Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19
Abstract
:1. Introduction
2. Coronavirus
3. Potential Mechanisms of Coronavirus Invasion
3.1. Molecular Mechanisms of Coronavirus Invasion
3.2. Factors Involved in Transcription and Release of Coronavirus Particles
4. Current Treatment of Coronavirus
5. Current Diagnosis and Treatment of COVID-19
5.1. Current Molecular Diagnostic Assays
5.2. Treatment of COVID-19 with Remdesivir
5.3. Pharmacological Therapeutics Targeting Proteinase of SARS-Cov-2
5.4. Broad-Spectrum Antiviral Compounds NHC and EIDD-2801
5.5. Application of Anti-Viral Natural Products for Treatment of COVID-19
6. Spike Glycoproteins of SARS-CoV-2 and ACE2
7. Clinical trials of Remdesivir for Treatment of COVID-19 in China
7.1. Treatment of Mild/Moderate Case of COVID-19 with Remdesivir RCT (ClinicalTrials.gov Identifier: NCT04252664)
7.2. Treatment of Severe Case of COVID-19 with Remdesivir RCT (ClinicalTrials.gov Identifier: NCT04257656)
8. Coronavirus and Host Interaction
9. Concluding Remarks
Author Contribution
Funding
Conflicts of Interest
Abbreviations
| ARDS | Acute respiratory distress syndrome |
| ADE | Antibody-dependent enhancement |
| SARS-Cov-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Novel Coronavirus disease-2019 |
| 3CLpro | Coronavirus main proteinase |
| ACE2 | Angiotensin-converting Enzyme 2 |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cov | Coronavirus |
| dsRNA | Double strand RNA |
| DPP4 | Dipeptidyl-peptidase 4 |
| ERGIC | ER-Golgi intermediate compartment |
| EBOV | Ebola virus |
| gRNA | Guide RNA |
| GSK3 | Glycogen synthase kinase 3 |
| hM-RNAP | Human mitochondrial RNA polymerase |
| Orf1 | Open reading frame 1 |
| PLpro | Papain-like protease |
| MERS | Middle East Respiratory Syndrome |
| MPO | Myeloperoxidase |
| NTD | N-terminal domain |
| NMBA | Neuromuscular blocking agents |
| NHC | β-D-N4-hydroxycytidine |
| N protein | Coronavirus nucleoproteins |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| RBD | Receptor-binding domain |
| RT-PCR | Reverse transcription PCR |
| SARS | Severe Acute Respiratory Syndrome |
| S protein | Spike glycoprotein |
| ssRNA | Single strand RNA |
| SPR | Surface plasmon resonance |
| SKP2 | S-phase kinase-associated protein 2 |
| SpO2 | Peripheral capillary oxygen saturation |
| RdRp | RNA-dependent RNA Polymerase |
| RBD | Receptor-binding domain |
| TMPSS2 | Transmembrane protease serine 2 |
| TATs | Trioxa-adamantane-triols |
| UPR | Unfolded protein response |
| VLPs | Virus-like particles |
References
- Service, R.F. Coronavirus epidemic snarls science worldwide. Science 2020, 367, 836–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Coronavirus Disease (COVID-19) Pandemic; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Park, M.; Cook, A.; Lim, J.T.; Sun, Y.; Dickens, B. A Systematic Review of COVID-19 Epidemiology Based on Current Evidence. J. Clin. Med. 2020, 9, 967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellberg, B.; Haddix, M.; Lee, R.; Butler-Wu, S.; Holtom, P.; Yee, H.; Gounder, P. Community Prevalence of SARS-CoV-2 Among Patients With Influenzalike Illnesses Presenting to a Los Angeles Medical Center in March 2020. JAMA 2020. [Google Scholar] [CrossRef] [PubMed]
