CTLA-4 Genetic Variants Predict Survival in Patients with Sepsis
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Data Collection
2.3. CTLA-4 Genotyping and Haplotyping
2.4. Statistical Analyses
2.5. Data Availability
3. Results
3.1. Baseline Characteristics
3.2. Outcomes
3.3. Cox Regression Analysis
3.4. Disease Severity
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Marshall, J.C.; Ñamendys-Silva, S.A.; François, B.; Martin-Loeches, I.; Lipman, J.; Reinhart, K.; Antonelli, M.; Pickkers, P.; Njimi, H.; et al. Assessment of the worldwide burden of critical illness: The Intensive Care Over Nations (ICON) audit. Lancet Respir. Med. 2014, 2, 380–386. [Google Scholar] [CrossRef]
- Gaieski, D.F.; Edwards, J.M.; Kallan, M.J.; Carr, B.G. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit. Care Med. 2013, 41, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Mansur, A.; Mulwande, E.; Steinau, M.; Bergmann, I.; Frederik Popov, A.; Ghadimi, M.; Beissbarth, T.; Bauer, M.; Hinz, J. Chronic kidney disease is associated with a higher 90-day mortality than other chronic medical conditions in patients with sepsis. Sci. Rep. 2015, 5, 10539. [Google Scholar] [CrossRef] [PubMed]
- Angus, D.C.; van der Poll, T. Severe Sepsis and Septic Shock. Available online: https://www.nejm.org/doi/10.1056/NEJMra1208623?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov (accessed on 14 October 2018).
- Namath, A.; Patterson, A.J. Genetic polymorphisms in sepsis. Crit. Care Nurs. Clin. N. Am. 2011, 23, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Mansur, A.; Liese, B.; Steinau, M.; Ghadimi, M.; Bergmann, I.; Tzvetkov, M.; Popov, A.F.; Beissbarth, T.; Bauer, M.; Hinz, J. The CD14 rs2569190 TT genotype is associated with an improved 30-day survival in patients with sepsis: A prospective observational cohort study. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Mansur, A.; von Gruben, L.; Popov, A.F.; Steinau, M.; Bergmann, I.; Ross, D.; Ghadimi, M.; Beissbarth, T.; Bauer, M.; Hinz, J. The regulatory toll-like receptor 4 genetic polymorphism rs11536889 is associated with renal, coagulation and hepatic organ failure in sepsis patients. J. Transl. Med. 2014, 12, 177. [Google Scholar] [CrossRef]
- Mansur, A.; Hinz, J.; Hillebrecht, B.; Bergmann, I.; Popov, A.F.; Ghadimi, M.; Bauer, M.; Beissbarth, T.; Mihm, S. Ninety-day survival rate of patients with sepsis relates to programmed cell death 1 genetic polymorphism rs11568821. J. Investig. Med. 2014, 62, 638–643. [Google Scholar] [CrossRef]
- Arcaroli, J.; Fessler, M.B.; Abraham, E. Genetic polymorphisms and sepsis. Shock 2005, 24, 300–312. [Google Scholar] [CrossRef]
- Man, M.; Close, S.L.; Shaw, A.D.; Bernard, G.R.; Douglas, I.S.; Kaner, R.J.; Payen, D.; Vincent, J.-L.; Fossceco, S.; Janes, J.M.; et al. Beyond single-marker analyses: Mining whole genome scans for insights into treatment responses in severe sepsis. Pharmacogenom. J. 2013, 13, 218–226. [Google Scholar] [CrossRef]
- Nobel Media AB 2018 All Nobel Prizes. Available online: https://www.nobelprize.org/prizes/lists/all-nobel-prizes/ (accessed on 15 October 2018).
