Nonstop Selection for High and Stable Crop Yield by Two Prognostic Equations to Reduce Yield Losses
Abstract
:1. Introduction
 , (ii) the stability, estimated by the standardized progeny mean yield
, (ii) the stability, estimated by the standardized progeny mean yield  , where
, where  and
and  represent the mean and the standard deviation of the entry/line where each plant belongs, and (iii) the adaptability or responsiveness to inputs, estimated by the standardized selection differential
represent the mean and the standard deviation of the entry/line where each plant belongs, and (iii) the adaptability or responsiveness to inputs, estimated by the standardized selection differential  . The independence among the three genetic components was key for predicting for the first time the possibility to develop monogenotypic cultivars that are density-neutral (i.e., capable to attain maximal yields under a wide range of planting densities), while simultaneously possessing high yields per hectare under standard crop stands. The existence of density-neutral cultivars, further verified in maize [21], a crop whose hybrid performance is known to be highly density-dependent, demonstrated conclusively that density ceases to represent a component of crop yield potential, an idea that had been a widespread working assumption during the recent past (e.g., [22]). The same article [19] first presented the concept of ‘nonstop’ selection as an essential component of a comprehensive breeding procedure.
. The independence among the three genetic components was key for predicting for the first time the possibility to develop monogenotypic cultivars that are density-neutral (i.e., capable to attain maximal yields under a wide range of planting densities), while simultaneously possessing high yields per hectare under standard crop stands. The existence of density-neutral cultivars, further verified in maize [21], a crop whose hybrid performance is known to be highly density-dependent, demonstrated conclusively that density ceases to represent a component of crop yield potential, an idea that had been a widespread working assumption during the recent past (e.g., [22]). The same article [19] first presented the concept of ‘nonstop’ selection as an essential component of a comprehensive breeding procedure.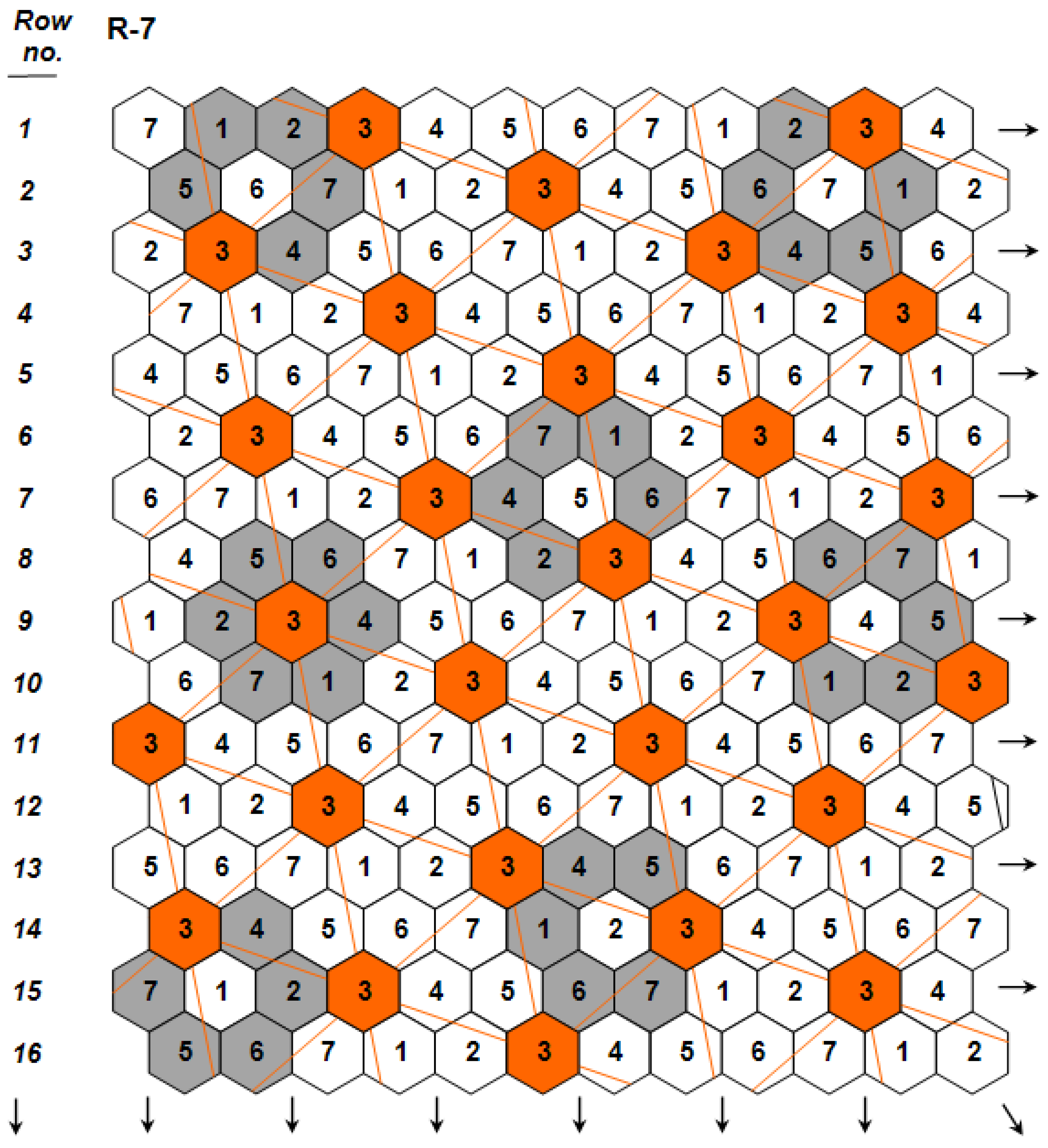
