Revealing Heavy Metal-Resistant Mechanisms and Bioremediation Potential in a Novel Croceicoccus Species Using Microbial-Induced Carbonate Precipitation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Culture Conditions
2.2. Resistance and Removal Ability to Mn2+
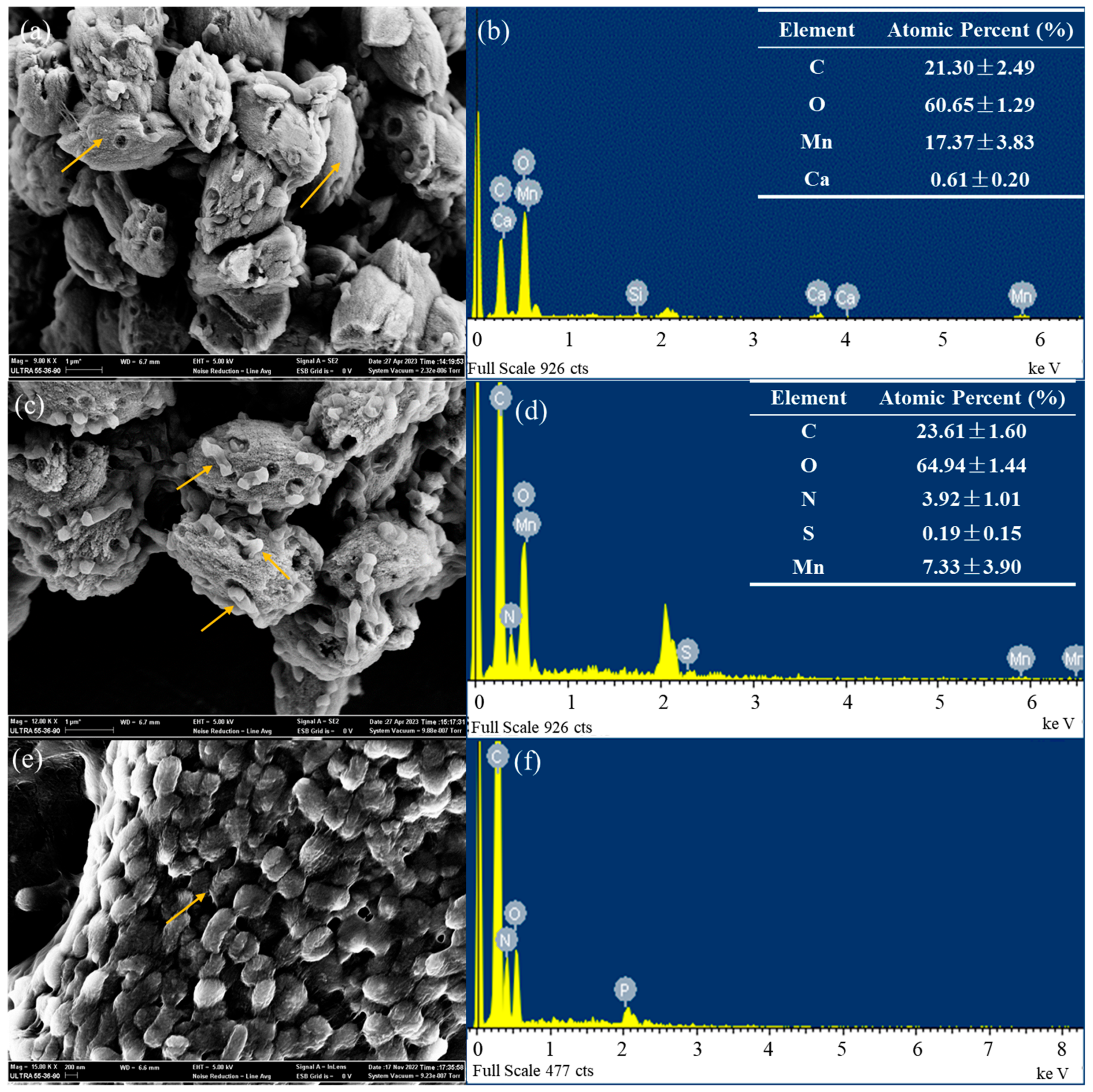
2.3. Synthesis and Characterization of Microbial-Induced Rhodochrosite
2.4. Heavy Metal Tolerance Ability, Motility and Biofilm Formation
2.5. Urea Hydrolytic Activity and EPS Production
2.6. Morphological, Physiological, and Chemotaxonomic Characteristics
2.7. Genome Sequencing and Analysis
3. Result
3.1. Heavy Metal Resistance and Removal Ability of Mn2+
3.2. Crystallographic Structure and Elemental Composition
3.3. Morphological, Physiological, and Chemotaxonomic Characteristics
3.4. General Genome Characteristics and Phylogenetic Properties
3.5. Genes Related to Motility, Chemotaxis, and Biofilm Formation
3.6. Genes Related to REDOX and Metal Transporters
3.7. Genes Related to Urea Hydrolysis Activity and Exopolysaccharide Production
4. Discussion
4.1. Strategies to Adapt to Deep-Sea Environments with High Metal Concentrations
4.2. Mechanism of Rhodochrosite Precipitation Formation
4.3. Proposal of Novel Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hein, J.R.; Koschinsky, A.; Kuhn, T. Deep-ocean polymetallic nodules as a resource for critical materials. Nat. Rev. Earth Environ. 2020, 1, 158–169. [Google Scholar] [CrossRef]
- Ren, J.B.; Deng, Y.N.; Lai, P.X.; He, G.W.; Wang, F.L.; Yao, H.Q.; Deng, X.G.; Liu, Y.G. Geochemical characteristics and genesis of the polymetallic nodules in the Pacific survey area. Earth Sci. Front. 2021, 28, 412–425, (English Translation). [Google Scholar]
- Wang, X.; Schloßmacher, U.; Wiens, M.; Schröder, H.C.; Müller, W.E. Biogenic origin of polymetallic nodules from the Clarion-Clipperton zone in the Eastern Pacific Ocean: Electron microscopic and EDX evidence. Mar. Biotechnol. 2009, 11, 99–108. [Google Scholar] [CrossRef]
- Van, D.C.L. Inactive sulfide ecosystems in the deep sea: A review. Front. Mar. Sci. 2019, 6, 461. [Google Scholar]
- Khripounoff, A.; Caprais, J.C.; Crassous, P.; Etoubleau, J. Geochemical and biological recovery of the disturbed seafloor in polymetallic nodule fields of the Clipperton-Clarion Fracture Zone (CCFZ) at 5000 m depth. Limnol. Oceanogr. 2006, 51, 2033–2041. [Google Scholar] [CrossRef]
- Yaqoob, A.A.; Ahmad, A.; Alshammari, M.B. Advanced Technologies for Wastewater Treatment. Green Chem. Sustain. Water Purif. 2023, 1, 179–202. [Google Scholar]
- Kang, B.; Zha, F.; Li, H.; Xu, L.; Sun, X.; Lu, Z. Bio-mediated method for immobilizing copper tailings sand contaminated with multiple heavy metals. Crystals 2022, 12, 522. [Google Scholar] [CrossRef]
- Chuo, S.C.; Mohamed, S.F.; Mohd Setapar, S.H.; Ahmad, A.; Jawaid, M.; Wani, W.A.; Yaqoob, A.A.; Mohamad Ibrahim, M.N. Insights into the current trends in the utilization of bacteria for microbially induced calcium carbonate Precipitation. Materials 2020, 13, 4993. [Google Scholar] [CrossRef]