- Fallon, E.A.; Biron-Girard, B.M.; Chung, C.-S.; Lomas-Neira, J.; Heffernan, D.S.; Monaghan, S.F.; Ayala, A. A novel role for coinhibitory receptors/checkpoint proteins in the immunopathology of sepsis. J. Leukoc. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Walunas, T.L.; Lenschow, D.J.; Bakker, C.Y.; Linsley, P.S.; Freeman, G.J.; Green, J.M.; Thompson, C.B.; Bluestone, J.A. Pillars article: CTLA-4 can function as a negative regulator of T cell activation. immunity. 1994. 1: 405–413. J. Immunol. 2011, 187, 3466–3474. [Google Scholar] [PubMed]
- Gardner, D.; Jeffery, L.E.; Sansom, D.M. Understanding the CD28/CTLA-4 (CD152) pathway and its implications for costimulatory blockade. Am. J. Transpl. 2014, 14, 1985–1991. [Google Scholar] [CrossRef] [PubMed]
- McCoy, K.D.; Le Gros, G. The role of CTLA-4 in the regulation of T cell immune responses. Immunol. Cell Biol. 1999, 77, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 pathways: Similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Stearns-Kurosawa, D.J.; Osuchowski, M.F.; Valentine, C.; Kurosawa, S.; Remick, D.G. The pathogenesis of sepsis. Annu. Rev. Pathol. 2011, 6, 19–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Lou, J.; Zhou, Y.; Bo, L.; Zhu, J.; Zhu, K.; Wan, X.; Cai, Z.; Deng, X. Upregulation of programmed death-1 on T cells and programmed death ligand-1 on monocytes in septic shock patients. Crit. Care 2011, 15. [Google Scholar] [CrossRef]
- Mewes, C.; Büttner, B.; Hinz, J.; Alpert, A.; Popov, A.F.; Ghadimi, M.; Beissbarth, T.; Tzvetkov, M.; Shen-Orr, S.; Bergmann, I.; et al. The CTLA-4 rs231775 GG genotype is associated with favorable 90-day survival in Caucasian patients with sepsis. Sci. Rep. 2018, 8, 15140. [Google Scholar] [CrossRef]
- Soskic, B.; Qureshi, O.S.; Hou, T.; Sansom, D.M. Chapter Four—A Transendocytosis Perspective on the CD28/CTLA-4 Pathway. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 124, pp. 95–136. [Google Scholar]
- Benhatchi, K.; Jochmanová, I.; Habalová, V.; Wagnerová, H.; Lazúrová, I. CTLA4 exon1 A49G polymorphism in Slovak patients with rheumatoid arthritis and Hashimoto thyroiditis—Results and the review of the literature. Clin. Rheumatol 2011, 30, 1319–1324. [Google Scholar] [CrossRef]
- Wang, X.; Pirskanen, R.; Giscombe, R.; Lefvert, A.K. Two SNPs in the promoter region of the CTLA-4 gene affect binding of transcription factors and are associated with human myasthenia gravis. J. Int. Med. 2008, 263, 61–69. [Google Scholar] [CrossRef]
- Torres, B.; Aguilar, F.; Franco, E.; Sánchez, E.; Sánchez-Román, J.; Alonso, J.J.; Núñez-Roldán, A.; Martín, J.; González-Escribano, M.F. Association of the CT60 marker of the CTLA4 gene with systemic lupus erythematosus. Arthritis Rheum. 2004, 50, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.; Rocca, K.; Song, Y.; Pandey, J. CTLA-4 gene polymorphisms in systemic lupus erythematosus: A highly significant association with a determinant in the promoter region. Hum. Genet. 2002, 111, 452–455. [Google Scholar] [CrossRef]
- Zaletel, K.; Krhin, B.; Gaberšček, S.; Biček, A.; Pajič, T.; Hojker, S. Association of CT60 cytotoxic T lymphocyte antigen-4 gene polymorphism with thyroid autoantibody production in patients with Hashimoto’s and postpartum thyroiditis. Clin. Exp. Immunol. 2010, 161, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Douroudis, K.; Prans, E.; Kisand, K.; Nemvalts, V.; Uibo, R. Cytotoxic T-lymphocyte antigen 4 gene polymorphisms are associated with latent autoimmune diabetes in adults. Clin. Chim. Acta 2009, 403, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Karabon, L.; Kosmaczewska, A.; Bilinska, M.; Pawlak, E.; Ciszak, L.; Jedynak, A.; Jonkisz, A.; Noga, L.; Pokryszko-Dragan, A.; Koszewicz, M.; et al. The CTLA-4 gene polymorphisms are associated with CTLA-4 protein expression levels in multiple sclerosis patients and with susceptibility to disease. Immunology 2009, 128, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Muro, M.; Rojas, G.; Botella, C.; Miras, M.; Campillo, J.A.; Minguela, A.; Sánchez-Bueno, F.; Bermejo, J.; Ramírez, P.; Álvarez-López, M.R. CT60 A/G marker of the 3′-UTR of the CTLA4 gene and liver transplant. Transpl. Immunol. 2008, 18, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.H.; Sibbald, W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Hinz, J.; Büttner, B.; Kriesel, F.; Steinau, M.; Frederik Popov, A.; Ghadimi, M.; Beissbarth, T.; Tzvetkov, M.; Bergmann, I.; Mansur, A. The FER rs4957796 TT genotype is associated with unfavorable 90-day survival in Caucasian patients with severe ARDS due to pneumonia. Sci. Rep. 2017, 7, 9887. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Stephens, M.; Scheet, P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005, 76, 449–462. [Google Scholar] [CrossRef]
- Submitted SNP(ss) Details: ss79540. Available online: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss79540 (accessed on 21 November 2018).
