Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Construction of Plasmids
2.3. Cells and Transfection
2.4. Fly Strains
2.5. MG132 (Carbobenzoxy-L-leucyl-L-leucyl-L-leucinal) Treatment
2.6. Ambroxol Treatment
2.7. RNA Preparation
2.8. RT-PCR
2.9. Quantitative Real Time PCR (qRT-PCR)
2.10. Whole Transcriptome Sequencing and Analysis
2.11. Detection of Spliced Xbp1 mRNA Processing
2.12. SDS–PAGE and Western Blotting
2.13. Total Lipid Extraction and Quantification of GlcCer and Ceramide
2.14. GlcCer and GlcSph Determination by LC-MS/MS
2.15. GCase Activity Assay
2.16. GCase Labeling with Activity-Based Probes
2.17. Endoglycosidase H (Endo-H) Sensitivity
2.18. Lysotracker and Confocal Imaging
2.19. Isolation of Hemolymph and FACS Analysis
2.20. Climbing Assay
2.21. Survival Assay
2.22. Statistics
3. Results
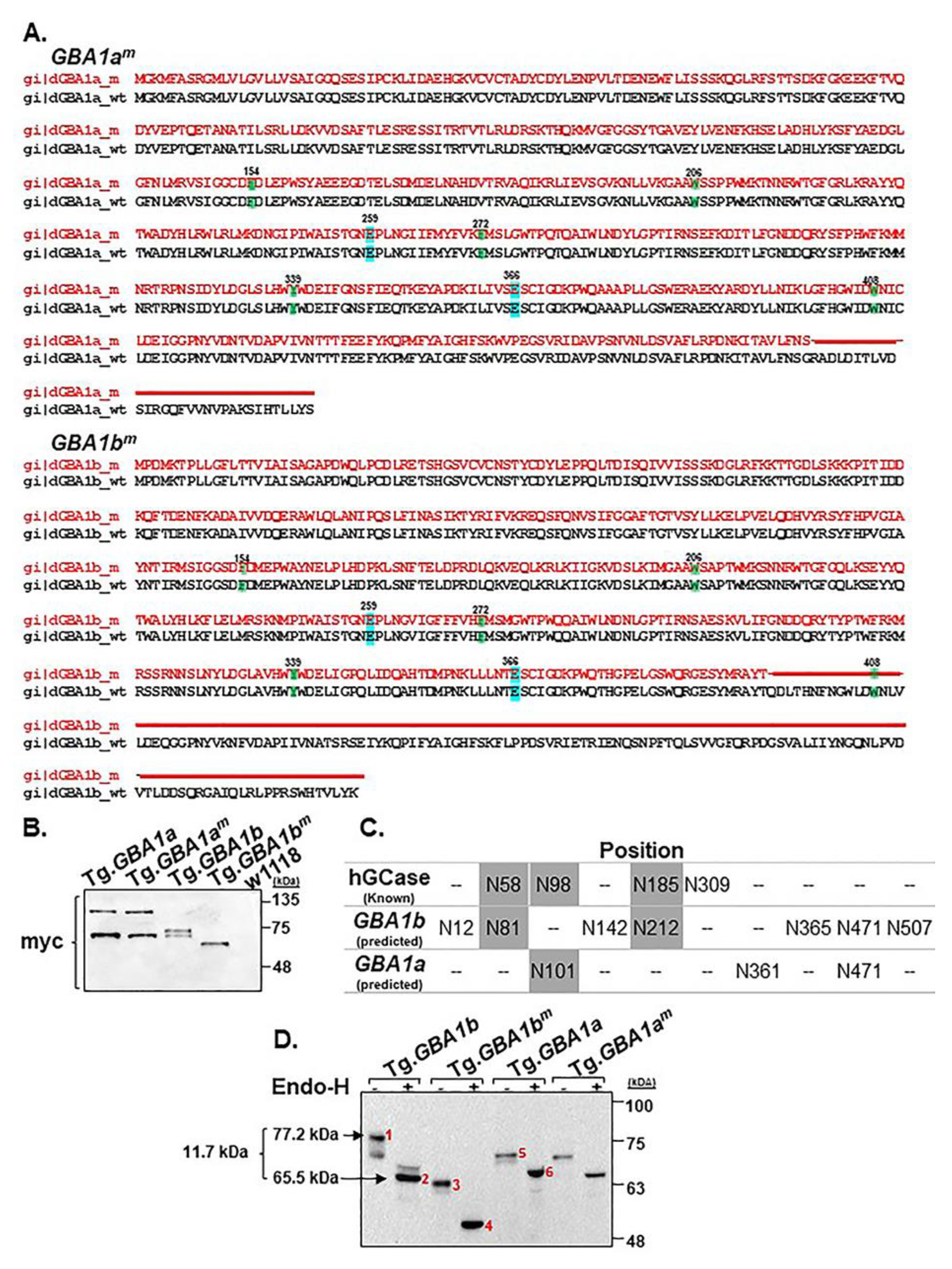
3.1. Expression of the Fly Normal and Mutant GBA1 Genes
3.2. GCase Activity and Substrate Accumulation
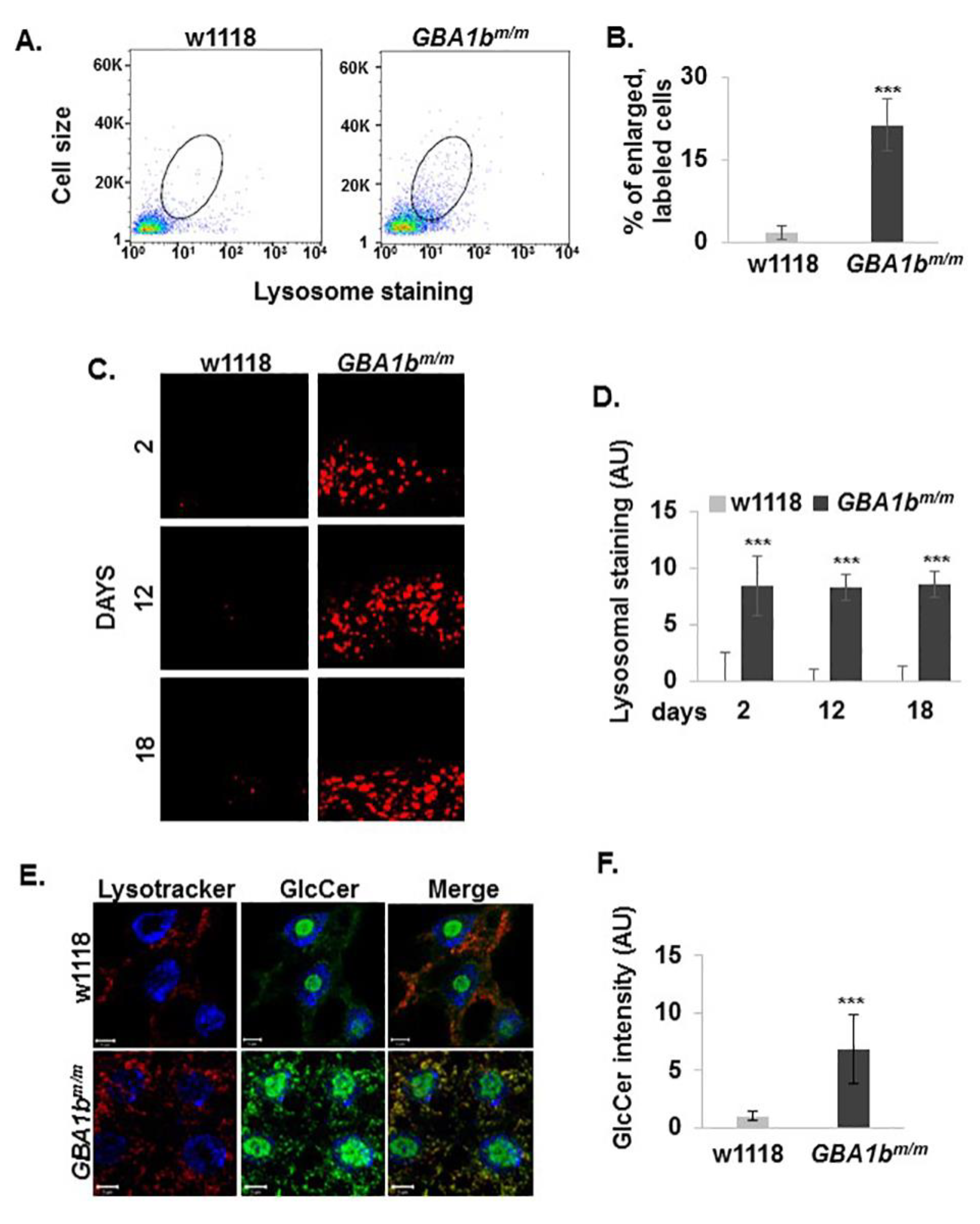
3.3. Changes in Lysosome Morphology in GBA1bm/m Mutant Flies
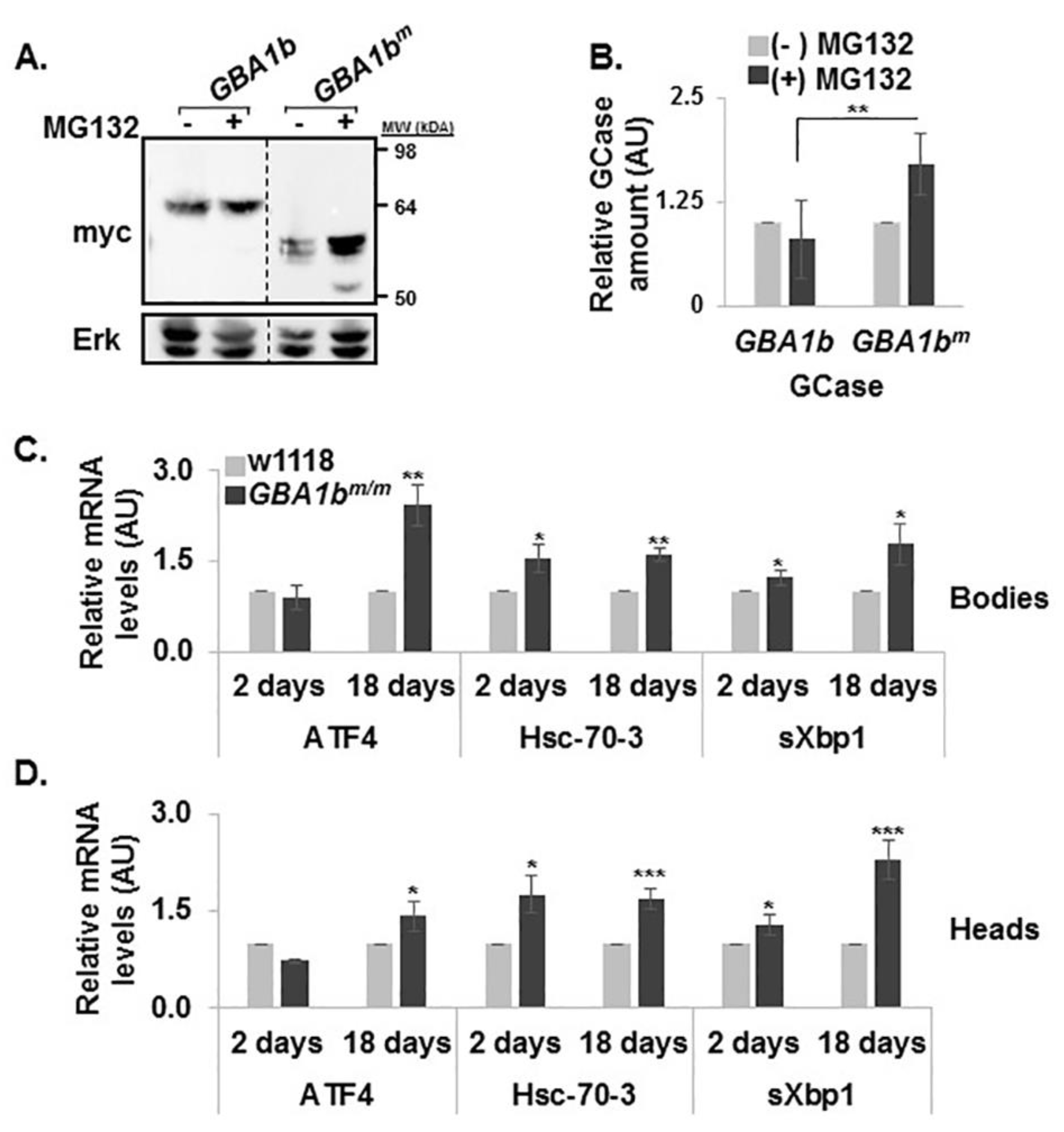
3.4. ERAD and UPR Activation in GBA1bm/m flies
3.5. Development of Inflammation and Neuroinflammation in GBA1bm/m flies
3.6. Partial Rescue of the GBA1bm/m Phenotype by Ambroxol
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brady, R.O.; Kanfer, J.N.; Shapiro, D. Metabolism of Glucocerebrosides. Ii. Evidence of an Enzymatic Deficiency in Gaucher’s Disease. Biochem. Biophys. Res. Commun. 1965, 18, 221–225. [Google Scholar] [CrossRef]
- Nilsson, O.; Svennerholm, L. Accumulation of Glucosylceramide and Glucosylsphingosine (Psychosine) in Cerebrum and Cerebellum in Infantile and Juvenile Gaucher Disease. J. Neurochem. 1982, 39, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Orviský, E.; Park, J.K.; LaMarca, M.; Ginns, E.I.; Martin, B.M.; Tayebi, N.; Sidransky, E. Glucosylsphingosine accumulation in tissues from patients with Gaucher disease: Correlation with phenotype and genotype. Mol. Genet. Metab. 2002, 76, 262–270. [Google Scholar] [CrossRef]
- Dekker, N.; Van Dussen, L.; Hollak, C.E.M.; Overkleeft, H.; Scheij, S.; Ghauharali, K.; Van Breemen, M.J.; Ferraz, M.J.; Groener, J.E.M.; Maas, M.; et al. Elevated plasma glucosylsphingosine in Gaucher disease: Relation to phenotype, storage cell markers, and therapeutic response. Blood 2011, 118, e118–e127. [Google Scholar] [CrossRef] [PubMed]
- Aerts, M.J.; Hollak, C.E. Plasma and metabolic abnormalities in Gaucher’s disease. Baillieres Clin. Haematol. 1997, 10, 691–709. [Google Scholar] [CrossRef]