- Khanjani, M.; Westenberg, D.J.; Kumar, A.; Ma, H. Tuning polymorphs and morphology of microbially induced calcium carbonate: Controlling factors and underlying mechanisms. ACS Omega 2021, 6, 11988–12003. [Google Scholar] [CrossRef]
- Xue, Z.F.; Cheng, W.C.; Wang, L.; Hu, W. Effects of bacterial inoculation and calcium source on microbial-induced carbonate precipitation for lead remediation. J. Hazard. Mater. 2022, 426, 128090. [Google Scholar] [CrossRef]
- Liu, Y.; Ali, A.; Su, J.F.; Li, K.; Hu, R.Z.; Wang, Z. Microbial-induced calcium carbonate precipitation: Influencing factors, nucleation pathways, and application in waste water remediation. Sci. Total Environ. 2023, 860, 160439. [Google Scholar] [CrossRef] [PubMed]
- Dewi, A.K.; Chen, T.H.; Lin, P.Y.; Sharma, R.K.; Huang, Y.H.; Lu, C.M.; Lu, C.K. Microbial-Induced Manganese Carbonate (MnCO3) Precipitation for Heavy Metal Removal from Water. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Xu, X.W.; Wu, Y.H.; Wang, C.S.; Wang, X.G.; Oren, A.; Wu, M. Croceicoccus marinus gen. nov., sp. nov., a yellow-pigmented bacterium from deep-sea sediment, and emended description of the family Erythrobacteraceae. Int. J. Syst. Evol. Microbiol. 2009, 59, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zeng, Y.; Feng, H.; Wu, Y.; Xu, X. Croceicoccus naphthovorans sp. nov., a polycyclic aromatic hydrocarbons-degrading and acylhomoserine-lactone-producing bacterium isolated from marine biofilm, and emended description of the genus Croceicoccus. Int. J. Syst. Evol. Microbiol. 2015, 65, 1531–1536. [Google Scholar] [CrossRef]
- Wu, Y.H.; Li, G.Y.; Jian, S.L.; Cheng, H.; Huo, Y.Y.; Wang, C.S.; Shao, Z.Z.; Xu, X.W. Croceicoccus pelagius sp. nov. and Croceicoccus mobilis sp. nov., isolated from marine environments. Int. J. Syst. Evol. Microbiol. 2016, 66, 4506–4511. [Google Scholar] [CrossRef]
- Shen, Y.; Li, Z.; Huo, Y.Y.; Bao, L.; Gao, B.; Xiao, P.; Hu, X.; Xu, X.W.; Li, J. Structural and functional insights into CmGH1, a novel GH39 family β-glucosidase from deep-sea bacterium. Front. Microbiol. 2019, 10, 2922. [Google Scholar] [CrossRef]
- Wu, Y.H.; Cheng, H.; Huo, Y.Y.; Xu, L.; Liu, Q.; Wang, C.S.; Xu, X.W. Complete genome sequence of esterase-producing bacterium Croceicoccus marinus E4A9T. Stand. Genom. Sci. 2017, 12, 88. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Feng, H.; Wang, J.; Yang, X.; Wang, Z. Genome-guided identification and characterization of bacteria for simultaneous degradation of polycyclic aromatic hydrocarbons and resistance to hexavalent chromium. Int. Biodeter. Biodegr. 2019, 138, 78–86. [Google Scholar] [CrossRef]
- Mbani, B.; Greinert, J. Analysis-ready optical underwater images of manganese-nodule covered seafloor of the Clarion-Clipperton Zone. Sci. Data 2023, 10, 316. [Google Scholar] [CrossRef]
- Sheng, M.; Peng, D.; Luo, S.; Ni, T.; Luo, H.; Zhang, R.; Wen, Y.; Xu, H. Micro-dynamic process of cadmium removal by microbial induced carbonate precipitation. Environ. Pollut. 2022, 308, 119585. [Google Scholar] [CrossRef]