- Submitted SNP(ss) Details: ss44315036. Available online: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss44315036 (accessed on 21 November 2018).
- Mansur, A.; Steinau, M.; Popov, A.F.; Ghadimi, M.; Beissbarth, T.; Bauer, M.; Hinz, J. Impact of statin therapy on mortality in patients with sepsis-associated acute respiratory distress syndrome (ARDS) depends on ARDS severity: A prospective observational cohort study. BMC Med. 2015, 13, 128. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Howson, J.M.M.; Esposito, L.; Heward, J.; Snook, H.; Chamberlain, G.; Rainbow, D.B.; Hunter, K.M.D.; Smith, A.N.; Di Genova, G.; et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003, 423, 506–511. [Google Scholar] [CrossRef] [PubMed]




| Parameter | All (n = 644) | rs3087243 | p-Value | |
|---|---|---|---|---|
| AA (n = 142) | GG/AG (n = 502) | |||
| Age (years) | 63 ± 15 | 67 ± 14 | 62 ± 15 | 0.0033 |
| Male (%) | 66 | 64 | 67 | 0.5560 |
| Body mass index | 28 ± 6 | 27 ± 6 | 28 ± 6 | 0.2881 |
| Sepsis severity | ||||
| Septic shock (%) | 51 | 52 | 51 | 0.8467 |
| SOFA score day 1 | 9.4 ± 3.9 | 9.4 ± 3.9 | 9.4 ± 3.9 | 0.8047 |
| APACHE II score day 1 | 22 ± 7 | 22 ± 7 | 21 ± 7 | 0.2844 |
| Comorbidities, n (%) | ||||
| Hypertension | 54 | 53 | 55 | 0.7094 |
| History of myocardial infarction | 5 | 6 | 5 | 0.9057 |
| Chronic obstructive pulmonary disease | 15 | 12 | 16 | 0.2436 |
| Renal dysfunction | 10 | 8 | 11 | 0.3879 |
| Non-insulin-dependent diabetes mellitus | 9 | 8 | 9 | 0.8492 |
| Insulin-dependent diabetes mellitus | 11 | 12 | 10 | 0.5349 |
| Chronic liver disease | 6 | 6 | 6 | 0.9875 |
| History of cancer | 16 | 15 | 16 | 0.7830 |
| History of stroke | 6 | 9 | 5 | 0.0623 |
| Recent surgical history, n (%) | ||||
| Elective surgery | 29 | 37 | 27 | |
| Emergency surgery | 53 | 46 | 55 | |
| No history of surgery | 18 | 17 | 18 | |
| Site of infection, n (%) | ||||
| Lung | 62 | 64 | 61 | |
| Abdomen | 20 | 20 | 20 | |
| Bone or soft tissue | 4 | 3 | 4 | |
| Surgical wound | 2 | 2 | 1 | |
| Urogenital | 2 | 1 | 3 | |