- Hruska, K.S.; Lamarca, M.E.; Scott, C.R.; Sidransky, E. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA). Hum. Mutat. 2008, 29, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E.; Gelbart, T.; Kuhl, W.; Zimran, A.; West, C. Mutations in Jewish patients with Gaucher disease. Blood 1992, 79, 1662–1666. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, M.; Pasmanik-Chor, M.; Borochowitz, Z.; Falik-Zaccai, T.; Heldmann, K.; Carmi, R.; Parvari, R.; Beit-Or, H.; Goldman, B.; Peleg, L.; et al. Prevalence of glucocerebrosidase mutations in the Israeli Ashkenazi Jewish population. Hum. Mutat. 1998, 12, 240–244. [Google Scholar] [CrossRef]
- Aharon-Peretz, J.; Rosenbaum, H.; Gershoni-Baruch, R. Mutations in the Glucocerebrosidase Gene and Parkinson’s disease in Ashkenazi Jews. N. Engl. J. Med. 2004, 351, 1972–1977. [Google Scholar] [CrossRef]
- Horowitz, M.; Elstein, D.; Zimran, A.; Goker-Alpan, O. New Directions in Gaucher Disease. Hum. Mutat. 2016, 37, 1121–1136. [Google Scholar] [CrossRef]
- Tayebi, N.; Cushner, S.R.; Kleijer, W.; Lau, E.K.; Damschroder-Williams, P.J.; Stubblefield, B.K.; Hollander, J.D.; Sidransky, E. Prenatal lethality of a homozygous null mutation in the human glucocerebrosidase gene. Am. J. Med Genet. 1997, 73, 41–47. [Google Scholar] [CrossRef]
- Bendikov-Bar, I.; Horowitz, M.; Bendikov-Bar, I. Gaucher disease paradigm: From ERAD to comorbidity. Hum. Mutat. 2012, 33, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Maor, G.; Rencus-Lazar, S.; Filocamo, M.; Steller, H.; Segal, D.; Horowitz, M. Unfolded protein response in Gaucher disease: From human to Drosophila. Orphanet J. Rare Dis. 2013, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Ron, I.; Horowitz, M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum. Mol. Genet. 2005, 14, 2387–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farfel-Becker, T.; Vitner, E.B.; Futerman, A.H. Animal models for Gaucher disease research. Dis. Model. Mech. 2011, 4, 746–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.Y.; Trinh, K.; Thomas, R.E.; Yu, S.; Germanos, A.A.; Whitley, B.N.; Sardi, S.P.; Montine, T.J.; Pallanck, L.J. Glucocerebrosidase Deficiency in Drosophila Results in α-Synuclein-Independent Protein Aggregation and Neurodegeneration. PLoS Genet. 2016, 12, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Kinghorn, K.J.; Grönke, S.; Castillo-Quan, J.I.; Woodling, N.S.; Li, L.; Sirka, E.; Gegg, M.; Mills, K.; Hardy, J.; Bjedov, I.; et al. A Drosophila Model of Neuronopathic Gaucher Disease Demonstrates Lysosomal-Autophagic Defects and Altered mTOR Signalling and Is Functionally Rescued by Rapamycin. J. Neurosci. 2016, 36, 11654–11670. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Fujikake, N.; Takeuchi, T.; Kohyama-Koganeya, A.; Nakajima, K.; Hirabayashi, Y.; Wada, K.; Nagai, Y. Glucocerebrosidase deficiency accelerates the accumulation of proteinase K-resistant α-synuclein and aggravates neurodegeneration in aDrosophilamodel of Parkinson’s disease. Hum. Mol. Genet. 2015, 24, 6675–6686. [Google Scholar] [CrossRef]
