Abstract
The development of low-cost, open-source Remotely Operated Vehicle (ROV) systems has provided almost unrestricted access for researchers looking to monitor the marine environment in ever greater resolution. Sampling microbial communities from the marine environment, however, still usually relies on Niskin-bottle sampling (ROV or Conductivity-Temperature-Depth sampler (CTD) based), a method which introduces an inaccuracy and variability that is incompatible with metatranscriptomic analysis, for example. Here, we describe a versatile, easily-replicated platform which achieves in situ mRNA preservation, via the addition of RNAlater to filtered microbial cells, to enhance ROV or CTD functionality.
Based on the modified Nansen bottle (invented in 1894); the Niskin bottle (1967), invented just a few years after the discovery and characterisation of mRNA, was developed for the retrieval of seawater samples to the surface (Hill, 1900; Niskin, 1966; Cobb, 1990) [1,2,3]. Traditional Niskin sampling still dominates oceanic analysis, while metatranscriptomic (whole community mRNA profiling) based techniques have revolutionised our understanding of the function of mixed community assemblages at the molecular level (Gilbert et al., 2011) [4]. Together, they have provided a much needed insight on the fundamental workings of global biogeochemical cycling. However, while metatranscriptomics suffers from a necessity to reduce technical variation as much as possible to allow meaningful interpretation of results, it is stifled by the inaccuracy and variability that is irrevocably associated with current Niskin-based sampling methods. Whilst cellular mRNA profiles can respond to environmental insults within milliseconds, the mandatory transcriptional alterations induced by Niskin sampling, which subjects samples to unavoidable exposure to differences in pressure, temperature and light, in addition to the inherent temporal delay, is difficult to circumvent. This irreconcilable observation has stimulated the development of many in situ profiling technologies for the marine environment (Feike et al., 2012; Taylor et al., 2015; McQuillan and Robidart, 2017) [5,6,7], however these solutions have not gained dominance or widespread use as yet, primarily due to cost restrictions.
In tandem to the dawning realisation that the majority of current marine transcriptomic and metatranscriptomic analyses are inherently inaccurate, the development of low cost open source ROV systems has provided easy access (to the top 100 m of the ocean at the very least) for researchers looking to monitor, and sample, the marine environment in ever greater resolution. Whilst utilising an ROV mounted Niskin system to study metatranscriptomic profiles, we were struck by the contrast between the antiquated nature of this traditional and inaccurate sampling technique, and the low cost, high-performance simplicity of the ROV system upon which is was mounted. To this end, we looked to develop a versatile, easily replicated RNA sampling platform (“RNA Automated Preservation in situ Device, RAPID”) inspired by low-cost, high-performance and simplicity. It is well established that in situ mRNA preservation can be achieved rapidly and simply through the addition of RNAlater (Ambion, Austin, TX, USA) to microbial cells (Ottesen et al., 2011) [8].
With this premise in mind, we looked to design a system that could both concentrate and preserve samples in a rapid, simple and low cost manner. Utilising off the shelf components we assembled and tested an Arduino (Leonardo) controlled dual pump system [9], capable of pushing seawater through a suitable filter unit, prior to the delivery of RNAlater (Figure 1). With motors and electronics encased and powered from 12 V supply (4 × AA batteries) within a permanently sealed waterproof junction box (Model: a16030800ux0347, Uxcell, Hong Kong, China) (Figure 2), and filtration units and RNAlater reservoir (saline drip bag (Model: GMEPN-UK-72813179, Amazlabs, Guandong, China)) external for easy replacement and retrieval, our system was mounted on an OpenROV (OpenROV, Berkeley, CA, USA) (rated to 100 m depth) for testing [10]. Pumps were mounted alongside each other and tubing joined via a T-junction with one-way values (Model: 1024989, Online Car Parts Ltd, Lancashire, UK) attached to the (external) Swinnex 25 mm filter assembly (Merck Millipore, Hertfordshire, UK). Initial trials with centrifugal pumps (adapted from a NERF Electrostorm water pistol; Hasbro, Pawtucket, RI, USA) revealed rapid degradation of internal components exposed to seawater and RNAlater, so we favoured a peristaltic pump option (Model: A518, ZJchao, Guandong, China). Any filter assembly (and filter type) capable of withstanding pressure can be used (we have utilised 25 mm and 47 mm filter assemblies, as well as the Sterivex system). The Arduino was mounted on the lid of the box, so that in the situation of structural integrity being lost, water damage to the circuit would be minimised (total immersion in silicon oil is another simple way to reduce pressure effects). Nevertheless, replacement of the junction box with a more robust structure may be necessary to go beyond 100 m depth. Whilst we developed here a single filter sample system, the addition of simple controlled distribution valves will provide the opportunity for numerous samples to be taken and preserved in procession. Following activation of pump 1, seawater is pushed though the filter assembly at a rate of ~2.5 mL/s (we achieved filtration of ~500 mL through a 0.22 µm Sterivex Filter in 4 min (Merck Millipore, Hertfordshire, UK)), applying different filters varies the rate of flow, as does biomass accumulation on the filter, until pump 2 is engaged for a 10 s flooding with ~27 mL of RNAlater. Although not instantaneous, the sample is not subjected to any temperature, pressure and/or light variation (unless the ROV is operated to specifically induce such conditions) and filtration/preservation is performed rapidly in situ. This potentially represents a significant improvement in both accuracy of transcript profiles and rapidity in comparison with current sampling procedures which usually rely on a delay for filtering on board ship following sample retrieval.
