhof, A.; Dou, Y.; Hsieh, J.; Bauer, U.M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007, 21, 3369–3380. [Google Scholar] [CrossRef] [PubMed]
- Migliori, V.; Muller, J.; Phalke, S.; Low, D.; Bezzi, M.; Mok, W.C.; Sahu, S.K.; Gunaratne, J.; Capasso, P.; Bassi, C.; et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012, 19, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, F.; Cai, Y.; Jin, J. The Functional Analysis of Histone Acetyltransferase MOF in Tumorigenesis. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.R.; Cayrou, C.; Huang, R.; Lane, W.S.; Cote, J.; Lucchesi, J.C. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell Biol. 2005, 25, 9175–9188. [Google Scholar] [CrossRef] [PubMed]
- Mendjan, S.; Taipale, M.; Kind, J.; Holz, H.; Gebhardt, P.; Schelder, M.; Vermeulen, M.; Buscaino, A.; Duncan, K.; Mueller, J.; et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 2006, 21, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Swanson, S.K.; Cole, M.D.; Choi, S.H.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 2010, 285, 4268–4272. [Google Scholar] [CrossRef] [PubMed]
- Raja, S.J.; Charapitsa, I.; Conrad, T.; Vaquerizas, J.M.; Gebhardt, P.; Holz, H.; Kadlec, J.; Fraterman, S.; Luscombe, N.M.; Akhtar, A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell 2010, 38, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Van Nguyen, N.; Georgiev, P.; Gaub, A.; Brettschneider, J.; Cusack, S.; Kadlec, J.; Akhtar, A. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014, 28, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Milne, T.A.; Tackett, A.J.; Smith, E.R.; Fukuda, A.; Wysocka, J.; Allis, C.D.; Chait, B.T.; Hess, J.L.; Roeder, R.G. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 2005, 121, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Su, J.; Wang, F.; Liu, D.; Ding, J.; Yang, Y.; Conaway, J.W.; Conaway, R.C.; Cao, L.; Wu, D.; et al. Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 di-methylation activity by MLL/SET complexes. PLoS Genet. 2013, 9, e1003940. [Google Scholar] [CrossRef] [PubMed]
- Basta, J.; Rauchman, M. The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res. 2015, 165, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Bode, D.; Yu, L.; Tate, P.; Pardo, M.; Choudhary, J. Characterization of Two Distinct Nucleosome Remodeling and Deacetylase (NuRD) Complex Assemblies in Embryonic Stem Cells. Mol. Cell. Proteom. 2016, 15, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Ee, L.S.; McCannell, K.N.; Tang, Y.; Fernandes, N.; Hardy, W.R.; Green, M.R.; Chu, F.; Fazzio, T.G. An Embryonic Stem Cell-Specific NuRD Complex Functions through Interaction with WDR5. Stem Cell Rep. 2017, 8, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Z.; Tsai, Y.P.; Yang, M.H.; Huang, C.H.; Chang, S.Y.; Chang, C.C.; Teng, S.C.; Wu, K.J. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol. Cell 2011, 43, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Malek, R.; Gajula, R.P.; Williams, R.D.; Nghiem, B.; Simons, B.W.; Nugent, K.; Wang, H.; Taparra, K.; Lemtiri-Chlieh, G.; Yoon, A.R.; et al. TWIST1-WDR5-Hottip Regulates Hoxa9 Chromatin to Facilitate Prostate Cancer Metastasis. Cancer Res. 2017, 77, 3181–3193. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, N. Set1/MLL complex is indispensable for the transcriptional ability of heat shock transcription factor 2. Biochem. Biophys. Res. Commun. 2015, 467, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.R.; Wang, Q.; Grieb, B.C.; Phan, J.; Foshage, A.M.; Sun, Q.; Olejniczak, E.T.; Clark, T.; Dey, S.; Lorey, S.; et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 2015, 58, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bell, J.L.; Carter, D.; Gherardi, S.; Poulos, R.C.; Milazzo, G.; Wong, J.W.; Al-Awar, R.; Tee, A.E.; Liu, P.Y.; et al. WDR5 Supports an N-Myc Transcriptional Complex That Drives a Protumorigenic Gene Expression Signature in Neuroblastoma. Cancer Res. 2015, 75, 5143–5154. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.A.; Tremblay, V.; Lin, G.; Bochar, D.A. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell Biol. 2008, 28, 3894–3904. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.A.; Menon, T.; Thompson, B.A.; Bochar, D.A. Regulation of HOXA2 gene expression by the ATP-dependent chromatin remodeling enzyme CHD8. FEBS Lett. 2010, 584, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Du, Y.; Ward, J.M.; Shimbo, T.; Lackford, B.; Zheng, X.; Miao, Y.L.; Zhou, B.; Han, L.; Fargo, D.C.; et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 2014, 14, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, T.; Gutierrez, J.L.; Li, B.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Abmayr, S.M.; Workman, J.L. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008, 15, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Faiola, F.; Xu, M.; Pan, S.; Martinez, E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008, 283, 33808–33815. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Whyte, W.A.; Anderssen, E.; Apostolou, E.; Chen, H.H.; Akbarian, S.; Bronson, R.T.; Hochedlinger, K.; Ramaswamy, S.; Young, R.A.; et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 2012, 11, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed]
- Vilhais-Neto, G.C.; Fournier, M.; Plassat, J.L.; Sardiu, M.E.; Saraf, A.; Garnier, J.M.; Maruhashi, M.; Florens, L.; Washburn, M.P.; Pourquie, O. The WHHERE coactivator complex is required for retinoic acid-dependent regulation of embryonic symmetry. Nat. Commun. 2017, 8, 728. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Y.; Sun, Z.; Mullokandov, G.; Bosch, A.; Qadeer, Z.A.; Cihan, E.; Rapp, Z.; Parsons, R.; Aguirre-Ghiso, J.A.; Farias, E.F.; et al. Cbx8 Acts Non-canonically with Wdr5 to Promote Mammary Tumorigenesis. Cell Rep. 2016, 16, 472–486. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife 2014, 3, e02046. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; He, J.; Liang, Q.; Ren, L.L.; Yan, T.T.; Yu, T.C.; Tang, J.Y.; Bao, Y.J.; Hu, Y.; Lin, Y.; et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016, 6, 784–801. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Chen, X.; Xie, R.; Han, J.; Xie, W.; Wang, B.; Dong, W.; Chen, C.; Yang, M.; Jiang, J.; et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol. Ther. 2017, 25, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Gadadhar, S.; Bodakuntla, S.; Natarajan, K.; Janke, C. The tubulin code at a glance. J. Cell Sci. 2017, 130, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Verhey, K.J.; Gaertig, J. The tubulin code. Cell Cycle 2007, 6, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Fields, A.T.; Cheng, K.; Lee, A.; Wagenaar, E.; Lagrois, R.; Schmidt, B.; Xia, B.; Ma, D. WD repeat-containing protein 5 (WDR5) localizes to the midbody and regulates abscission. J. Biol. Chem. 2015, 290, 8987–9001. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Veeranki, S.N.; Chinchole, A.; Tyagi, S. MLL/WDR5 Complex Regulates Kif2A Localization to Ensure Chromosome Congression and Proper Spindle Assembly during Mitosis. Dev. Cell 2017, 41, 605–622. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Veeranki, S.N.; Tyagi, S. A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia. Nucleic Acids Res. 2014, 42, 7611–7624. [Google Scholar] [CrossRef] [PubMed]
- Meunier, S.; Shvedunova, M.; Van Nguyen, N.; Avila, L.; Vernos, I.; Akhtar, A. An epigenetic regulator emerges as microtubule minus-end binding and stabilizing factor in mitosis. Nat. Commun. 2015, 6, 7889. [Google Scholar] [CrossRef] [PubMed]
- Orpinell, M.; Fournier, M.; Riss, A.; Nagy, Z.; Krebs, A.R.; Frontini, M.; Tora, L. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010, 29, 2381–2394. [Google Scholar] [CrossRef] [PubMed]