- Sardiello, M.; Palmieri, M.; Di Ronza, A.; Valenza, M.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; Parenti, G.; Cattaneo, E.; et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef] [Green Version]
- Bouché, V.; Espinosa, A.P.; Leone, L.; Sardiello, M.; Ballabio, A.; Botas, J. Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 2016, 12, 484–498. [Google Scholar] [CrossRef]
- Kawasaki, H.; Suzuki, T.; Ito, K.; Takahara, T.; Goto-Inoue, N.; Setou, M.; Sakata, K.; Ishida, N. Minos-insertion mutant of the Drosophila GBA gene homologue showed abnormal phenotypes of climbing ability, sleep and life span with accumulation of hydroxy-glucocerebroside. Gene 2017, 614, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Pekar, O.; Benjamin, S.; Weidberg, H.; Smaldone, S.; Ramirez, F.; Horowitz, M. EHD2 shuttles to the nucleus and represses transcription. Biochem. J. 2012, 444, 383–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Statistical Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Ferraz, M.J.; Kallemeijn, W.W.; Mirzaian, M.; Moro, D.H.; Marques, A.R.A.; Wisse, P.; Boot, R.G.; Willems, L.I.; Overkleeft, H.; Aerts, J.; et al. Gaucher disease and Fabry disease: New markers and insights in pathophysiology for two distinct glycosphingolipidoses. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2014, 1841, 811–825. [Google Scholar] [CrossRef]
- Groener, J.E.; Poorthuis, B.J.; Kuiper, S.; Helmond, M.T.; Hollak, C.E.; Aerts, J.M. HPLC for Simultaneous Quantification of Total Ceramide, Glucosylceramide, and Ceramide Trihexoside Concentrations in Plasma. Clin. Chem. 2007, 53, 742–747. [Google Scholar] [CrossRef] [Green Version]
- Mirzaian, M.; Wisse, P.; Ferraz, M.J.; Gold, H.; Donker-Koopman, W.E.; Verhoek, M.; Overkleeft, H.S.; Boot, R.G.; Kramer, G.; Dekker, N.; et al. Mass spectrometric quantification of glucosylsphingosine in plasma and urine of type 1 Gaucher patients using an isotope standard. Blood Cells Mol. Dis. 2015, 54, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Mirzaian, M.; Wisse, P.; Ferraz, M.; Marques, A.R.A.; Gaspar, P.J.M.D.S.; Oussoren, S.; Kytidou, K.; Codée, J.; Van Der Marel, G.; Overkleeft, H.; et al. Simultaneous quantitation of sphingoid bases by UPLC-ESI-MS/MS with identical 13 C-encoded internal standards. Clin. Chim. Acta 2017, 466, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Witte, M.D.; Kallemeijn, W.W.; Aten, J.; Li, K.-Y.; Strijland, A.; Donker-Koopman, W.E.; Nieuwendijk, A.M.C.H.V.D.; Bleijlevens, B.; Kramer, G.; Florea, B.I.; et al. Ultrasensitive in situ visualization of active glucocerebrosidase molecules. Nat. Methods 2010, 6, 907–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, H.K.; Kamikouchi, A.; Ito, K. Methods for quantifying simple gravity sensing in Drosophila melanogaster. Nat. Protoc. 