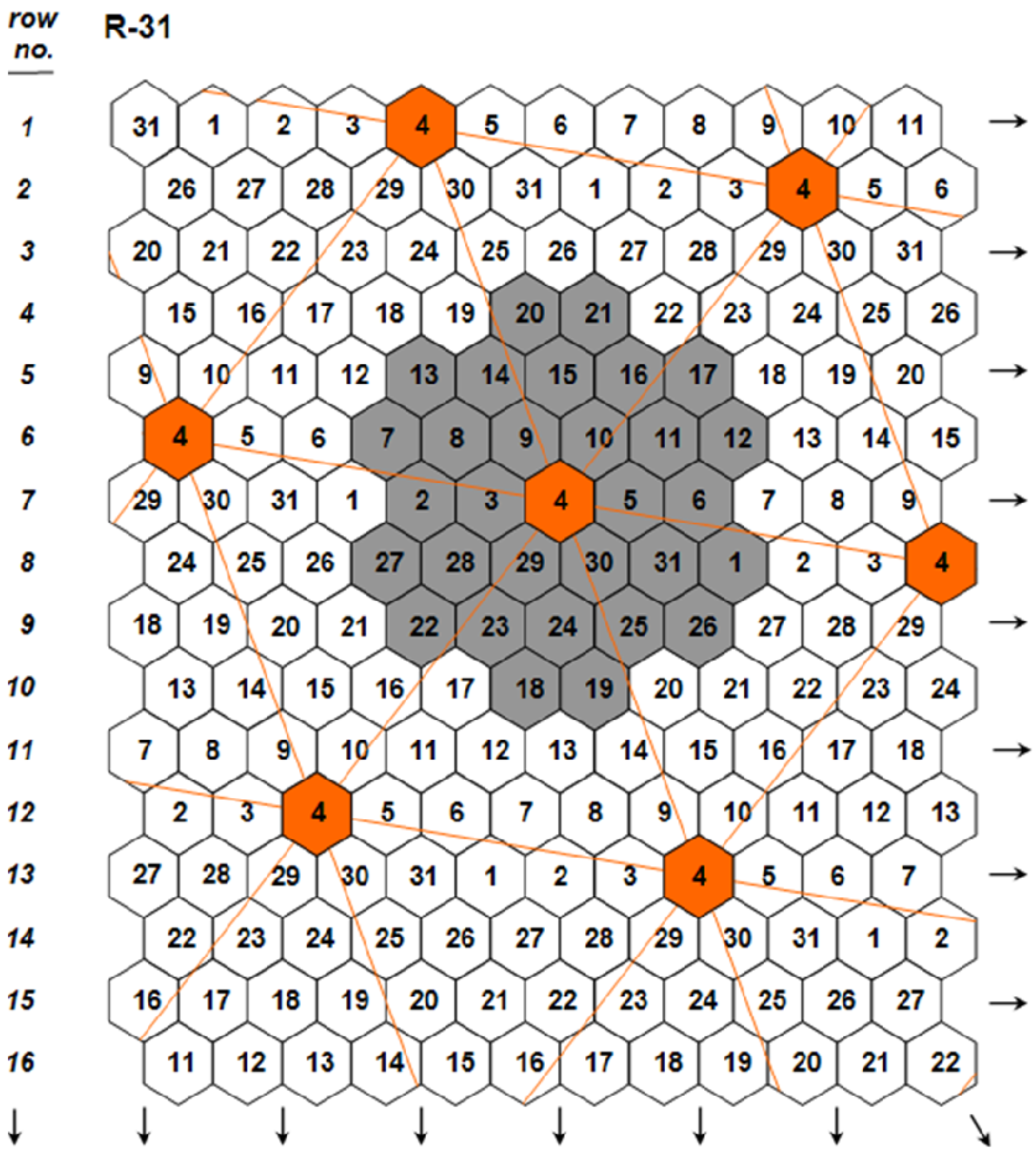
 (Figure 2; Design Code 4 exemplifies plants whose yield per plant
(Figure 2; Design Code 4 exemplifies plants whose yield per plant  is compared to the mean yield
is compared to the mean yield  of the surrounding plants of the 30 remaining entries in the grey area), measuring with accuracy individual plant yield potential, and the Coefficient of Homeostasis CH =
of the surrounding plants of the 30 remaining entries in the grey area), measuring with accuracy individual plant yield potential, and the Coefficient of Homeostasis CH =  , measuring yield stability or homeostasis. The CH incorporates every factor affecting stability of performance, including heritability, heterozygosity, biotic and abiotic stresses, high planting densities, and any other source contributing to yield variability. The product of the two components measures the plant’s crop yield potential and constitutes the first of two novel equations used in the selection process.
, measuring yield stability or homeostasis. The CH incorporates every factor affecting stability of performance, including heritability, heterozygosity, biotic and abiotic stresses, high planting densities, and any other source contributing to yield variability. The product of the two components measures the plant’s crop yield potential and constitutes the first of two novel equations used in the selection process.
 , where
, where  is the mean yield of the progeny line (lines 1 to 31 in this case) and
is the mean yield of the progeny line (lines 1 to 31 in this case) and  the mean yield of all plants in the trial:
the mean yield of all plants in the trial: 
2. Key Attributes Underlying the Supremacy of the Two Prognostic Equations
- 1. The choice of the individual plant grown under ultra-wide plant spacings as unit of evaluation and selection to eliminate the confounding effects of density and competition on single-plant yields.