- Sinegani, A.A.S.; Younessi, N. Antibiotic resistance of bacteria isolated from heavy metal-polluted soils with different land uses. J. Glob. Antimicrob. Resist. 2017, 10, 247–255. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Usman, J.; Kaleem, F.; Omair, M.; Khalid, A.; Iqbal, M. Evaluation of different detection methods of biofilm formation in the clinical isolates. J. Infect. Dis. 2011, 15, 305–311. [Google Scholar]
- Dong, X.Z.; Cai, M.Y. Determinative Manual for Routine Bacteriology; Scientific Press: Beijing, China, 2001; (English Translation). [Google Scholar]
- Hildebrand, D.C.; Palleroni, N.J.; Hendson, M.; Toth, J.; Johnson, J.L. Pseudomonas flavescens sp. nov., isolated from walnut blight cankers. Int. J. Syst. Evol. Microbiol. 1994, 44, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J.J., III; Janda, J.M.; Brenner, F.W.; Cameron, D.N.; Birkhead, K.M. Genus I. Vibrio Pacini 1854, 411AL. In Bergey’s Manual of Systematic Bacteriology, the Proteobacteria, Part B, the Gammaproteobacteria, 2nd ed.; Garrity, G.M., Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; Volume 2, pp. 494–546. [Google Scholar]
- Leifson, E.J. Determination of carbohydrate metabolism of marine bacteria. J. Bacteriol. 1963, 85, 1183. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.D.; Baik, K.S.; Yi, H.; Bae, K.S.; Chun, J. Pseudoalteromonas byunsanensis sp. nov., isolated from tidal flat sediment in Korea. Int. J. Syst. Evol. Microbiol. 2005, 55, 2519–2523. [Google Scholar] [CrossRef]
- Wu, Y.H.; Xu, L.; Zhou, P.; Wang, C.S.; Oren, A.; Xu, X.W. Brevirhabdus pacifica gen. nov., sp. nov., isolated from deep-sea sediment in a hydrothermal vent field. Int. J. Syst. Evol. Microbiol. 2015, 65, 3645–3651. [Google Scholar] [CrossRef]
- Komagata, K.; Suzuki, K.I. 4 Lipid and cell-wall analysis in bacterial systematics. Method Microbiol. 1988, 19, 161–207. [Google Scholar]
- Minnikin, D.E.; O’donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Fang, M.X.; Zhang, W.W.; Zhang, Y.Z.; Tan, H.Q.; Zhang, X.Q.; Wu, M.; Zhu, X.F. Brassicibacter mesophilus gen. nov., sp. nov., a strictly anaerobic bacterium isolated from food industry wastewater. Int. J. Syst. Evol. Microbiol. 2012, 62, 3018–3023. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Lechner, M.; Hernandez-Rosales, M.; Doerr, D.; Wieseke, N.; Thévenin, A.; Stoye, J.; Hartmann, R.K.; Prohaska, S.J.; Stadler, P.F. Orthology detection combining clustering and synteny for very large datasets. PLoS ONE 2014, 9, e105015. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von, H.A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera, A.R.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Hatayama, K.J.G.J. Manganese carbonate precipitation induced by calcite-forming bacteria. Geomicrobiol. J. 2020, 37, 603–609. [Google Scholar] [CrossRef]