2010, 5, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Dvir, H.; Harel, M.; McCarthy, A.A.; Toker, L.; Silman, I.; Futerman, A.H.; Sussman, J.L. X-ray structure of human acid-β-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003, 4, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Kohyama-Koganeya, A.; Hirabayashi, Y. New insights on glucosylated lipids: Metabolism and functions. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 1475–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beutler, E. Gaucher’s disease. Compr. Ther. 1980, 6, 65–68. [Google Scholar] [PubMed]
- Bessler, H.; Djaldetti, M.; Fishman, P. The Surface Ultrastructure of Gaucher Cells. Am. J. Clin. Pathol. 1979, 71, 146–150. [Google Scholar]
- Wang, L.; Kounatidis, I.; Ligoxygakis, P. Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer. Front. Microbiol. 2014, 3, 113. [Google Scholar] [CrossRef]
- Gözdaşoğlu, S. Gaucher Disease and Gaucher Cells. Turk. J. Hematol. 2015, 32, 187–188. [Google Scholar] [CrossRef]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Selective Inhibitors of the Proteasome-dependent and Vacuolar Pathways of Protein Degradation inSaccharomyces cerevisiae. J. Boil. Chem. 1996, 271, 27280–27284. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.M.; Mehta, A.D.; Zhu, J.; Shoham, S.; Chen, X.; Wells, Q.R.; Palter, K.B. Genomic structure and sequence analysis of Drosophila melanogaster HSC70 genes. Gene 1993, 128, 155–163. [Google Scholar] [CrossRef]
- Chan, S.M.; Zhao, X.; Elfowiris, A.; Ratnam, C.; Herbert, T.P. The role of de novo protein synthesis and SIRT1 in ER stress-induced Atf4 and Chop mRNA expression in mammalian cells. Biochime 2017, 138, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, H.D.; Domingos, P.M.; Kang, M.J.; Steller, H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007, 26, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Verhoek, M.; De Fost, M.; Hollak, C.E.M.; Maas, M.; Bleijlevens, B.; Van Breemen, M.J.; Van Meurs, M.; Boven, L.A.; Laman, J.D.; et al. Marked elevation of the chemokine CCL18/PARC in Gaucher disease: A novel surrogate marker for assessing therapeutic intervention. Blood 2004, 103, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Farfel-Becker, T.; Vitner, E.B.; Pressey, S.N.R.; Eilam, R.; Cooper, J.D.; Futerman, A.H. Spatial and temporal correlation between neuron loss and neuroinflammation in a mouse model of neuronopathic Gaucher disease. Hum. Mol. Genet. 2011, 20, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Aflaki, E.; Moaven, N.; Borger, D.K.; Lopez, G.; Westbroek, W.; Chae, J.J.; Marugan, J.; Patnaik, S.; Maniwang, E.; Gonzalez, A.N.; et al. Lysosomal storage and impaired autophagy lead to inflammasome activation in Gaucher macrophages. Aging Cell 2016, 15, 77–88. [Google Scholar] [CrossRef]