- 2. The partition of the plant crop yield potential into two components: (1) the plant yield potential, and (2) the plant stability of performance. The invention of the honeycomb selection designs permits the formulation of two parameters measuring the two components with high precision and accuracy.
- 3. The realization of accurate and precision whole-plant field phenotyping for crop yield potential.
- 4. The possibility of applying ultra-high selection pressures (1 to 0.5%, meaning less than 1% plants retained after the comparative evaluation) and the resulting attainment of high advance through selection without sacrificing accuracy of selection. Although in theory [43], the higher the selection pressures, the higher the expected progress through selection, in conventional breeding practice, the working selection pressures are commonly at the level of 10% or more.
- 5. The growing of plants in the absence of competition cancels the undesirable correlation between the CV and the mean that confounds predictions and permits the use of CV to select for stability of performance.
- 6. The predictive power of the two equations is further increased due to the possibility to select in the same generation plants characterized by high and stable crop yield potential, as opposed to progeny testing requiring at least two successive generations.
- 7. The increase of the genetic gain per annum from the average 1% genetic gain attained by conventional breeding, to a two-digit percentage.
- 8. The production of density-neutral cultivars possessing a number of advantages, including avoidance of reseeding in case of non-satisfactory germination and increased tolerance to drought stress through use of lower seeding rates.
- 9. The increase of the effectiveness of molecular breeding by enabling to rank an unlimited number of plants for crop yield potential with equal accuracy of assessment.
- 10. The unit of selection in honeycomb breeding is the individual plant, while the unit of analysis in molecular breeding is also the individual plant genome. This offers the possibility to successfully marry phenotyping with genotyping and to bridge the so-called genotype-phenotype gap [44].