- Zhu, Y.; Mam, N.; Jin, W.; Wu, S.; Sun, C. Genomic and transcriptomic insights into calcium carbonate biomineralization by marine actinobacterium Brevibacterium linens BS258. Front. Microbiol. 2017, 8, 602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ju, Y.; Zong, Y.; Qi, H.; Zhao, K. In situ real-time study on dynamics of microbially induced calcium carbonate precipitation at a single-cell level. Environ. Sci. Technol. 2018, 52, 9266–9276. [Google Scholar] [CrossRef]
- Xu, L.; Sun, C.; Fang, C.; Oren, A.; Xu, X.W. Genomic-based taxonomic classification of the family Erythrobacteraceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 4470. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Danese, P.N.; Pratt, L.A.; Dove, S.L.; Kolter, R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 2000, 37, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Prigent-Combaret, C.; Prensier, G.; Le, T.T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Haagensen, J.A.; Schembri, M.A.; Zechner, E.L.; Molin, S. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 2003, 48, 933–946. [Google Scholar] [CrossRef]
- Luo, G.; Huang, L.; Su, Y.; Qin, Y.; Xu, X.; Zhao, L.; Yan, Q. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes. Infect. 2016, 5, e85. [Google Scholar] [CrossRef] [PubMed]
- Drummelsmith, J.; Whitfield, C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000, 19, 57–66. [Google Scholar] [CrossRef]
- Nesper, J.; Hill, C.M.; Paiment, A.; Harauz, G.; Beis, K.; Naismith, J.H.; Whitfield, C. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30: Structural and functional analysis of the outer membrane lipoprotein Wza. J. Biol. Chem. 2003, 278, 49763–49772. [Google Scholar] [CrossRef]
- Whitfield, C.; Paiment, A. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 2003, 338, 2491–2502. [Google Scholar] [CrossRef]
- Beis, K.; Collins, R.F.; Ford, R.C.; Kamis, A.B.; Whitfield, C.; Naismith, J.H. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J. Biol. Chem. 2004, 279, 28227–28232. [Google Scholar] [CrossRef]
- Reid, A.N.; Whitfield, C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: Evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 2005, 187, 5470–5481. [Google Scholar] [CrossRef]
- Yong, Y.C.; Zhong, J.J. Impacts of quorum sensing on microbial metabolism and human health. Trends Biotechnol. 2013, 131, 25–61. [Google Scholar]
- Monteiro, C.; Papenfort, K.; Hentrich, K.; Ahmad, I.; Le, G.S.; Reimann, R.; Grantcharova, N.; Römling, U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012, 9, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Rybtke, M.; Steiner, E.; Weidel, E.; Berthelsen, J.; Groizeleau, J.; Bin, W.; Zhi, B.Z. Regulation of Burkholderia cenocepacia biofilm formation by RpoN and the c-di-GMP effector BerB. Microbiol. Open 2017, 6, e00480. [Google Scholar] [CrossRef] [PubMed]
- Hausrath, A.C.; Ramirez, N.A.; Ly, A.T.; McEvoy, M.M. The bacterial copper resistance protein CopG contains a cysteine-bridged tetranuclear copper cluster. J. Biol. Chem. 2020, 295, 11364–11376. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, C.N.; Soldatova, A.V.; Lee, S.W.; Spiro, T.G.; Tebo, B.M. Mn(II, III) oxidation and MnO2 mineralization by an expressed bacterial multicopper oxidase. Proc. Natl. Acad. Sci. USA 2013, 110, 11731–11735. [Google Scholar] [CrossRef]