- Bourouiba, L. Turbulent Gas Clouds and Respiratory Pathogen Emissions: Potential Implications for Reducing Transmission of COVID-19. JAMA 2020. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, C.; Malani, P.N. 2019 Novel Coronavirus—Important Information for Clinicians. JAMA 2020, 323, 1039. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Heymann, D.L. Data sharing and outbreaks: Best practice exemplified. Lancet 2020, 395, 469–470. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.-C.; Deng, Q.-X.; Dai, S.-X. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Cao, Q.; Qin, L.; Wang, X.; Cheng, Z.; Pan, A.; Dai, J.; Sun, Q.; Zhao, F.; Qu, J.; et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):A multi-center study in Wenzhou city, Zhejiang, China. J. Infect. 2020, 80, 388–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Li, J.; Li, X.; Qi, X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology 2020, 295, 18. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Duan, F.; Luo, C.; Liu, Q.; Qu, X.; Liang, L.; Wu, K. Characteristics of Ocular Findings of Patients With Coronavirus Disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, X.; Tong, Q.; Li, W.; Wang, B.; Sutter, K.; Trilling, M.; Lu, M.; Dittmer, U.; Yang, D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med.Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.-W.; Wu, X.-X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Krüger, N.; Müller, M.; Drosten, C.; Pöhlmann, S.; A Muller, M. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Mullard, A. Ebola outbreak prompts experimental drug rollout. Nat. Rev. Drug Discov. 2018, 17, 460. [Google Scholar] [CrossRef]
- Zuñiga, S.; Cruz, J.L.G.; Sola, I.; Mateos-Gómez, P.; Palacio, L.; Enjuanes, L. Coronavirus Nucleocapsid Protein Facilitates Template Switching and Is Required for Efficient Transcription. J. Virol. 2009, 84, 2169–2175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, K.-A.; Dee, M.; Britton, P.; Hiscox, J.A. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology 2008, 370, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuñiga, S.; Sola, I.; Moreno, J.L.; Sabella, P.; Plana-Durán, J.; Enjuanes, L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 2007, 357, 215–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-H.; Yeh, S.-H.; Tsay, Y.-G.; Shieh, Y.-H.; Kao, C.-L.; Chen, Y.-S.; Wang, S.-H.; Kuo, T.-J.; Chen, P.-J.; Chen, P.-J. Glycogen Synthase Kinase-3 Regulates the Phosphorylation of Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein and Viral Replication. J. Boil. Chem. 2008, 284, 5229–5239. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Chen, Q.; Chen, J.; Chen, K.; Shen, X.; Jiang, H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005, 579, 2623–2628. [Google Scholar] [CrossRef] [Green Version]
- Nakauchi, M.; Kariwa, H.; Kon, Y.; Yoshii, K.; Maeda, A.; Takashima, I. Analysis of severe acute respiratory syndrome coronavirus structural proteins in virus-like particle assembly. Microbiol. Immunol. 2008, 52, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.M.; et al. The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.P.; Liu, D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001, 276, 17515–17523. [Google Scholar] [CrossRef] [Green Version]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [Green Version]