3. Results and Discussion
3.1. Intra-Cultivar Selection for Yield and Stability within Elite Cotton Cultivars
| Line | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank no. | Design code no. | CH Value | Equation A value | Rank no. | Design code no. | CH Value | Equation A value | Rank no. | Design code no. | CH Value | Equation A value |
| 1 | 5 | 23.7 | 45 | 11 | 22 | 11.2 | 22 | 21 | 10 | 7.2 | 13 |
| 2 | 3 | 20.8 | 49 | 12 | 7 | 10 | 14 | 22 | 14 | 7.2 | 8 |
| 3 | 16 | 20.3 | 39 | 13 | 21 | 9.7 | 16 | 23 | 12 | 7.1 | 16 |
| 4 | 6 | 16.3 | 33 | 14 | 9 | 9.4 | 18 | 24 | 27 | 6.8 | 13 |
| 5 | 8 | 16.1 | 26 | 15 | 4 | 9.3 | 19 | 25 | 11 | 6.1 | 8 |
| 6 | 19 | 14.5 | 27 | 16 | 29 | 9.2 | 21 | 26 | 26 | 5.8 | 4 |
| 7 | 1 | 14.4 | 25 | 17 | 25 | 8.8 | 16 | 27 | 13 | 5.2 | 9 |
| 8 | 23 | 12.7 | 20 | 18 | 30 | 8.4 | 20 | 28 | 17 | 4.9 | 8 |
| 9 | 28 | 12.1 | 20 | 19 | 18 | 7.8 | 21 | 29 | 24 | 4.8 | 10 |
| 10 | 15 | 11.2 | 21 | 20 | Original cultivar | 7.5 | 12 | 30 | 2 | 3.8 | 7 |
| 31 | 20 | 3.2 | 17 | ||||||||
| Line no. of top plants | Equation A Values | CH
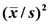 | PYI
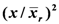 |
|---|---|---|---|
| 3 | 65 | 20.8 | 3.13 |
| 5 | 55 | 23.7 | 2.31 |
| 3 | 55 | 20.8 | 2.66 |
| 3 | 55 | 20.8 | 2.62 |
| 6 | 51 | 16.3 | 3.17 |
| 5 | 49 | 23.7 | 2.04 |
| 5 | 46 | 23.7 | 1.96 |
| 16 | 46 | 20.3 | 2.28 |
| 16 | 40 | 20.3 | 1.96 |
| 5 | 39 | 23.7 | 1.63 |
| 3 | 37 | 20.8 | 1.80 |
| 5 | 36 | 23.7 | 1.53 |
| 5 | 36 | 23.7 | 1.51 |
| 5 | 36 | 23.7 | 1.51 |
| 16 | 36 | 20.3 | 1.77 |
| Line code no. | Replicates | |||||
|---|---|---|---|---|---|---|
| I | II | III | IV | Mean | Gain | |
| 5 | 6.58 | 6.50 | 6.82 | 6.70 | 6.65 a | 118.5 |
| 3 | 6.60 | 5.50 | 6.90 | 5.80 | 6.18 ab | 110.2 |
| 16 | 6.24 | 5.82 | 6.50 | 5.26 | 5.96 ab | 106.2 |
| 19 | 6.60 | 5.50 | 4.70 | 5.74 | 5.64 b | 100.5 |
| Original cultivar | 5.84 | 5.10 | 6.10 | 5.40 | 5.61 b | 100.0 |
| 1 | 5.26 | 6.10 | 5.70 | 4.60 | 5.42 b | 96.6 |
| 6 | 5.38 | 5.16 | 5.30 | 5.80 | 5.41 b | 96.4 |
| Line code no. | Replicates | |||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | Mean | Gain | |
| 5 | 5.88 | 5.38 | 5.88 | 5.50 | 6.75 | 7.00 | 6.07 a | 121.6 |
| 3 | 4.63 | 4.88 | 5.63 | 4.38 | 5.00 | 5.88 | 5.07 b | 101.6 |
| Original cultivar | 5.20 | 4.37 | 5.25 | 5.00 | 4.88 | 5.25 | 4.99 b | 100.0 |
3.2. Intra-Cultivar Selection for Yield and Resistance to Cotton Verticillium Wilt
4. Experimental Section
4.1. Intra-Cultivar Selection for Yield Stability and Crop Yield Potential
4.2. Intra-Cultivar Selection for Yield and Resistance to Cotton Verticillium Wilt
5. Conclusions
Acknowledgments
References
- Lobell, D.B.; Cassman, K.G.; Field, C.B. Crop yield gaps: Their importance, magnitudes, and causes. Annu. Rev. Environ. Resour. 2009, 34, 179–204. [Google Scholar] [CrossRef]
- Licker, R.; Johnston, M.; Foley, J.A.; Barford, C.; Kucharik, C.J.; Monfreda, C.; Ramankutty, N. Mind the gap: How do climate and agricultural management explain the “yield gap” of croplands around the world? Glob. Ecol. Biogeogr. 2010, 19, 769–782. [Google Scholar] [CrossRef]
- Edmeades, G.; Fisher, T.; Byerlee, D. Can We Feed the World in 2050? Food Security from Sustainable Agriculture, Proceedings of the 15th ASA Conference, Lincoln, New Zealand, 15–19 November 2010. 2010. Available online: http://www.regional.org.au/au/asa/2010/plenary/climate-change/7384_edmeades.htm (accessed on 31 August 2012).