- Vitner, E.B.; Farfel-Becker, T.; Ferreira, N.S.; Leshkowitz, D.; Sharma, P.; Lang, K.S.; Futerman, A.H. Induction of the type I interferon response in neurological forms of Gaucher disease. J. Neuroinflamm. 2016, 13, 559. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Cao, Y.; Chtarbanova, S.; Petersen, A.J.; Ganetzky, B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. USA 2013, 110, E1752–E1760. [Google Scholar] [CrossRef]
- Hollak, C.E.; van Weely, S.; van Oers, M.H.; Aerts, J.M. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J. Clin. Investig. 1994, 93, 1288–1292. [Google Scholar] [CrossRef] [PubMed]
- Jung, O.; Patnaik, S.; Marugan, J.; Sidransky, E.; Westbroek, W. Progress and potential of non-inhibitory small molecule chaperones for the treatment of Gaucher disease and its potential implications for Parkinson disease. Expert Rev. Proteom. 2016, 13, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Babajani, G.; Tropak, M.B.; Mahuran, D.J.; Kermode, A.R. Pharmacological chaperones facilitate the post-ER transport of recombinant N370S mutant β-glucocerebrosidase in plant cells: Evidence that N370S is a folding mutant. Mol. Genet. Metab. 2012, 106, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Maegawa, G.H.B.; Tropak, M.B.; Buttner, J.D.; Rigat, B.A.; Fuller, M.; Pandit, D.; Tang, L.; Kornhaber, G.J.; Hamuro, Y.; Clarke, J.T.R.; et al. Identification and Characterization of Ambroxol as an Enzyme Enhancement Agent for Gaucher Disease*. J. Boil. Chem. 2009, 284, 23502–23516. [Google Scholar] [CrossRef]
- Bendikov-Bar, I.; Ron, I.; Filocamo, M.; Horowitz, M. Characterization of the ERAD process of the L444P mutant glucocerebrosidase variant. Blood Cells, Mol. Dis. 2011, 46, 4–10. [Google Scholar] [CrossRef]
- Narita, A.; Shirai, K.; Itamura, S.; Matsuda, A.; Ishihara, A.; Matsushita, K.; Fukuda, C.; Kubota, N.; Takayama, R.; Shigematsu, H.; et al. Ambroxol chaperone therapy for neuronopathic Gaucher disease: A pilot study. Ann. Clin. Transl. Neurol. 2016, 3, 200–215. [Google Scholar] [CrossRef]
- Suzuki, T.; Shimoda, M.; Ito, K.; Hanai, S.; Aizawa, H.; Kato, T.; Kawasaki, K.; Yamaguchi, T.; Ryoo, H.D.; Goto-Inoue, N.; et al. Expression of Human Gaucher Disease Gene GBA Generates Neurodevelopmental Defects and ER Stress in Drosophila Eye. PLoS ONE 2013, 8, e69147. [Google Scholar] [CrossRef]