- Migliori, V.; Mapelli, M.; Guccione, E. On WD40 proteins: Propelling our knowledge of transcriptional control? Epigenetics 2012, 7, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Muller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Licht, J.D. Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Carugo, A.; Genovese, G.; Seth, S.; Nezi, L.; Rose, J.L.; Bossi, D.; Cicalese, A.; Shah, P.K.; Viale, A.; Pettazzoni, P.F.; et al. In Vivo Functional Platform Targeting Patient-Derived Xenografts Identifies WDR5-Myc Association as a Critical Determinant of Pancreatic Cancer. Cell Rep. 2016, 16, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Townsend, E.C.; Karatas, H.; Xu, J.; Li, L.; Lee, S.; Liu, L.; Chen, Y.; Ouillette, P.; Zhu, J.; et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 2014, 53, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Thiel, A.T.; Blessington, P.; Zou, T.; Feather, D.; Wu, X.; Yan, J.; Zhang, H.; Liu, Z.; Ernst, P.; Koretzky, G.A.; et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 2010, 17, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Anastassiadis, K.; Kranz, A.; Stewart, A.F.; Arndt, K.; Waskow, C.; Yokoyama, A.; Jones, K.; Neff, T.; Lee, Y.; et al. MLL2, Not MLL1, Plays a Major Role in Sustaining MLL-Rearranged Acute Myeloid Leukemia. Cancer Cell 2017, 31, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Bolshan, Y.; Getlik, M.; Kuznetsova, E.; Wasney, G.A.; Hajian, T.; Poda, G.; Nguyen, K.T.; Wu, H.; Dombrovski, L.; Dong, A.; et al. Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction. ACS Med. Chem. Lett. 2013, 4, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Senisterra, G.; Wu, H.; Allali-Hassani, A.; Wasney, G.A.; Barsyte-Lovejoy, D.; Dombrovski, L.; Dong, A.; Nguyen, K.T.; Smil, D.; Bolshan, Y.; et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 2013, 449, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Grebien, F.; Vedadi, M.; Getlik, M.; Giambruno, R.; Grover, A.; Avellino, R.; Skucha, A.; Vittori, S.; Kuznetsova, E.; Smil, D.; et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol. 2015, 11, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Chen, W.L.; Wang, Z.H.; Xie, Y.Y.; Xu, X.L.; Jiang, Z.Y.; Zhang, X.J.; You, Q.D.; Guo, X.K. High-affinity small molecular blockers of mixed lineage leukemia 1 (MLL1)-WDR5 interaction inhibit MLL1 complex H3K4 methyltransferase activity. Eur. J. Med. Chem. 2016, 124, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Wang, Z.H.; Chen, W.L.; Xie, Y.Y.; You, Q.D.; Guo, X.K. Structure-based design of ester compounds to inhibit MLL complex catalytic activity by targeting mixed lineage leukemia 1 (MLL1)-WDR5 interaction. Bioorg. Med. Chem. 2016, 24, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Li, D.D.; Wang, Z.H.; Xu, X.L.; Zhang, X.J.; Jiang, Z.Y.; Guo, X.K.; You, Q.D. Design, synthesis, and initial evaluation of affinity-based small molecular probe for detection of WDR5. Bioorg. Chem. 2017, 76, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Sammons, M.A.; Donahue, G.; Dou, Z.; Vedadi, M.; Getlik, M.; Barsyte-Lovejoy, D.; Al-awar, R.; Katona, B.W.; Shilatifard, A.; et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015, 525, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guarnaccia, A.D.; Tansey, W.P. Moonlighting with WDR5: A Cellular Multitasker. J. Clin. Med. 2018, 7, 21. https://doi.org/10.3390/jcm7020021
Guarnaccia AD, Tansey WP. Moonlighting with WDR5: A Cellular Multitasker. Journal of Clinical Medicine. 2018; 7(2):21. https://doi.org/10.3390/jcm7020021
Chicago/Turabian StyleGuarnaccia, Alissa DuPuy, and William Patrick Tansey. 2018. "Moonlighting with WDR5: A Cellular Multitasker" Journal of Clinical Medicine 7, no. 2: 21. https://doi.org/10.3390/jcm7020021
APA StyleGuarnaccia, A. D., & Tansey, W. P. (2018). Moonlighting with WDR5: A Cellular Multitasker. Journal of Clinical Medicine, 7(2), 21. https://doi.org/10.3390/jcm7020021