- Fok, A.C.M. Cotton yield stagnation: Addressing a Common Effect of Various Causes. In New Frontiers in Cotton Research, Proceedings of the 2nd World Cotton Research Conference, Athens, Greece, 6–12 September 1998; Gillham, F.M., Ed.; Petridis: Thessaloniki, Greece, 2000; pp. 38–45. [Google Scholar]
- Peltonen-Sainio, P.; Jauhiainen, L.; Laurila, I.P. Cereal yield trends in northern European conditions: Changes in yield potential and its realization. Field Crop. Res. 2009, 110, 85–90. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Peterson, C.J. Genetic improvement in winter wheat yields in the great plains of North America, 1959-2008. Crop Sci. 2010, 50, 1882–1890. [Google Scholar] [CrossRef]
- Brisson, N.; Gate, P.; Gouache, D.; Charmet, G.; Oury, F.X.; Huard, F. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crop. Res. 2010, 119, 201–212. [Google Scholar] [CrossRef]
- Gregory, P.J.; George, T.S. Feeding nine billion: The challenge to sustainable crop production. J. Exp. Bot. 2011, 62, 5233–5239. [Google Scholar] [CrossRef]
- Gornall, J.; Betts, R.; Burke, E.; Clark, R.; Camp, J.; Willett, K.; Wiltshire, A. Implications of climate change for agricultural productivity in the early twenty-first century. Philos. Trans. R. Soc. B 2010, 365, 2973–2989. [Google Scholar]
- Jaggard, K.W.; Qi, A.; Ober, E.S. Possible changes to arable crop yields by 2050. Philos. Trans. R. Soc. B 2010, 365, 2835–2851. [Google Scholar]
- Luck, J.; Spackman, M.; Freeman, A.; Tre˛bicki, P.; Griffiths, W.; Finlay, K.; Chakraborty, S. Climate Change and Diseases of Food Crops. Plant Pathol. 2011, 60, 113–121. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Fasoulas, A.C.; Fasoula, V.A. Honeycomb selection designs. Plant Breed. Rev. 1995, 13, 87–139. [Google Scholar]
- Kyriakou, D.T.; Fasoulas, A.C. Effects of competition and selection pressure on yield response in winter rye (Secale cereale L.). Euphytica 1985, 34, 833–895. [Google Scholar] [CrossRef]
- Fasoula, D.A. Correlations between auto-, all-, and nil-competition and their implications in plant breeding. Euphytica 1990, 50, 57–62. [Google Scholar] [CrossRef]
- Fasoula, D.A.; Fasoula, V.A. Competitive ability and plant breeding. Plant Breed. Rev. 1997, 14, 89–138. [Google Scholar]
- Janick, J. Exploitation of heterosis: Uniformity and stability. In The Genetics and Exploitation of Heterosis in Crops; ASA-CSSA-SSA: Madison, WI, USA, 1999; pp. 319–333. [Google Scholar]
- Tollenaar, M.; Wu, J. Yield improvement in temperate maize is attributable to greater stress tolerance. Crop Sci. 1999, 39, 1597–1604. [Google Scholar] [CrossRef]
- Fasoula, V.A.; Fasoula, D.A. Honeycomb breeding: Principles and applications. Plant Breed. Rev. 2000, 18, 177–250. [Google Scholar]
- Fasoula, V.A.; Fasoula, D.A. Principles underlying genetic improvement for high and stable crop yield potential. Field Crop. Res. 2002, 75, 191–209. [Google Scholar] [CrossRef]
- Tokatlidis, I.S.; Koutsika-Sotiriou, M.; Fasoulas, A.C. The development of density-independent hybrids in maize. Maydica 2001, 46, 21–25. [Google Scholar]
- Yan, W.; Wallace, D.H. Breeding for negatively associated traits. Plant Breed. Rev. 1995, 13, 141–177. [Google Scholar]
- Mauromoustakos, A.; Fasoula, V.A.; Thompson, K. Honeycomb Designs Computing and Analysis. In Proceedings of International Biometric Society: Eastern North American Region, Tampa, FL, USA, 26-29 March 2006.