- Rolfs, A.; Giese, A.-K.; Grittner, U.; Mascher, D.; Elstein, D.; Zimran, A.; Böttcher, T.; Lukas, J.; Hübner, R.; Gölnitz, U.; et al. Glucosylsphingosine Is a Highly Sensitive and Specific Biomarker for Primary Diagnostic and Follow-Up Monitoring in Gaucher Disease in a Non-Jewish, Caucasian Cohort of Gaucher Disease Patients. PLoS ONE 2013, 8, 79732. [Google Scholar] [CrossRef]
- Uemura, N.; Koike, M.; Ansai, S.; Kinoshita, M.; Ishikawa-Fujiwara, T.; Matsui, H.; Naruse, K.; Sakamoto, N.; Uchiyama, Y.; Todo, T.; et al. Viable Neuronopathic Gaucher Disease Model in Medaka (Oryzias latipes) Displays Axonal Accumulation of Alpha-Synuclein. PLoS Genet. 2015, 11, e1005065. [Google Scholar] [CrossRef]
- Boven, L.A.; Boot, R.G.; Mehta, A.; Boon, L.; Aerts, J.M.; Laman, J.D.; Van Meurs, M. Gaucher Cells Demonstrate a Distinct Macrophage Phenotype and Resemble Alternatively Activated Macrophages. Am. J. Clin. Pathol. 2004, 122, 359–369. [Google Scholar] [CrossRef]
- Vitner, E.B.; Farfel-Becker, T.; Eilam, R.; Biton, I.; Futerman, A.H. Contribution of brain inflammation to neuronal cell death in neuronopathic forms of Gaucher’s disease. Brain 2012, 135, 1724–1735. [Google Scholar] [CrossRef]
- Pandey, M.K.; Jabre, N.A.; Xu, Y.-H.; Zhang, W.; Setchell, K.D.; Grabowski, G.A. Gaucher disease: Chemotactic factors and immunological cell invasion in a mouse model. Mol. Genet. Metab. 2014, 111, 163–171. [Google Scholar] [CrossRef]
- Keatinge, M.; Bui, H.; Menke, A.; Chen, Y.-C.; Sokol, A.M.; Bai, Q.; Ellett, F.; Da Costa, M.; Burke, D.; Gegg, M.; et al. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum. Mol. Genet. 2015, 24, 6640–6652. [Google Scholar] [CrossRef]
- Watson, L.; Keatinge, M.; Gegg, M.; Bai, Q.; Sandulescu, M.C.; Vardi, A.; Futerman, A.H.; Schapira, A.H.; Burton, E.A.; Bandmann, O. Ablation of the pro-inflammatory master regulator miR-155 does not mitigate neuroinflammation or neurodegeneration in a vertebrate model of Gaucher’s disease. Neurobiol. Dis. 2019, 127, 563–569. [Google Scholar] [CrossRef]
- Liu, J.; Halene, S.; Yang, M.; Iqbal, J.; Yang, R.; Mehal, W.Z.; Chuang, W.-L.; Jain, D.; Yuen, T.; Sun, L.; et al. Gaucher disease gene GBA functions in immune regulation. Proc. Natl. Acad. Sci. USA 2012, 109, 10018–10023. [Google Scholar] [CrossRef]
- Schiffmann, R.; FitzGibbon, E.J.; Harris, C.; Devile, C.; Davies, E.H.; Abel, L.; Van Schaik, I.N.; Benko, W.S.; Timmons, M.; Ries, M.; et al. Randomized, Controlled Trial of Miglustat in Gaucher’s Disease Type 3. Ann. Neurol. 2008, 64, 514–522. [Google Scholar] [CrossRef]
- De La Mata, M.; Cotán, D.; Oropesa-Ávila, M.; Garrido-Maraver, J.; Cordero, M.D.; Paz, M.V.; Pavón, A.D.; Alcocer-Gómez, E.; De Lavera, I.; Ybot-Gonzalez, P.; et al. Pharmacological Chaperones and Coenzyme Q10 Treatment Improves Mutant β-Glucocerebrosidase Activity and Mitochondrial Function in Neuronopathic Forms of Gaucher Disease. Sci. Rep. 2015, 5, 10903. [Google Scholar] [CrossRef]