| Primary bacteremia | 7 | 6 | 7 | |
| Other | 3 | 4 | 4 | |
| Organ support (%) | ||||
| Used during observation period | ||||
| Mechanical ventilation | 93 | 96 | 93 | 0.1850 |
| Use of vasopressor | 79 | 82 | 78 | 0.4063 |
| Renal replacement therapy | 21 | 20 | 21 | 0.7173 |
| Used on sepsis onset | ||||
| Mechanical ventilation | 86 | 86 | 86 | 0.9381 |
| Use of vasopressor | 67 | 66 | 67 | 0.9048 |
| Renal replacement therapy | 9 | 9 | 9 | 0.8851 |
| Parameter | All (n = 644) | H1: TAA | p-Value | |
|---|---|---|---|---|
| Positive (n = 447) | Negative (n = 197) | |||
| Age (years) | 63 ± 15 | 64 ± 15 | 62 ± 14 | 0.1014 |
| Male (%) | 66 | 67 | 64 | 0.4356 |
| Body mass index | 28 ± 6 | 28 ± 6 | 28 ± 7 | 0.2388 |
| Sepsis severity | ||||
| Septic shock (%) | 51 | 51 | 53 | 0.6384 |
| SOFA score day 1 | 9.4 ± 3.9 | 9.3 ± 3.9 | 9.7 ± 3.8 | 0.1802 |
| APACHE II score day 1 | 22 ± 7 | 21 ± 7 | 22 ± 7 | 0.1474 |
| Comorbidities, n (%) | ||||
| Hypertension | 54 | 54 | 54 | 0.9671 |
| History of myocardial infarction | 5 | 6 | 3 | 0.0758 |
| Chronic obstructive pulmonary disease | 15 | 15 | 16 | 0.5779 |
| Renal dysfunction | 10 | 10 | 11 | 0.6734 |
| Non-insulin-dependent diabetes mellitus | 9 | 9 | 8 | 0.6654 |
| Insulin-dependent diabetes mellitus | 11 | 9 | 13 | 0.1480 |
| Chronic liver disease | 6 | 6 | 8 | 0.3893 |
| History of cancer | 16 | 12 | 17 | 0.0731 |
| History of stroke | 6 | 6 | 6 | 0.8209 |
| Recent surgical history, n (%) | ||||
| Elective surgery | 29 | 31 | 26 | |
| Emergency surgery | 53 | 52 | 54 | |
| No history of surgery | 18 | 17 | 20 | |
| Site of infection, n (%) | ||||
| Lung | 62 | 64 | 58 | |
| Abdomen | 20 | 19 | 21 | |
| Bone or soft tissue | 4 | 3 | 6 | |
| Surgical wound | 2 | 2 | 2 | |
| Urogenital | 2 | 2 | 3 | |
| Primary bacteremia | 7 | 7 | 6 | |
| Other | 3 | 3 | 4 | |
| Organ support (%) | ||||
| Used during observation period | ||||
| Mechanical ventilation | 93 | 93 | 93 | 0.9580 |
| Use of vasopressor | 79 | 79 | 80 | 0.6749 |
| Renal replacement therapy | 21 | 18 | 26 | 0.0204 |