- Kaur, K.; Sharma, A.; Capalash, N.; Sharma, P. Multicopper oxidases: Biocatalysts in microbial pathogenesis and stress management. Microbiol. Res. 2019, 222, 1–13. [Google Scholar] [CrossRef]
- Geszvain, K.; Smesrud, L.; Tebo, B.M. Identification of a third Mn(II) oxidase enzyme in Pseudomonas putida GB-1. Appl. Environ. Microb. 2016, 82, 3774–3782. [Google Scholar] [CrossRef]
- Zhou, H.; Fu, C. Manganese-oxidizing microbes and biogenic manganese oxides: Characterization, Mn (II) oxidation mechanism and environmental relevance. Rev. Environ. Sci. Bio 2020, 19, 489–507. [Google Scholar] [CrossRef]
- Wu, H.X.; Lai, P.Y.; Lee, O.O.; Zhou, X.J.; Miao, L.; Wang, H.; Qian, P.Y. Erythrobacter pelagi sp. nov., a member of the family Erythrobacteraceae isolated from the Red Sea. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 6, 1348–1353. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Yutin, N.; Suzuki, M.T.; Teeling, H.; Weber, M.; Venter, J.C.; Rusch, D.B.; Béjà, O. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 2007, 9, 1464–1475. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Rosen, B.P. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 2002, 110, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Cheng, K.; Li, Z.; Ma, X.; Wei, Y.; Zhang, L.; Wang, Y. Biosorption of cadmium and manganese using free cells of Klebsiella sp. isolated from waste water. PLoS ONE 2015, 10, e0140962. [Google Scholar] [CrossRef]
- Dey, S.; Paul, A.K. Assessment of heavy metal tolerance and hexavalent chromium reducing potential of Corynebacterium paurometabolum SKPD 1204 isolated from chromite mine seepage. AIMS Bioeng. 2016, 3, 337–351. [Google Scholar] [CrossRef]
- Xu, L.; Xu, X.W.; Meng, F.X.; Huo, Y.Y.; Oren, A.; Yang, J.Y.; Wang, C.S. Halomonas zincidurans sp. nov., a heavy-metal-tolerant bacterium isolated from the deep-sea environment. Int. J. Syst. Evol. Microbiol. 2013, 63, 4230–4236. [Google Scholar] [CrossRef]
- Wang, X.; Lin, D.; Jing, X.; Zhu, S.; Yang, J.; Chen, J. Complete genome sequence of the highly Mn(II) tolerant Staphylococcus sp. AntiMn-1 isolated from deep-sea sediment in the Clarion-Clipperton Zone. J. Biotechnol. 2018, 266, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Gillard, B.; Chatzievangelou, D.; Thomsen, L.; Ullrich, M.S. Heavy-metal-resistant microorganisms in deep-sea sediments disturbed by mining activity: An application toward the development of experimental in vitro systems. Front. Mar. Sci. 2019, 6, 462. [Google Scholar] [CrossRef]
- Wu, Y.H.; Fang, C.; Zhou, P.; Wang, C.S.; Xu, X.W. Complete genome sequence of a heavy metal resistant bacterium Maribacter cobaltidurans B1T, isolated from the deep-sea sediment of the South Atlantic Ocean. Mar. Genom. 2018, 39, 19–21. [Google Scholar] [CrossRef]