- Brandizzi, F.; Barlowe, C. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Boil. 2013, 14, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Genet. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, K.; Berkhout, B.; Van Der Hoek, L. Antiviral strategies against human coronaviruses. Infect. Disord.-Drug Targets 2007, 7, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Tetro, J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020, 22, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Peeples, L. News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA 2020, 202005456. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Chudhary, S.A.; Imtiaz, S.; Iqbal, N. Laboratory Detection of Novel Corona Virus 2019 using Polymerase Chain Reaction. Int. J. Front. Sci. 2020, 4. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Seletsky, A.; O’Connell, M.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222–224. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, eabb3405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadam, R.U.; Wilson, I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA 2016, 114, 206–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, S.G.V.; Santos, W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Pública 2020, 44. [Google Scholar] [CrossRef]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Venkataraman, S.; Prasad, B.V.L.S.; Selvaraj, V. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundin, A.; Dijkman, R.; Bergström, T.; Kann, N.; Adamiak, B.; Hannoun, C.; Kindler, E.; Jonsdottir, H.; Muth, D.; Kint, J.; et al. Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus. PLoS Pathog. 2014, 10, e1004166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv 2020. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [Green Version]
- de Wit, E.; Feldmann, F.; Cronin, J.; Jordan, R.; Okumura, A.; Thomas, T.; Scott, D.; Cihlar, T.; Feldmann, H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2020, 117, 6771–6776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaeppler, U.; Stiefl, N.; Schiller, M.; Vicik, R.; Breuning, A.; Schmitz, W.; Rupprecht, D.; Schmuck, C.; Baumann, K.; Ziebuhr, J.; et al. A New Lead for Nonpeptidic Active-Site-Directed Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus Main Protease Discovered by a Combination of Screening and Docking Methods‖. J. Med. Chem. 2005, 48, 6832–6842. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.; Mukherjee, P.; Desai, P.; Avery, M. Computational approaches for the discovery of cysteine protease inhibitors against malaria and SARS. Curr. Comput. Drug Des. 2010, 6, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Martina, E.; Stiefl, N.; Degel, B.; Schulz, F.; Breuning, A.; Schiller, M.; Vicik, R.; Baumann, K.; Ziebuhr, J.; Schirmeister, T. Screening of electrophilic compounds yields an aziridinyl peptide as new active-site directed SARS-CoV main protease inhibitor. Bioorganic Med. Chem. Lett. 2005, 15, 5365–5369. [Google Scholar] [CrossRef]
- Jacobs, J.; Grum-Tokars, V.; Zhou, Y.; Turlington, M.; Saldanha, S.A.; Chase, P.; Eggler, A.; Dawson, E.S.; Baez-Santos, Y.M.; Tomar, S.; et al. Discovery, Synthesis, And Structure-Based Optimization of a Series ofN-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease. J. Med. Chem. 2013, 56, 534–546. [Google Scholar]
- Shie, J.J.; Fang, J.M.; Kuo, T.H.; Kuo, C.J.; Liang, P.H.; Huang, H.J.; Wu, Y.T.; Jan, J.T.; Cheng, Y.S.; Wong, C.H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic alpha,beta-unsaturated esters. Bioorg. Med. Chem. 2005, 13, 5240–5252. [Google Scholar] [CrossRef]
- Zhang, H.-Z.; Zhang, H.; Kemnitzer, W.; Tseng, B.; Cinatl, J.; Michaelis, M.; Doerr, H.W.; Cai, S.X. Design and Synthesis of Dipeptidyl Glutaminyl Fluoromethyl Ketones as Potent Severe Acute Respiratory Syndrome Coronovirus (SARS-CoV) Inhibitors. J. Med. Chem. 2006, 49, 1198–1201. [Google Scholar] [CrossRef]