| Used on sepsis onset | ||||
| Mechanical ventilation | 86 | 86 | 85 | 0.8341 |
| Use of vasopressor | 67 | 66 | 69 | 0.4944 |
| Renal replacement therapy | 9 | 6 | 15 | 0.0005 |
| Haplotype CTLA-4-1772 T>C/+49 A>G/+6230 G>A | Number of Haplotypes | n (%) |
|---|---|---|
| H1: TAA | 0 | 197 (30.59) |
| 1 | 305 (47.36) | |
| 2 | 142 (22.05) | |
| H2: TGG | 0 | 316 (49.07) |
| 1 | 263 (40.84) | |
| 2 | 65 (10.09) | |
| H3: TAG | 0 | 451 (70.03) |
| 1 | 178 (27.64) | |
| 2 | 15 (2.33) | |
| H4: CGG | 0 | 550 (85.40) |
| 1 | 90 (13.98) | |
| 2 | 4 (0.62) |
| Variable | 90 Days | |||||
| Univariate Analysis | Multivariate Analysis | |||||
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Age | 1.032 | 1.020–1.043 | 0.000000 | 1.025 | 1.013–1.038 | 0.000047 |
| Male gender | 1.093 | 0.810–1.474 | 0.558922 | 1.076 | 0.796–1.454 | 0.633436 |
| Body mass index | 0.985 | 0.961–1.009 | 0.218173 | 0.979 | 0.954–1.005 | 0.109574 |
| SOFA score day 1 | 1.104 | 1.065–1.143 | 0.000000 | 1.072 | 1.025–1.122 | 0.002432 |
| APACHE II score day 1 | 1.069 | 1.046–1.092 | 0.000000 | 1.033 | 1.004–.1062 | 0.026207 |
| Statin therapy | 1.114 | 0.807–1.536 | 0.511644 | 0.979 | 0.702–1.366 | 0.901706 |
| CTLA-4 rs3087243 G allele | 0.612 | 0.450–0.833 | 0.001760 | 0.667 | 0.489–0.909 | 0.010307 |
| 28 Days | ||||||
| Univariate Analysis | Multivariate Analysis | |||||
| Age | 1.029 | 1.015–1.043 | 0.000035 | 1.023 | 1.009–1.039 | 0.001824 |
| Male gender | 1.278 | 0.875–1.866 | 0.204154 | 1.270 | 0.868–1.860 | 0.218759 |
| Body mass index | 0.982 | 0.953–1.013 | 0.252686 | 0.976 | 0.945–1.008 | 0.135751 |
| SOFA score day 1 | 1.119 | 1.071–1.169 | 0.000000 | 1.085 | 1.026–1.146 | 0.003854 |
| APACHE II score day 1 | 1.073 | 1.045–1.102 | 0.000000 | 1.033 | 0.998–1.070 | 0.063787 |
| Statin therapy | 0.964 | 0.641–1.451 | 0.861906 | 0.841 | 0.552–1.281 | 0.420505 |
| CTLA-4 rs3087243 G allele | 0.624 | 0.428–0.908 | 0.013778 | 0.676 | 0.463–0.987 | 0.042323 |
| Variable | 90 Days | |||||
| Univariate Analysis | Multivariate Analysis | |||||