- Wu, Y.H.; Cheng, H.; Zhou, P.; Huo, Y.Y.; Wang, C.S.; Xu, X.W. Complete genome sequence of the heavy metal resistant bacterium Altererythrobacter atlanticus 26DY36T, isolated from deep-sea sediment of the North Atlantic Mid-ocean ridge. Mar. Genom. 2015, 24, 289–292. [Google Scholar] [CrossRef]
- Wang, W.; Shao, Z.; Liu, Y.; Wang, G. Removal of multi-heavy metals using biogenic manganese oxides generated by a deep-sea sedimentary bacterium–Brachybacterium sp. strain Mn32. Microbiol. Res. 2009, 155, 1989–1996. [Google Scholar] [CrossRef]
- Du, R.; Gao, D.; Wang, Y.; Liu, L.; Cheng, J.; Liu, J.; Zhang, X.H.; Yu, M. Heterotrophic sulfur oxidation of Halomonas titanicae SOB56 and its habitat adaptation to the hydrothermal environment. Front. Microbiol. 2022, 13, 888833. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Yin, K.; Wang, Q.; Lv, M.; Chen, L. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 2019, 360, 1553–1563. [Google Scholar] [CrossRef]
- Radisky, D.; Kaplan, J. Regulation of transition metal transport across the yeast plasma membrane. J. Biol. Chem. 1999, 274, 4481–4484. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.E.; Muller-Moule, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef] [PubMed]
- Frankel, R.B.; Bazylinski, D.A. Bazylinski, and geochemistry, Biologically induced mineralization by bacteria. Rev. Mineral. Geochem. 2003, 54, 95–114. [Google Scholar] [CrossRef]
- Ercole, C.; Bozzelli, P.; Altieri, F.; Cacchio, P.; Del, G.M. Calcium carbonate mineralization: Involvement of extracellular polymeric materials isolated from calcifying bacteria. Microsc. Microanal. 2012, 18, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Gutierrez, T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef] [PubMed]
- Picard, A.; Gartman, A.; Clarke, D.R.; Girguis, P.R. Sulfate-reducing bacteria influence the nucleation and growth of mackinawite and greigite. Geochim. Cosmochim. Acta. 2018, 220, 367–384. [Google Scholar] [CrossRef]
- Achal, V.; Pan, X. Characterization of urease and carbonic anhydrase producing bacteria and their role in calcite precipitation. Curr. Microbiol. 2011, 62, 894–902. [Google Scholar] [CrossRef]
- Teng, Z.; Shao, W.; Zhang, K.; Huo, Y.; Zhu, J.; Li, M. Pb biosorption by Leclercia adecarboxylata: Protective and immobilized mechanisms of extracellular polymeric substances. Chem. Eng. J. 2019, 375, 122113. [Google Scholar] [CrossRef]
- Han, L.; Li, J.; Xue, Q.; Chen, Z.; Zhou, Y.; Poon, C.S. Bacterial-induced mineralization (BIM) for soil solidification and heavy metal stabilization: A critical review. Sci. Total Environ. 2020, 746, 140967. [Google Scholar] [CrossRef] [PubMed]





| Characteristics | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Color | Yellow | Cream | Yellow | Yellow | Yellow |
| Nitrate reduction | − | − | − | + | − |
| H2S production | + | − | + | − | − |
| Urea hydrolysis | + | + | − | − | + |
| Tryptophane deaminase | + | + | + | w | + |
| Utilization of | |||||
| L-Arabinose | + | − | + | − | − |