- Severson, W.E.; Shindo, N.; Sosa, M.; Fletcher, T.; White, E.L.; Ananthan, S.; Jonsson, C.B. Development and Validation of a High-Throughput Screen for Inhibitors of SARS CoV and Its Application in Screening of a 100,000-Compound Library. J. Biomol. Screen. 2007, 12, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Konno, S.; Thanigaimalai, P.; Yamamoto, T.; Nakada, K.; Kakiuchi, R.; Takayama, K.; Yamazaki, Y.; Yakushiji, F.; Akaji, K.; Kiso, Y.; et al. Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorganic Med. Chem. 2013, 21, 412–424. [Google Scholar] [CrossRef]
- Shie, J.-J.; Fang, J.-M.; Kuo, C.-J.; Kuo, T.-H.; Liang, P.-H.; Huang, H.-J.; Yang, W.-B.; Lin, C.-H.; Chen, J.-L.; Wu, Y.-T.; et al. Discovery of Potent Anilide Inhibitors against the Severe Acute Respiratory Syndrome 3CL Protease. J. Med. Chem. 2005, 48, 4469–4473. [Google Scholar] [CrossRef]
- Kesel, A.J. Synthesis of novel test compounds for antiviral chemotherapy of severe acute respiratory syndrome (SARS). Curr. Med. Chem. 2005, 12, 2095–2162. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, J.E.; Elowe, N.H.; Huitema, C.; Fortin, P.D.; Cechetto, J.D.; Eltis, L.D.; Brown, E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Boil. 2004, 11, 1445–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Fang, L.; Yang, Z.; Xu, S.; Lv, M.; Sun, Z.; Chen, J.; Wang, D.; Gao, J.; Xiao, S. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J. 2019, 33, 14575–14587. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2020, 1–7. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, Q.; Wang, Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatic 2020. [Google Scholar] [CrossRef]
- Yoon, J.-J.; Toots, M.; Lee, S.; Lee, M.-E.; Ludeke, B.; Luczo, J.; Ganti, K.; Cox, R.M.; Sticher, Z.M.; Edpuganti, V.; et al. Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, eabb5883. [Google Scholar] [CrossRef] [Green Version]
- Kaul, T.N.; Middleton, E.; Ogra, P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985, 15, 71–79. [Google Scholar] [CrossRef]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of Chymotrypsin-like Protease Inhibitors of SARS-CoV-2 Via Integrated Computational Approach. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Gautret, P.; Lagier, J.-C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 105949. [Google Scholar] [CrossRef]
- Weinreb, S.M. Synthetic lessons from quinine. Nature 2001, 411, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Keyaerts, E.; Vijgen, L.; Maes, P.; Neyts, J.; Van Ranst, M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004, 323, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-C.; Chen, S.-Y.; Liang, P.-H.; Lu, I.-L.; Mahindroo, N.; Hsieh, H.-P.; Chao, Y.-S.; Liu, L.; Liu, N.; Lien, W.; et al. Discovery of a Novel Family of SARS-CoV Protease Inhibitors by Virtual Screening and 3D-QSAR Studies. J. Med. Chem. 2006, 49, 3485–3495. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Valle, C.; De Lamballerie, X.; Canard, B.; Seidah, N.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Tong, T.R. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infect. Disord. Drug Targets 2009, 9, 223–245. [Google Scholar] [CrossRef]
- Mukherjee, P.; Desai, P.; Ross, L.; White, E.L.; Avery, M.A. Structure-based virtual screening against SARS-3CL(pro) to identify novel non-peptidic hits. Bioorg. Med. Chem. 2008, 16, 4138–4149. [Google Scholar] [CrossRef]