- Luan, Z.; Li, L.; Higaki, K.; Nanba, E.; Suzuki, Y.; Ohno, K. The chaperone activity and toxicity of ambroxol on Gaucher cells and normal mice. Brain Dev. 2013, 35, 317–322. [Google Scholar] [CrossRef]
- Beeh, K.M.; Beier, J.; Esperester, A.; Paul, L.D. Antiinflammatory properties of ambroxol. Eur. J. Med Res. 2008, 13, 557–562. [Google Scholar]
- Bianchi, M.; Mantovani, A.; Erroi, A.; Dinarello, C.A.; Ghezzi, P. Ambroxol inhibits interleukin 1 and tumor necrosis factor production in human mononuclear cells. Inflamm. Res. 1990, 31, 275–279. [Google Scholar] [CrossRef]
- Su, X.; Wang, L.; Song, Y.; Bai, C. Inhibition of inflammatory responses by ambroxol, a mucolytic agent, in a murine model of acute lung injury induced by lipopolysaccharide. Intensiv. Care Med. 2004, 30, 133–140. [Google Scholar] [CrossRef]




| Name | Primers for Construction of Plasmids |
| GBA1a | F: 5′-CTGAATAGGGAATTGGGAATTCGTATGGGAAAAATGTTCGC-3′ |
| R: 5′-GAGGTACCCTCGAGCATTTCAAGCACTTATTGAAGAGAACGGCGG-3′ | |
| GBA1b | F: 5′-CTGAATAGGGAATTGGGAATTCGTATGCCAGATATGAAGACAC-3′ |
| R: 5′-CCCTCTAGAGGTACCCTCGAGTAGGCCCTCATATAGCTTTCACC-3′ | |
| Name | Primers for qRT-PCR |
| GBA1a | F: 5′-GAGTGGTTCCTTATCTCCAGTT-3′ |
| R: 5′-ACGGTAAACTTCTCCTCCTTAC-3′ | |
| GBA1b | F: 5′-AAGAACTTCCGGTGGAGCTA-3′ |
| R: 5′-CAATTCATTGTATGCCCAGGGT-3′ | |
| sXbp1 | F: 5′-CCGAACTGAAGCAGCAACAGC-3′ |
| R: 5′-GTATACCCTGCGGCAGATCC-3′ | |
| HSC-70-3 | F: 5′-GCTGGTGTTATTGCCGGTCTGC-3′ |
| R: 5′-GATGCCTCGGGATGGTTCCTTGC-3′ | |
| ATF4 | F: 5′-AGACGCTGCTTCGCTTCCTTC-3′ |
| R: 5′-GCCCGTAAGTGCGAGTACGCT-3′ | |
| ATTC | F: 5′-CTGCACTGGACTACTCCCACATCA-3′ |
| R: 5′-CGATCCTGCGACTGCCAAAGATTG-3′ | |
| Cec | F: 5′-CATTGGACAATCGGAAGCTGGGTG-3′ |
| R: 5′-TAATCATCGTGGTCAACCTCGGGC-3′ | |
| Drs | F: 5′-AGTACTTGTTCGCCCTCTTCGCTG-3′ |
| R: 5′-CCTTGTATCTTCCGGACAGGCAGT-3′ | |
| Mtk | F: 5′-CATCAATCAATTCCCGCCACCGAG-3′ |
| R: 5′-AAATGGGTCCCTGGTGACGATGAG-3′ | |
| RP49 | F: 5′-TAAGAAGCGCACAAAGCACT-3′ |
| R: 5′-GGGCATCAGATATTGTCCCT-3′ |
| Biological Process | All Known Participating Genes (D. Mel.) | Expected | Observed | Fold Enrichment | p-Value | FDR |
|---|---|---|---|---|---|---|
| Response to external biotic stimulus | 363 | 1.37 | 18 | 13.13 | 1.03 × 10−15 | 8.04 × 10−12 |
| Response to bacterium | 256 | 0.97 | 15 | 15.57 | 3.88 × 10−14 | 7.56 × 10−11 |
| Defense response | 367 | 1.39 | 15 | 10.82 | 5.65 × 10−12 | 8.81 × 10−9 |
| Defense response to Gram-positive bacterium | 56 | 0.21 | 8 | 37.82 | 8.63 × 10−11 | 1.12 × 10−7 |
| Response to external stimulus | 912 | 3.44 | 19 | 5.52 | 4.18 × 10−10 | 4.66 × 10−7 |
| Defense response to bacterium | 224 | 0.85 | 11 | 13 | 8.90 × 10−10 | 8.68 × 10−7 |
| Multi-organism process | 1431 | 5.41 | 22 | 4.07 | 3.10 × 10−9 | 2.69 × 10−6 |