| Capric acid | + | + | + | + | + |
| Glucose | − | w | + | + | + |
| Maltose | − | − | + | + | + |
| Malate | − | − | − | − | + |
| Mannitol | − | − | − | + | + |
| Acid production: | |||||
| Ethanol | − | + | − | − | − |
| D-Galactose | − | − | + | − | − |
| D-Xylose | − | − | + | − | − |
| Enzyme activities: | |||||
| α-Chymotrypsin | − | − | − | − | + |
| Cystine arylamidase | + | − | + | w | + |
| Esterase (C4) | + | + | + | + | − |
| Esterase lipase (C8) | + | + | + | − | + |
| α-Galactosidase | − | − | + | − | − |
| β-Galactosidase | − | − | + | − | − |
| α-Glucosidase | − | − | + | + | + |
| β-Glucosidase | − | − | + | − | − |
| β-Glucuronidase | + | − | + | − | − |
| Trypsin | − | − | − | − | + |
| Valine arylamidase | + | + | + | + | + |
| Antibiotic susceptibility: | |||||
| Ampicillin (10 μg) | w | − | + | + | − |
| Erythromycin (10 μg) | + | + | + | + | + |
| Neomycin (30 μg) | + | − | + | w | + |
| Nitrofurantoin (300 μg) | + | − | + | + | + |
| Penicillin (10 IU) | − | − | + | + | − |
| Tetracyline (30 μg) | − | − | + | w | w |
| DNA G+C content (%) | 62.5 | 62.2 | 64.0 | 62.8 | 62.5 |
| Fatty Acid (%) | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Straight-chain | |||||
| C14:0 | 1.3 | 1.6 | 2.1 | 1.4 | 1.4 |
| C15:0 | tr | tr | 2.1 | 1.0 | tr |
| C16:0 | 4.5 | 4.3 | 2.7 | 11.8 | 2.1 |
| Unsaturated | |||||
| C15:1ω6c | - | - | 0.9 | - | - |
| C16:1ω5c | 1.5 | 1.1 | 0.7 | 0.9 | 2.0 |
| C17:1ω8c | 0.8 | 1.2 | 1.8 | - | - |
| C17:1ω6c | 4.6 | 6.3 | 16.9 | 6.1 | 1.9 |
| C18:1ω7c | 56.6 | 49.4 | 26.0 | 45.8 | 24.4 |
| C18:1ω5c | 2.0 | 2.3 | 3.3 | 1.5 | 1.7 |
| 11 methyl C18:1ω7c | 1.8 | 1.8 | - | 0.7 | 0.6 |
| Hydroxy | |||||
| C13:0 2OH | - | - | 0.6 | - | - |
| C14:0 2OH | 13.3 | 15.7 | 22.2 | 16.9 | 41.9 |
| C15:0 2OH | 1.6 | 2.2 | 5.5 | 2.1 | 2.7 |
| C16:1 2OH | - | - | - | - | 1.6 |
| iso-C16:0 3OH | 1.2 | 0.6 | - | 0.5 | 2.5 |
| C16:0 2OH | 2.5 | 1.6 | - | - | 5.8 |
| C18:1 2OH | 0.8 | tr | - | - | 1.4 |
| Cyclic | |||||
| C19:0ω8c cyclo | tr | 1.9 | 4.6 | - | - |
| Summed Feature * | |||||
| 3 | 7.0 | 9.4 | 10.7 | 11.1 | 9.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, X.; Zhong, Y.; Fu, G.; Wu, Y.; Xu, X. Revealing Heavy Metal-Resistant Mechanisms and Bioremediation Potential in a Novel Croceicoccus Species Using Microbial-Induced Carbonate Precipitation. J. Mar. Sci. Eng. 2023, 11, 2195. https://doi.org/10.3390/jmse11112195
Lv X, Zhong Y, Fu G, Wu Y, Xu X. Revealing Heavy Metal-Resistant Mechanisms and Bioremediation Potential in a Novel Croceicoccus Species Using Microbial-Induced Carbonate Precipitation. Journal of Marine Science and Engineering. 2023; 11(11):2195. https://doi.org/10.3390/jmse11112195
Chicago/Turabian StyleLv, Xuya, Yingwen Zhong, Geyi Fu, Yuehong Wu, and Xuewei Xu. 2023. "Revealing Heavy Metal-Resistant Mechanisms and Bioremediation Potential in a Novel Croceicoccus Species Using Microbial-Induced Carbonate Precipitation" Journal of Marine Science and Engineering 11, no. 11: 2195. https://doi.org/10.3390/jmse11112195