- Lee, T.-W.; Cherney, M.M.; Liu, J.; James, K.E.; Powers, J.C.; Eltis, L.D.; James, M.N. Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase. J. Mol. Boil. 2007, 366, 916–932. [Google Scholar] [CrossRef]
- Wang, D.; Huang, J.; Gui, T.; Yang, Y.; Feng, T.; Tzvetkov, N.T.; Xu, T.; Gai, Z.; Zhou, Y.; Zhang, J.; et al. SR-BI as a target of natural products and its significance in cancer. Semin. Cancer Boil. 2020. [Google Scholar] [CrossRef]
- Goetz, D.H.; Choe, Y.; Hansell, E.; Chen, Y.T.; McDowell, M.; Jonsson, C.B.; Roush, W.R.; McKerrow, J.; Craik, C.S. Substrate Specificity Profiling and Identification of a New Class of Inhibitor for the Major Protease of the SARS Coronavirus. Biochemistry 2007, 46, 8744–8752. [Google Scholar] [CrossRef]
- Dooley, A.J.; Shindo, N.; Taggart, B.; Park, J.G.; Pang, Y.P. From genome to drug lead: Identification of a small-molecule inhibitor of the SARS virus. Bioorg. Med. Chem. Lett. 2006, 16, 830–833. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Mott, B.T.; Rosenthal, A.S.; Genna, D.T.; Posner, G.H.; Arav-Boger, R. An artemisinin-derived dimer has highly potent anti-cytomegalovirus (CMV) and anti-cancer activities. PLoS ONE 2011, 6, e24334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Alessandro, S.; Scaccabarozzi, D.; Signorini, L.; Perego, F.; Ilboudo, D.; Ferrante, P.; Delbue, S. The Use of Antimalarial Drugs against Viral Infection. Microorganisms 2020, 8, 85. [Google Scholar] [CrossRef] [Green Version]
- Reiter, C.; Fröhlich, T.; Gruber, L.; Hutterer, C.; Marschall, M.; Voigtländer, C.; Friedrich, O.; Kappes, B.; Efferth, T.; Tsogoeva, S.B. Highly potent artemisinin-derived dimers and trimers: Synthesis and evaluation of their antimalarial, antileukemia and antiviral activities. Bioorg. Med. Chem. 2015, 23, 5452–5458. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Fallah, F.; Setzer, W.N.; Heravi, R.E.; Sharifi-Rad, M. Tordylium persicum Boiss. & Hausskn extract: A possible alternative for treatment of pediatric infectious diseases. Cell Mol. Biol. 2016, 62, 20–26. [Google Scholar]
- Park, I.-W.; Han, C.; Song, X.-P.; A Green, L.; Wang, T.; Liu, Y.; Cen, C.; Song, X.; Yang, B.; Chen, G.; et al. Inhibition of HIV-1 entry by extracts derived from traditional Chinese medicinal herbal plants. BMC Complement. Altern. Med. 2009, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- Kilianski, A.; Baker, S.C. Cell-based antiviral screening against coronaviruses: Developing virus-specific and broad-spectrum inhibitors. Antivir. Res. 2013, 101, 105–112. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Nazaruk, J.; Polito, L.; Morais-Braga, M.F.B.; Rocha, J.E.; Coutinho, H.; Salehi, B.; Tabanelli, G.; Montanari, C.; Contreras, M.D.M.; et al. Matricaria genus as a source of antimicrobial agents: From farm to pharmacy and food applications. Microbiol. Res. 2018, 215, 76–88. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Luo, C.; Liu, H.; Xu, W.; Chen, G.; Liew, O.W.; Zhu, W.; Puah, C.M.; Shen, X.; et al. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): Structure-activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006, 14, 8295–8306. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Rolain, J.-M.; Lagier, J.-C.; Brouqui, P.; Raoult, D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 2020, 105932. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Iriti, M.; Setzer, W.N.; Sharifi-Rad, M.; Roointan, A.; Salehi, B. Antiviral activity of Veronica persica Poir. on herpes virus infection. Cell. Mol. Boil. 2018, 64, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Coronavirus: Chinese Scientists to Test 30 Therapeutic Candidates 2020. Available online: https://www.pharmaceutical-technology.com/news/china-tests-30-drug-candidates/ (accessed on 21 February 2020).