| Response to stress | 1122 | 4.24 | 19 | 4.48 | 1.24 × 10−8 | 8.78 × 10−6 |
| Defense response to other organism | 291 | 1.1 | 11 | 10.01 | 1.23 × 10−8 | 9.61 × 10−6 |
| Humoral immune response | 87 | 0.33 | 7 | 21.3 | 5.60 × 10−8 | 3.64 × 10−5 |
| Antimicrobial humoral response | 72 | 0.27 | 6 | 22.06 | 4.41 × 10−7 | 2.64 × 10−4 |
| Immune response | 192 | 0.73 | 8 | 11.03 | 7.30 × 10−7 | 4.06 × 10−4 |
| Biological Process | All Known Participating Genes (D. Mel.) | Expected | Observed | Fold Enrichment | p-Value | FDR |
|---|---|---|---|---|---|---|
| Antibacterial humoral response | 28 | 0.09 | 6 | 68.61 | 7.99 × 10−10 | 6.23 × 10−6 |
| Antimicrobial humoral response | 72 | 0.22 | 7 | 31.13 | 4.17 × 10−9 | 1.08 × 10−5 |
| Immune system process | 316 | 0.99 | 11 | 11.14 | 3.24 × 10−9 | 1.26 × 10−5 |
| Humoral immune response | 87 | 0.27 | 7 | 25.76 | 1.42 × 10−9 | 1.59 × 10−5 |
| Response to biotic stimulus | 363 | 1.13 | 11 | 9.7 | 1.31 × 10−8 | 1.70 × 10−5 |
| Defense response to Gram-positive bacterium | 56 | 0.17 | 6 | 34.3 | 3.42 × 10−8 | 3.34 × 10−5 |
| Response to bacterium | 256 | 0.8 | 9 | 11.26 | 9.82 × 10−8 | 8.51 × 10−5 |
| Immune response | 192 | 0.6 | 8 | 13.34 | 1.58 × 10−7 | 1.23 × 10−4 |
| Defense response | 367 | 1.15 | 10 | 8.72 | 1.75 × 10−7 | 1.24 × 10−4 |
| Defense response to other organism | 291 | 0.91 | 9 | 9.9 | 2.82 × 10−7 | 1.83 × 10−4 |
| Defense response to bacterium | 224 | 0.7 | 8 | 11.43 | 4.91 × 10−7 | 2.95 × 10−4 |
| Response to external stimulus | 912 | 2.85 | 13 | 4.56 | 2.78 × 10−6 | 1.55 × 10−3 |
| Response to stimulus | 2640 | 8.25 | 21 | 2.55 | 1.14 × 10−5 | 5.94 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabasso, O.; Paul, S.; Dorot, O.; Maor, G.; Krivoruk, O.; Pasmanik-Chor, M.; Mirzaian, M.; Ferraz, M.; Aerts, J.; Horowitz, M. Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease. J. Clin. Med. 2019, 8, 1420. https://doi.org/10.3390/jcm8091420
Cabasso O, Paul S, Dorot O, Maor G, Krivoruk O, Pasmanik-Chor M, Mirzaian M, Ferraz M, Aerts J, Horowitz M. Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease. Journal of Clinical Medicine. 2019; 8(9):1420. https://doi.org/10.3390/jcm8091420
Chicago/Turabian StyleCabasso, Or, Sumit Paul, Orly Dorot, Gali Maor, Olga Krivoruk, Metsada Pasmanik-Chor, Mina Mirzaian, Maria Ferraz, Johannes Aerts, and Mia Horowitz. 2019. "Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease" Journal of Clinical Medicine 8, no. 9: 1420. https://doi.org/10.3390/jcm8091420
APA StyleCabasso, O., Paul, S., Dorot, O., Maor, G., Krivoruk, O., Pasmanik-Chor, M., Mirzaian, M., Ferraz, M., Aerts, J., & Horowitz, M. (2019). Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease. Journal of Clinical Medicine, 8(9), 1420. https://doi.org/10.3390/jcm8091420