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Age | 1.032 | 1.020–1.043 | 0.000000 | 1.025 | 1.013–1.038 | 0.000049 |
| Male gender | 1.093 | 0.810–1.474 | 0.558922 | 1.079 | 0.798–1.459 | 0.620112 |
| BMI | 0.985 | 0.961–1.009 | 0.218173 | 0.975 | 0.950–1.001 | 0.061158 |
| SOFA score day 1 | 1.104 | 1.065–1.143 | 0.000000 | 1.038 | 0.988–1.091 | 0.139057 |
| APACHE II score day 1 | 1.069 | 1.046–1.092 | 0.000000 | 1.027 | 0.997–1.057 | 0.078734 |
| Statin therapy | 1.114 | 0.807–1.536 | 0.511644 | 1.086 | 0.777–1.519 | 0.629022 |
| Renal replacement therapy during observation period | 2.854 | 2.134–3.817 | 0.000000 | 2.724 | 1.867–3.974 | 0.000000 |
| Renal replacement therapy upon sepsis onset | 1.565 | 1.021–2.399 | 0.040064 | 1.973 | 1.192–3.268 | 0.008241 |
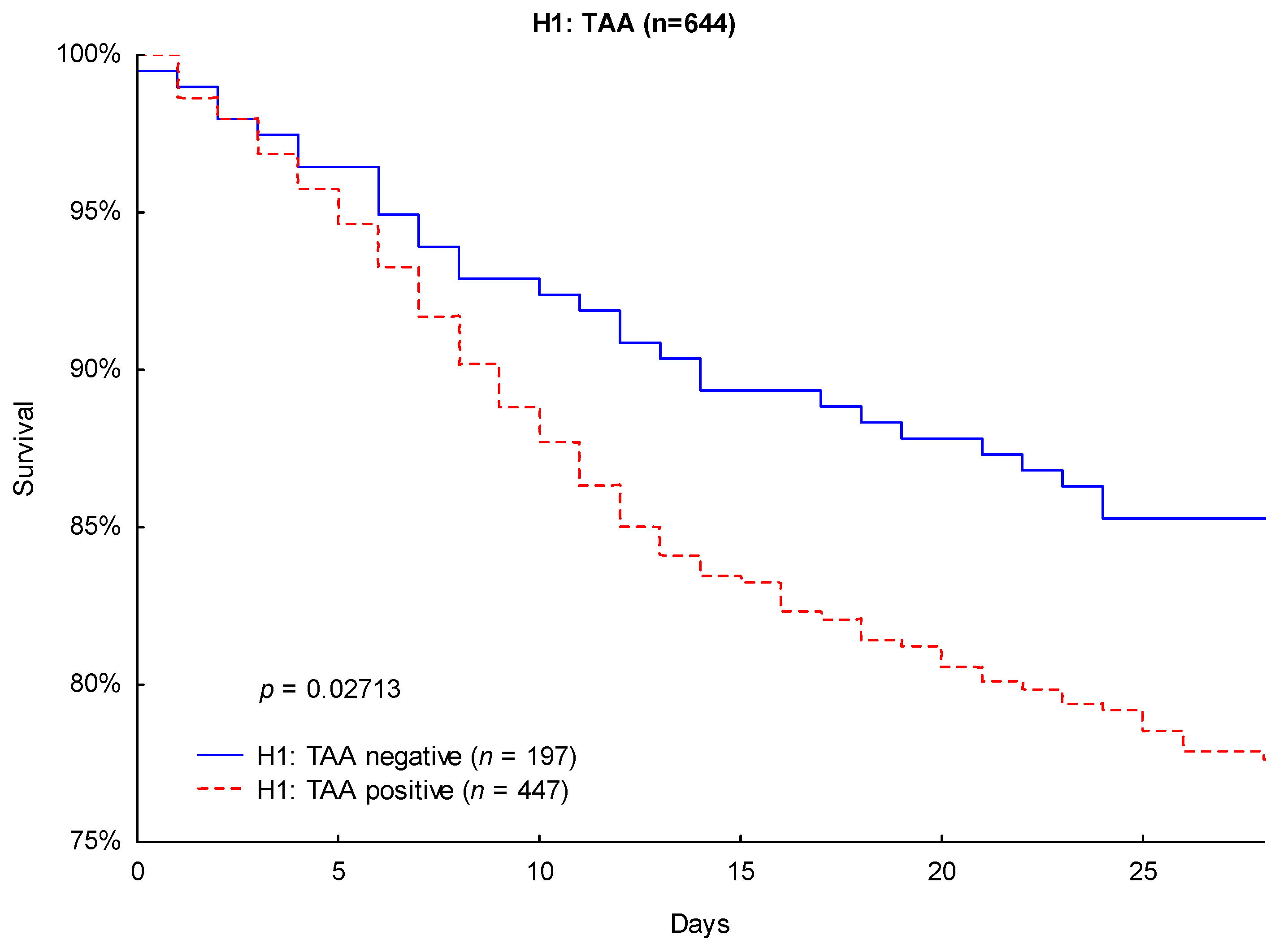
| H1: TAA negative | 0.698 | 0.504–0.967 | 0.030812 | 0.685 | 0.491–0.956 | 0.026202 |
| 28 Days | ||||||
| Univariate Analysis | Multivariate Analysis | |||||
| Age | 1.029 | 1.015–1.043 | 0.000035 | 1.023 | 1.009–1.039 | 0.001984 |
| Male gender | 1.278 | 0.875–1.866 | 0.204154 | 1.269 | 0.866–1.859 | 0.221286 |
| BMI | 0.982 | 0.953–1.013 | 0.252686 | 0.972 | 0.940–1.004 | 0.089311 |
| SOFA score day 1 | 1.119 | 1.071–1.169 | 0.000000 | 1.050 | 0.988–1.116 | 0.118403 |
| APACHE II score day 1 | 1.073 | 1.045–1.102 | 0.000000 | 1.026 | 0.990–1.064 | 0.155294 |
| Statin therapy | 0.964 | 0.641–1.451 | 0.861906 | 0.790 | 0.517–1.209 | 0.278234 |
| Renal replacement therapy during observation period | 3.028 | 2.131–4.301 | 0.000000 | 2.772 | 1.769–4.343 | 0.000009 |
| Renal replacement therapy upon sepsis onset | 1.616 | 0.970–2.691 | 0.065366 | 1.933 | 1.060–3.524 | 0.031464 |
| H1: TAA negative | 0.632 | 0.418–0.955 | 0.029340 | 0.621 | 0.407–0.947 | 0.027008 |
| Variable | All (n = 644) | rs3087243 | p-Value | |
|---|---|---|---|---|
| AA (n = 142) | GG/AG (n = 502) | |||
| SOFA | 7.0 ± 3.5 | 7.4 ± 3.8 | 6.9 ± 3.5 | 0.3005 |
| SOFA-Respiratory score | 2.0 ± 0.8 | 2.0 ± 0.8 | 2.0 ± 0.8 | 0.6043 |