- Fasoula, V.A. A novel equation paves the way for an everlasting revolution with cultivars characterized by high and stable crop yield and quality. In Proceedings of the 11th National Hellenic Conference in Genetics and Plant Breeding, Orestiada, Greece, 31 October-2 November 2006.
- Fasoula, V.A. Two Novel Whole-Plant Field Phenotyping Equations Maximize Selection Efficiency. In Modern Variety Breeding for Present and Future Needs, Proceedings of the 18th Eucarpia General Congress, Valencia, Spain, 9–12 September 2008; Prohens, J., Badenes, M., Eds.; Editorial Universidad Politecnica de Valencia: Valencia, Spain, 2008; pp. 361–365. [Google Scholar]
- Fasoula, V.A. Selection of High Yielding Plants Belonging to Entries of High Homeostasis Maximizes Efficiency in Maize Breeding. In Proceedings of the XXI International Eucarpia Conference in Maize and Sorghum Breeding in the Genomics Era, Bergamo, Italy, 21-24 June 2009.
- McClintock, B. The significance of the responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar]
- Rasmusson, D.C.; Phillips, R.L. Plant breeding progress and genetic diversity from de novo variation and elevated epistasis. Crop Sci. 1997, 37, 303–310. [Google Scholar] [CrossRef]
- Stokes, T.L.; Kunkel, B.N.; Richards, E.J. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002, 16, 171–182. [Google Scholar] [CrossRef]
- Lolle, S.; Victor, J.L.; Young, J.M.; Pruitt, R.E. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 2005, 434, 505–509. [Google Scholar] [CrossRef]
- Grant-Downton, R.T.; Dickinson, H.G. Epigenetics and its implications for plant biology 2. The “Epigenetic Epiphany”: Epigenetics, evolution and beyond. Ann. Bot. 2006, 97, 11–27. [Google Scholar] [CrossRef]
- Richards, E.J. Inherited epigenetic variation-revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagen. 2007, 49, 61–72. [Google Scholar]
- King, G.J.; Amoah, S.; Kurup, S. Exploring and exploiting epigenetic variation in crops. Genome 2010, 53, 856–868. [Google Scholar] [CrossRef]
- Zhang, M.; Kimatu, J.N.; Xu, K.; Liu, B. DNA cytosine methylation in plant development. J. Genet. Genomics 2010, 37, 1–12. [Google Scholar] [CrossRef]
- Hauser, M.T.; Aufsatz, W.; Jonak, C.; Luschnig, C. Transgenerational epigenetic inheritance in plants. Biochim. Biophys. Acta 1809, 459–468. [Google Scholar]
- Dworkin, I. Canalization, cryptic variation, and developmental buffering: A critical examination and analytical perspective, 2005. Available online: https://www.msu.edu/idworkin/publications/Dworkin%20Book%20chapter%208in%20VARIATIOn.pdf (accessed on 31 August 2012).