- Huang, J.; Wang, D.; Huang, L.-H.; Huang, H. Huang Roles of Reconstituted High-Density Lipoprotein Nanoparticles in Cardiovascular Disease: A New Paradigm for Drug Discovery. Int. J. Mol. Sci. 2020, 21, 739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, U.N. Can Bioactive Lipids Inactivate Coronavirus (COVID-19)? Arch. Med. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ohe, M.; Shida, H.; Jodo, S.; Kusunoki, Y.; Seki, M.; Furuya, K.; Goudarzi, H. Macrolide treatment for COVID-19: Will this be the way forward? Biosci. Trends 2020, 2020.03058. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Luo, H.; Chen, L.; Chen, J.; Shen, J.; Zhu, W.; Chen, K.; Shen, X.; Jiang, H. An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: Structures, functions and drug development. Curr. Pharm. Des. 2006, 12, 4539–4553. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020. [Google Scholar] [CrossRef]
- Park, Y.-J.; Walls, A.C.; Wang, Z.; Sauer, M.M.; Li, W.; Tortorici, M.A.; Bosch, B.-J.; DiMaio, F.; Veesler, D. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Boil. 2019, 26, 1151–1157. [Google Scholar] [CrossRef] [Green Version]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A. BStabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef] [Green Version]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 5570. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.-C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, eabb7269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, J.T.; Serrano, M.L.; Pujol, F.H.; Rangel, H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020, 19, 410–417. [Google Scholar]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, J.; Lu, Y.; Jin, X.; Zhang, L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Alhazzani, W.; Møller, M.H.; Arabi, Y.M.; Loeb, M.; Gong, M.N.; Fan, E.; Oczkowski, S.; Levy, M.M.; Derde, L.; Dzierba, A.; et al. Surviving Sepsis Campaign: Guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med. 2020, 1–34. [Google Scholar] [CrossRef]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; A Schneider, M.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020. [Google Scholar] [CrossRef]
- Brake, S.; Barnsley, K.; Lu, W.; McAlinden, K.; Eapen, M.S.; Sohal, S. Smoking Upregulates Angiotensin-Converting Enzyme-2 Receptor: A Potential Adhesion Site for Novel Coronavirus SARS-CoV-2 (Covid-19). J. Clin. Med. 2020, 9, 841. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-I.; Postema, P.G.; Arbelo, E.; Behr, E.R.; Bezzina, C.R.; Napolitano, C.; Robyns, T.; Probst, V.; Schulze-Bahr, E.; Remme, C.A.; et al. SARS-CoV-2, COVID-19 and inherited arrhythmia syndromes. Heart Rhythm 2020. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kuenze, G.; Smith, J.; Taylor, K.C.; Duran, A.M.; Hadziselimovic, A.; Meiler, J.; Vanoye, C.G.; George, A.L.; Sanders, C.R. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci. Adv. 2018, 4, eaar2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarma, P.; Prajapat, M.; Avti, P.; Kaur, H.; Kumar, S.; Medhi, B. Therapeutic options for the treatment of 2019-novel coronavirus: An evidence-based approach. Indian J. Pharmacol. 2020, 52, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-C.; Chen, M.-Y.; Chang, Y.-L. Potential therapeutic agents against COVID-19: What we know so far. J. Chin. Med. Assoc. 2020. [Google Scholar] [CrossRef]