| SOFA-Cardiovascular score | 1.5 ± 1.0 | 1.7 ± 1.0 | 1.5 ± 1.0 | 0.1284 |
| SOFA-Central Nervous System score | 2.0 ± 1.1 | 2.2 ± 1.0 | 1.9 ± 1.1 | 0.0094 |
| SOFA-Renal score | 0.8 ± 1.2 | 0.7 ± 1.1 | 0.8 ± 1.2 | 0.6669 |
| SOFA-Coagulation score | 0.4 ± 0.6 | 0.4 ± 0.7 | 0.3 ± 0.6 | 0.5521 |
| SOFA-Hepatic score | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.9780 |
| Organ support | ||||
| Ventilation days/observation days (%) | 66 ± 32 | 71 ± 32 | 65 ± 32 | 0.0280 |
| Vasopressor days/observation days (%) | 34 ± 30 | 37 ± 33 | 33 ± 29 | 0.2919 |
| Dialysis days/observation days (%) | 9 ± 23 | 9 ± 23 | 10 ± 22 | 0.8287 |
| Inflammatory values | ||||
| Leucocytes (1000/µL) | 13 ± 5 | 13 ± 5 | 13 ± 5 | 0.1026 |
| CRP (mg/L) | 152 ± 87 | 137 ± 81 | 156 ± 88 | 0.1356 |
| Procalcitonin (ng/dL) | 4.0 ± 9.3 | 3.5 ± 10.1 | 4.1 ± 9.1 | 0.0655 |
| Kidney values | ||||
| Urine output (mL/day) | 2978 ± 1337 | 2973 ± 1388 | 2979 ± 1324 | 0.9387 |
| Urine output (mL/kg/day) | 1.6 ± 0.8 | 1.6 ± 0.8 | 1.6 ± 0.78 | 0.9051 |
| Creatinine (mg/dL) | 1.2 ± 0.9 | 1.2 ± 0.9 | 1.3 ± 1.0 | 0.4218 |
| Liver values | ||||
| AST (GOT) (IU/L) | 169 ± 598 | 127 ± 238 | 182 ± 672 | 0.7476 |
| ALT (GPT) (IU/L) | 94 ± 188 | 101 ± 193 | 92 ± 187 | 0.2135 |
| Bilirubin (mg/dL) | 1.2 ± 2.1 | 1.2 ± 1.8 | 1.2 ± 2.1 | 0.5972 |
| Variable | All (n = 644) | H1: TAA | p-Value | |
|---|---|---|---|---|
| Positive (n = 447) | Negative (n = 197) | |||
| SOFA | 7.0 ± 3.5 | 7.0 ± 3.6 | 7.1 ± 3.5 | 0.5088 |
| SOFA-Respiratory score | 2.0 ± 0.8 | 2.0 ± 0.8 | 1.9 ± 0.8 | 0.9796 |
| SOFA-Cardiovascular score | 1.5 ± 1.0 | 1.5 ± 1.0 | 1.5 ± 1.0 | 0.7476 |
| SOFA-Central Nervous System score | 2.0 ± 1.1 | 2.0 ± 1.1 | 1.9 ± 1.1 | 0.2731 |
| SOFA-Renal score | 0.8 ± 1.2 | 0.7 ± 1.1 | 0.9 ± 1.3 | 0.1230 |
| SOFA-Coagulation score | 0.4 ± 0.6 | 0.4 ± 0.6 | 0.3 ± 0.6 | 0.6741 |
| SOFA-Hepatic score | 0.4 ± 0.7 | 0.4 ± 0.7 | 0.4 ± 0.8 | 0.4264 |
| Organ support | ||||
| Ventilation days/observation days (%) | 66 ± 32 | 67 ± 32 | 65 ± 32 | 0.3607 |
| Vasopressor days/observation days (%) | 34 ± 30 | 33 ± 30 | 35 ± 30 | 0.6474 |