- Ronnegard, L.; Valdar, W. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 2011, 188, 435–447. [Google Scholar] [CrossRef]
- Fasoula, V.A.; Boerma, H.R. Divergent selection at ultra-low plant density for seed protein and oil content within soybean cultivars. Field Crop. Res. 2005, 91, 217–229. [Google Scholar] [CrossRef]
- Fasoula, V.A.; Boerma, H.R. Intra-cultivar variation for seed weight and other agronomic traits within three elite soybean cultivars. Crop Sci. 2007, 47, 367–373. [Google Scholar] [CrossRef]
- Fasoula, D.A. An overlooked source of seed degradation and its implications in the efficient exploitation of plant genetic resources. Plant Genet. Resour. 2011, 9, 321–323. [Google Scholar] [CrossRef]
- Yates, J.L.; Boerma, H.R.; Fasoula, V.A. SSR-marker analysis of the intracultivar phenotypic variation discovered within 3 soybean cultivars. J. Hered. 2012. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics; John Wiley and Sons: New York, NY, USA, 1989. [Google Scholar]
- Fasoula, D.A. Accurate whole-plant phenotyping: An important component for successful marker assisted selection (MAS). In Genetic variation for Plant Breeding, Proceedings of 17th EUCARPIA General Congress, Tulln, Austria, 8–11 September 2004; Vollmann, J., Grausgruber, H., Ruckenbauer, P., Eds.; BOKU-University of Natural Resources and Applied Life Sciences: Vienna, Austria, 2004; pp. 203–206. [Google Scholar]
- Wych, R.D.; Rasmusson, D.C. Genetic improvement in malting barley cultivars since 1920. Crop Sci. 1983, 23, 1037–1040. [Google Scholar]
- Wych, R.D.; Stuthman, D.D. Genetic improvement in Minnesota-adapted oat cultivars released since 1923. Crop Sci. 1983, 23, 879–881. [Google Scholar] [CrossRef]
- Duvick, D.N. Genetic Contributions to Yield Gains of US Hybrid Maize, 1930-1980. In Genetic Contributions to Yield Gains of Five Major Crop Plants; Fehr, W.R., Ed.; Crop Science Society of America, American Society of Agronomy: Madison, WI, USA, 1984; pp. 15–47. [Google Scholar]
- Meredith, W.R.; Bridge, R.R. Genetic Contributions to Yield Changes in Upland Cotton. In Genetic Contributions to Yield Gains of Five Major Crop Plants; Fehr, W.R., Ed.; Crop Science Society of America, American Society of Agronomy: Madison, WI, USA, 1984; pp. 75–87. [Google Scholar]
- Schmidt, J.W. Genetic Contributions to Yield Gains in Wheat. In Genetic Contributions to Yield Gains of Five Major Crop Plants; Fehr, W.R., Ed.; Crop Science Society of America, American Society of Agronomy: Madison, WI, USA, 1984; pp. 89–101. [Google Scholar]
- Waddington, S.R.; Ransom, J.K.; Osmanzai, M.; Saunders, D.A. Improvement in the yield potential of bread wheat adapted to Northwest Mexico. Crop Sci. 1986, 26, 698–703. [Google Scholar] [CrossRef]
- Feil, B. Breeding progress in small grain cereals-A comparison of old and modern cultivars. Plant Breed. 1992, 108, 1–11. [Google Scholar] [CrossRef]
- Douches, D.S.; Maas, D.; Jastrzebski, K.; Chase, R.W. Assessment of potato breeding progress in the USA over the last century. Crop Sci. 1996, 36, 1544–1552. [Google Scholar]
- Rajaram, S. Approaches for breaching yield stagnation in wheat. Genome 1999, 42, 629–634. [Google Scholar] [CrossRef]
- Peng, S.; Cassman, K.G.; Virmani, S.S.; Sheehy, J.; Khush, G.S. Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci. 1999, 39, 1552–1559. [Google Scholar]
- Fasoulas, A.C.; Thessaloniki, P. Building up Resistance to Verticillium Wilt in Cotton through Honeycomb Breeding. In New Frontiers in Cotton Research, Proceedings of the 2nd World Cotton Research Conference, Athens, Greece, 6–12 September 1998; Gillham, F.M., Ed.; Petridis Publishers: Thessaloniki, Greece, 2000; pp. 120–124. [Google Scholar]
- Fasoula, D.A.; Lithourgidis, A. Selection within Cotton Cultivars (Gossypium hirsutum L.) under the Hellenic Climatic Conditions Maximizes Crop Yield. In Proceedings of the 12th National Hellenic Conference in Genetics and Plant Breeding, Naousa, Greece, 8-10 October 2008; pp. 51–55.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fasoula, D.A. Nonstop Selection for High and Stable Crop Yield by Two Prognostic Equations to Reduce Yield Losses. Agriculture 2012, 2, 211-227. https://doi.org/10.3390/agriculture2030211
Fasoula DA. Nonstop Selection for High and Stable Crop Yield by Two Prognostic Equations to Reduce Yield Losses. Agriculture. 2012; 2(3):211-227. https://doi.org/10.3390/agriculture2030211
Chicago/Turabian StyleFasoula, Dionysia A. 2012. "Nonstop Selection for High and Stable Crop Yield by Two Prognostic Equations to Reduce Yield Losses" Agriculture 2, no. 3: 211-227. https://doi.org/10.3390/agriculture2030211