- Narasaraju, T.; Yang, E.; Samy, R.P.; Ng, H.H.; Poh, W.P.; Liew, A.-A.; Phoon, M.C.; Van Rooijen, N.; Chow, V.T. Excessive Neutrophils and Neutrophil Extracellular Traps Contribute to Acute Lung Injury of Influenza Pneumonitis. Am. J. Pathol. 2011, 179, 199–210. [Google Scholar] [CrossRef]
- Sugamata, R.; Dobashi, H.; Nagao, T.; Yamamoto, K.-I.; Nakajima, N.; Sato, Y.; Aratani, Y.; Oshima, M.; Sata, T.; Kobayashi, K.; et al. Contribution of neutrophil-derived myeloperoxidase in the early phase of fulminant acute respiratory distress syndrome induced by influenza virus infection. Microbiol. Immunol. 2012, 56, 171–182. [Google Scholar] [CrossRef]
- Camp, J.V.; Jonsson, C.B. A Role for Neutrophils in Viral Respiratory Disease. Front. Immunol. 2017, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Smith, F.; Panizzi, J.R.; Goodwin, D.C.; Panizzi, P. Inactivation of myeloperoxidase by benzoic acid hydrazide. Arch. Biochem. Biophys. 2015, 570, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Milton, A.; Arnold, R.; Huang, H.; Smith, F.; Panizzi, J.R.; Panizzi, P. Methods for measuring myeloperoxidase activity toward assessing inhibitor efficacy in living systems. J. Leukoc. Boil. 2016, 99, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Favreau, D.J.; Meessen-Pinard, M.; Desforges, M.; Talbot, P.J. Human Coronavirus-Induced Neuronal Programmed Cell Death Is Cyclophilin D Dependent and Potentially Caspase Dispensable. J. Virol. 2011, 86, 81–93. [Google Scholar] [CrossRef] [Green Version]
- Chu, H.; Zhou, J.; Wong, B.H.-Y.; Li, C.; Chan, J.F.-W.; Cheng, Z.-S.; Yang, D.; Wang, D.; Lee, A.C.-Y.; Li, C.; et al. Middle East Respiratory Syndrome Coronavirus Efficiently Infects Human Primary T Lymphocytes and Activates the Extrinsic and Intrinsic Apoptosis Pathways. J. Infect. Dis. 2015, 213, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem. Rev. 2019, 119, 5537–5606. [Google Scholar] [CrossRef]
- Hung, A.M.; Tsuchida, Y.; Nowak, K.L.; Sarkar, S.; Chonchol, M.; Whitfield, V. IL-1 Inhibition and Function of the HDL-Containing Fraction of Plasma in Patients with Stages 3 to 5 CKD. Clin. J. Am. Soc. Nephrol. 2019, 14, 702–711. [Google Scholar] [CrossRef]
- Fung, T.S.; Liao, Y.; Liu, D.X. The Endoplasmic Reticulum Stress Sensor IRE1α Protects Cells from Apoptosis Induced by the Coronavirus Infectious Bronchitis Virus. J. Virol. 2014, 88, 12752–12764. [Google Scholar] [CrossRef] [Green Version]
- Fung, T.S.; Liu, D.X. Activation of the c-Jun NH2-terminal kinase pathway by coronavirus infectious bronchitis virus promotes apoptosis independently of c-Jun. Cell Death Dis. 2017, 8, 3215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, T.S.; Liu, D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014, 5, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, T.S.; Liao, Y.; Liu, D.X. Regulation of Stress Responses and Translational Control by Coronavirus. Viruses 2016, 8, 18. [Google Scholar] [CrossRef]
- Shi, C.-S.; Nabar, N.; Huang, N.-N.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019, 5, 101. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Baker, A.J. Recombinant human ACE2: Acing out angiotensin II in ARDS therapy. Crit. Care 2017, 21, 305. [Google Scholar] [CrossRef] [Green Version]