| Dialysis days/observation days (%) | 9 ± 23 | 9 ± 22 | 11 ± 23 | 0.1246 |
| Inflammatory values | ||||
| Leucocytes (1000/µL) | 13 ± 5 | 13 ± 5 | 13 ± 4 | 0.4087 |
| CRP (mg/L) | 152 ± 87 | 147 ± 81 | 162 ± 99 | 0.4562 |
| Procalcitonin (ng/dL) | 4.0 ± 9.3 | 3.5 ± 9.1 | 5.1 ± 9.7 | 0.0002 |
| Kidney values | ||||
| Urine output (mL/day) | 2978 ± 1337 | 3060 ± 1370 | 2792 ± 1242 | 0.0229 |
| Urine output (mL/kg/day) | 1.6 ± 0.8 | 1.6 ± 0.8 | 1.5 ± 0.8 | 0.0529 |
| Creatinine (mg/dL) | 1.2 ± 0.9 | 1.2 ± 0.9 | 1.4 ± 1.1 | 0.0902 |
| Liver values | ||||
| AST (GOT) (IU/L) | 169 ± 598 | 184 ± 687 | 131 ± 272 | 0.9910 |
| ALT (GPT) (IU/L) | 94 ± 188 | 99 ± 206 | 81 ± 142 | 0.1307 |
| Bilirubin (mg/dL) | 1.2 ± 2.1 | 1.2 ± 1.8 | 1.3 ± 2.5 | 0.9584 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mewes, C.; Büttner, B.; Hinz, J.; Alpert, A.; Popov, A.-F.; Ghadimi, M.; Beissbarth, T.; Tzvetkov, M.; Jensen, O.; Runzheimer, J.; et al. CTLA-4 Genetic Variants Predict Survival in Patients with Sepsis. J. Clin. Med. 2019, 8, 70. https://doi.org/10.3390/jcm8010070
Mewes C, Büttner B, Hinz J, Alpert A, Popov A-F, Ghadimi M, Beissbarth T, Tzvetkov M, Jensen O, Runzheimer J, et al. CTLA-4 Genetic Variants Predict Survival in Patients with Sepsis. Journal of Clinical Medicine. 2019; 8(1):70. https://doi.org/10.3390/jcm8010070
Chicago/Turabian StyleMewes, Caspar, Benedikt Büttner, José Hinz, Ayelet Alpert, Aron-Frederik Popov, Michael Ghadimi, Tim Beissbarth, Mladen Tzvetkov, Ole Jensen, Julius Runzheimer, and et al. 2019. "CTLA-4 Genetic Variants Predict Survival in Patients with Sepsis" Journal of Clinical Medicine 8, no. 1: 70. https://doi.org/10.3390/jcm8010070
APA StyleMewes, C., Büttner, B., Hinz, J., Alpert, A., Popov, A.-F., Ghadimi, M., Beissbarth, T., Tzvetkov, M., Jensen, O., Runzheimer, J., Quintel, M., Shen-Orr, S., Bergmann, I., & Mansur, A. (2019). CTLA-4 Genetic Variants Predict Survival in Patients with Sepsis. Journal of Clinical Medicine, 8(1), 70. https://doi.org/10.3390/jcm8010070