- Batlle, D.; Wysocki, J.; Satchell, K. Soluble angiotensin-converting enzyme 2: A potential approach for coronavirus infection therapy? Clin. Sci. 2020, 134, 543–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 2020, eabb3221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poland, G.A. SARS-CoV-2: A time for clear and immediate action. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Thompson, R. Novel Coronavirus Outbreak in Wuhan, China, 2020: Intense Surveillance Is Vital for Preventing Sustained Transmission in New Locations. J. Clin. Med. 2020, 9, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiura, H.; Linton, N.M.; Akhmetzhanov, A.R. Initial Cluster of Novel Coronavirus (2019-nCoV) Infections in Wuhan, China Is Consistent with Substantial Human-to-Human Transmission. J. Clin. Med. 2020, 9, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiura, H.; Kobayashi, T.; Yang, Y.; Hayashi, K.; Miyama, T.; Kinoshita, R.; Linton, N.M.; Jung, S.-M.; Yuan, B.; Suzuki, A.; et al. The Rate of Underascertainment of Novel Coronavirus (2019-nCoV) Infection: Estimation Using Japanese Passengers Data on Evacuation Flights. J. Clin. Med. 2020, 9, 419. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020. [Google Scholar] [CrossRef] [Green Version]
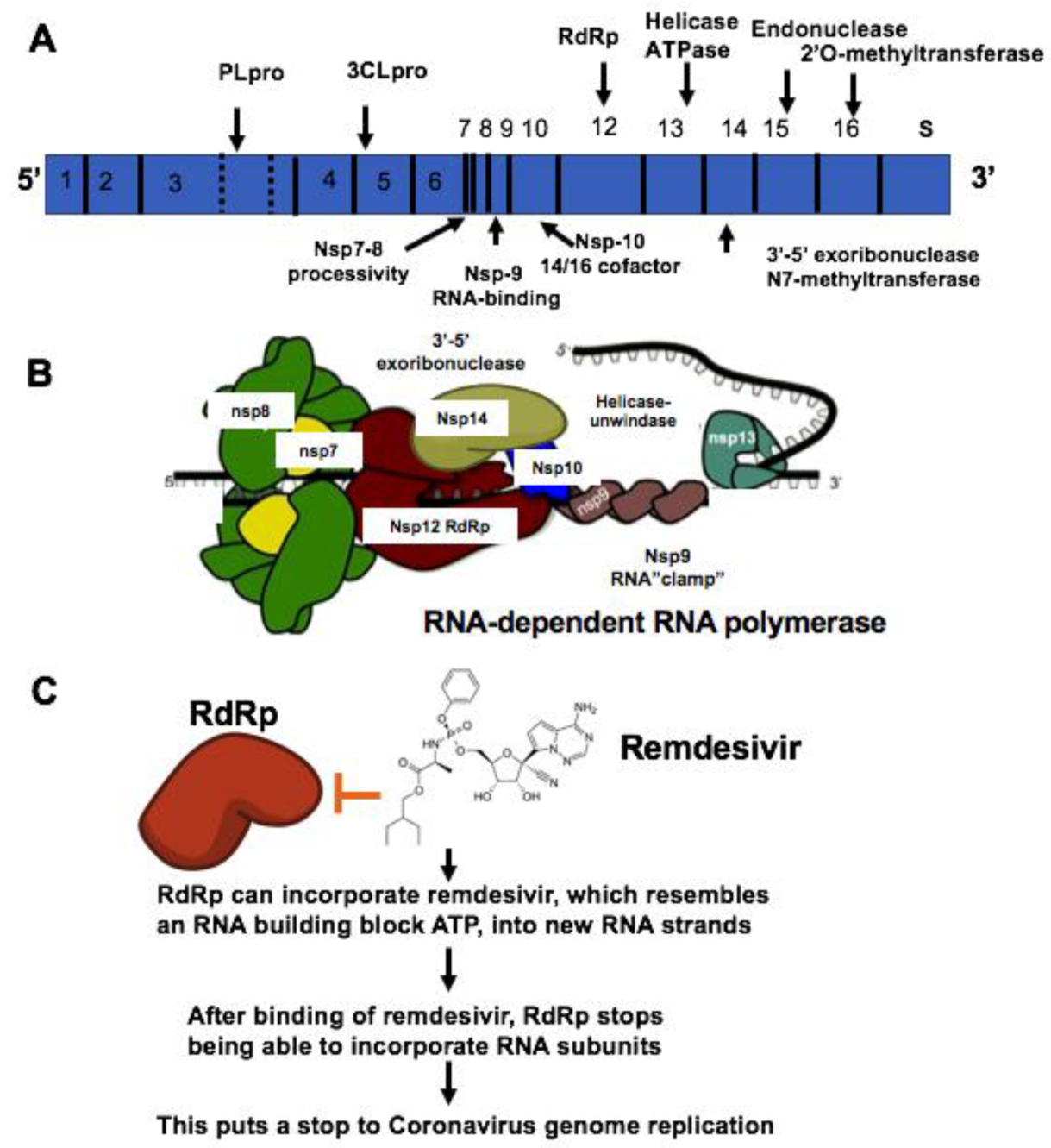




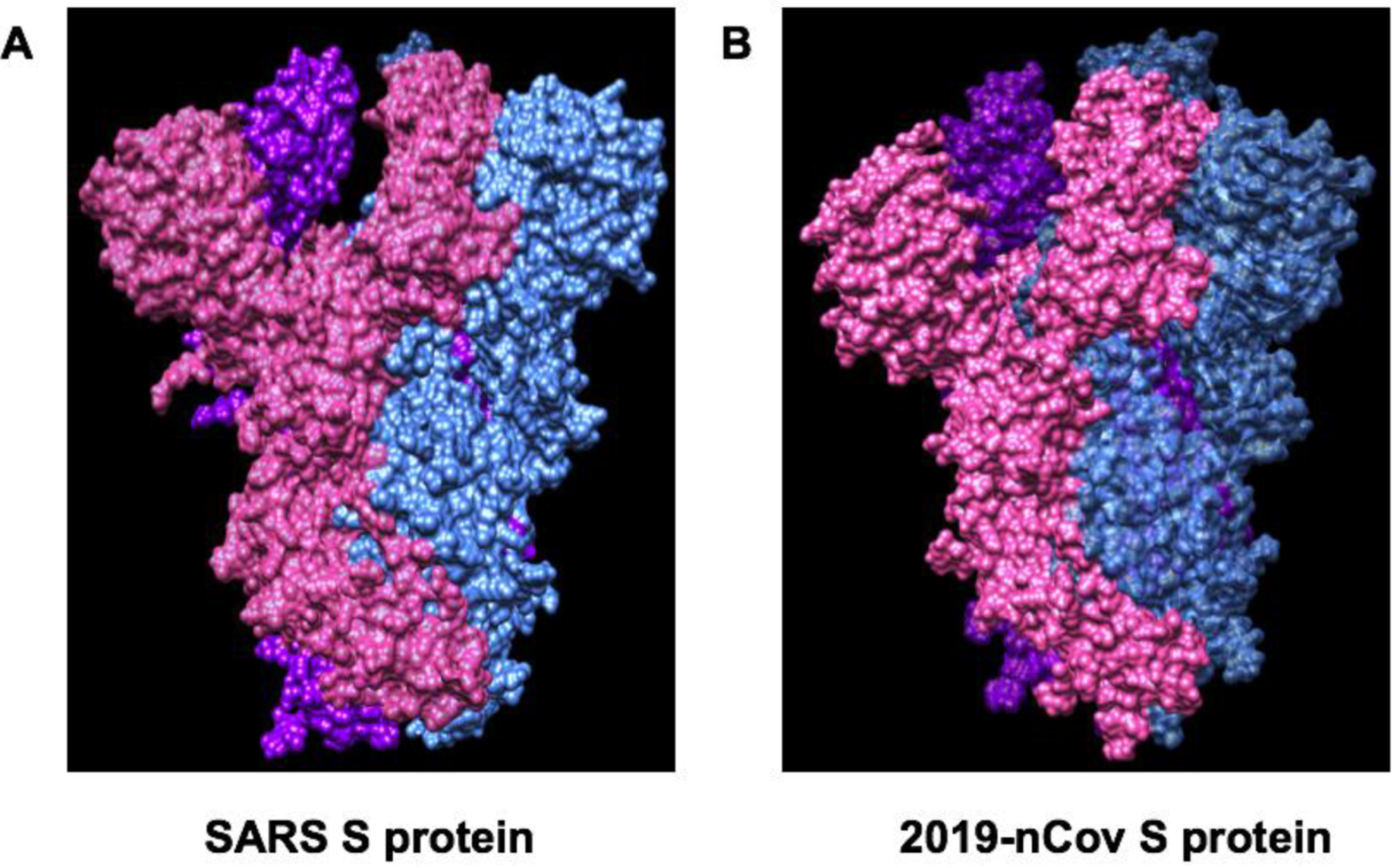
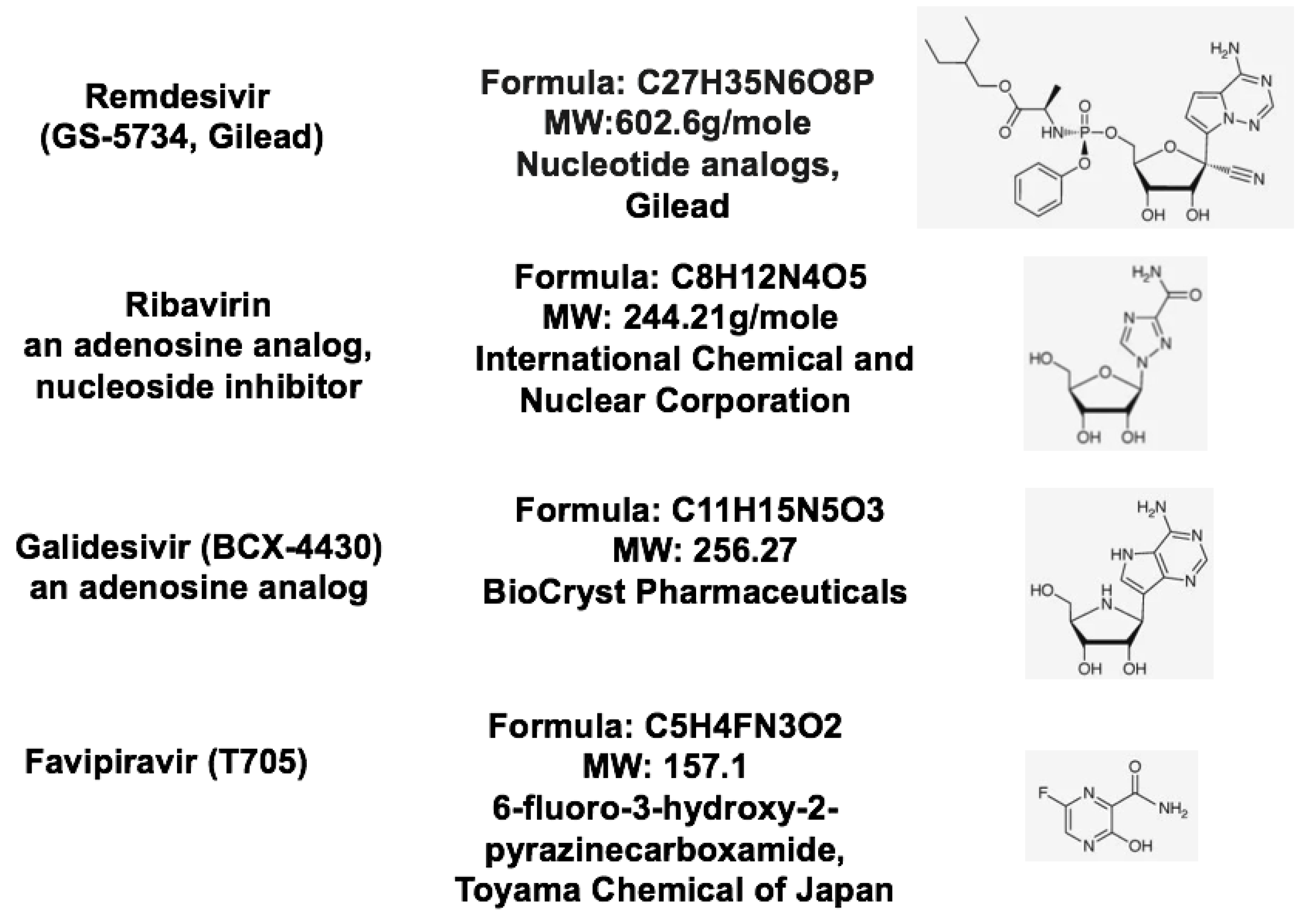
| Therapeutic Targets | Functions | Potential Drug Candidates | References |
|---|---|---|---|
| RNA-dependent RNA polymerase, RdRp | An RNA-dependent RNA polymerase for replicating coronavirus genome | Remdesivir, Ribavirin and Favipiravir can inhibit viral RdRp | [24] |
| Papain-like protease, PLpro | A protease for the conversion of viral polyprotein into functional enzyme | Lopinavir, protease inhibitors that may inhibit the viral proteases: 3CLpro or Plpro. Darunavir inhibits the proteolysis activity of 3-chymotrypsin-like protease. | [50] |
| Main protease 3CLproteinase, 3CLpro | A protease for the conversion of viral polyprotein into functional protein | Lopinavir, protease inhibitors that may inhibit the viral proteases: 3CLpro or Plpro. | [51,52] |
| S protein and TMPRSS2 | A viral surface protein for binding to host cell receptor ACE2. TMPRSS2, a host cell-produced protease that primes S protein to facilitate its binding to ACE2 | Arbidol can prevent S protein/ACE2 interaction and inhibit membrane fusion of the viral envelope by preventing the binding of viral envelope protein to host cells and preventing viral entry to the target cell. Camostat mesylate inhibits TMPRSS2 and viral cell entry | [28,53] |
| ACE2 | A viral receptor protein on the host cells which binds to viral S protein | Chloroquine and hydroxychloroquine can inhibits vial entry and endocytosis by increasing endosomal pH, interfere with ACE2 glycosylation as well as host immunomodulatory effects | [54,55] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Song, W.; Huang, H.; Sun, Q. Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19. J. Clin. Med. 2020, 9, 1131. https://doi.org/10.3390/jcm9041131
Huang J, Song W, Huang H, Sun Q. Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19. Journal of Clinical Medicine. 2020; 9(4):1131. https://doi.org/10.3390/jcm9041131
Chicago/Turabian StyleHuang, Jiansheng, Wenliang Song, Hui Huang, and Quancai Sun. 2020. "Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19" Journal of Clinical Medicine 9, no. 4: 1131. https://doi.org/10.3390/jcm9041131