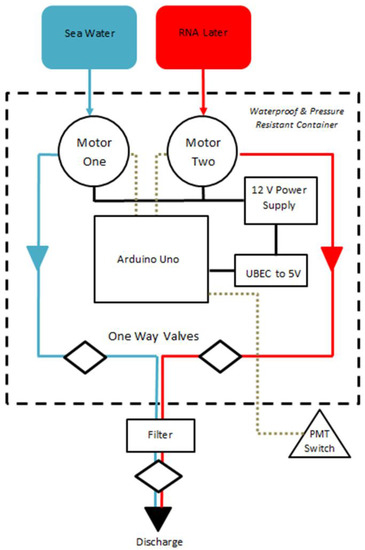
Figure 1.
Configuration of Nucleic Acid Preservation device. RNAlater is stored within a saline drip bag to minimise pressure effects. Proximal and distal one way valves serve to ensure filter remains immersed in RNAlater following preservation. Dashed line denotes components contained within pressure and water resistant shell.
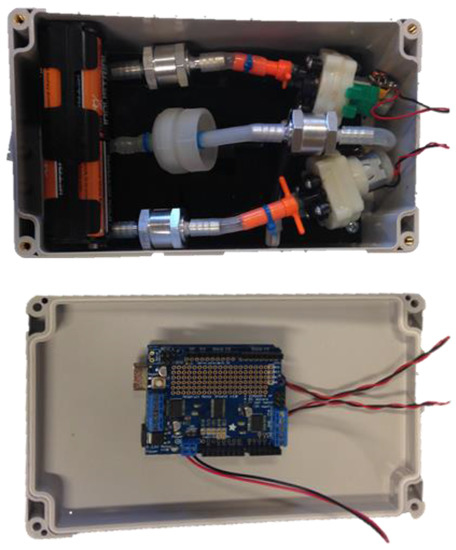
Figure 2.
Components within the casing and their configuration (Prototype 1, with NERF Electrostorm motors). L200 × W120 × D76 mm.
For samples where it is crucial to preserve the transcriptional profile immediately, pumps 1 and 2 can be run simultaneously to bring RNAlater into contact with the seawater immediately prior to filtration or bag collection. Following retrieval of the ROV to the surface and RNA extraction in the laboratory, no difference was observed in quality or quantity of total RNA obtained by Niskin or the on board system (Figure 3), thereby proving the principle that sampling via systems of this type can provide sufficient and suitable RNA, which is by virtue of its processing more representative of the natural environment from which it is taken. In addition to costing less than £50 to build and being small enough to mount on low-cost, entry-level ROV systems (which provide visualisation, easy maneuverability, and often accurate depth and temperature data, in real time and therefore with the opportunity for responsive action), such a system can also be utilised in conjunction with more established CTD instrumentation. In the spirit of the open source ethos, we invite others to join us and take up the challenge in testing and developing improved versions of this versatile system that has the potential to revolutionise the molecular analysis of the marine environment [11].
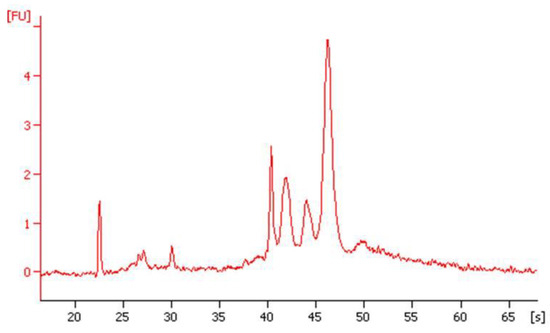
Figure 3.
Total RNA (112 ng/µL; RIN 28S:18S score, 8.0) extracted from approximately 500 mL of natural seawater from Plymouth Sound preserved by RAPID sampling, analysed by 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
Acknowledgments
We would like to acknowledge the internally funded PML Research Program (awarded to M.J.A.) and Cabot Institute Innovation Fund (awarded to T.S. and M.J.A.) for providing the funding for this study.
Author Contributions
M.J.A. and J.T. conceived and designed the experiments; J.T. performed the experiments; M.J.A., J.T., S.S., G.G. and T.S. analysed the data; M.J.A., J.T., G.G., S.S. and T.S. contributed reagents/materials/analysis tools; M.J.A. and J.T. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Niskin, S.J. Water Sampler Device. U.S. Patent 3,489,012A, 1967. [Google Scholar]
- Mill, H.R. The Pettersson-Nansen Insulating Water-Bottle. Geogr. J. 1900, 16, 469–471. [Google Scholar] [CrossRef]
- Cobb, M. Who discovered messenger RNA? Curr. Biol. 2015, 25, R526–R532. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Field, D.; Huang, Y.; Edwards, R.; Li, W.; Gilna, P.; Joint, I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS ONE 2008, 3, e3042. [Google Scholar] [CrossRef] [PubMed]
- Feike, J.; Jürgens, K.; Hollibaugh, J.T.; Krüger, S.; Jost, G.; Labrenz, M. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 2012, 6, 461–470. [Google Scholar] [CrossRef] [PubMed]
- McQuillan, J.S.; Robidart, J.C. Molecular-biological sensing in aquatic environments: Recent developments and emerging capabilities. Curr. Opin. Biotechnol. 2017, 45, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.D.; Edgcomb, V.P.; Doherty, K.W.; Engstrom, I.; Shanahan, T.; Pachiadaki, M.G.; Molyneaux, S.J.; Honjo, S. Fixation filter, device for the rapid in situ preservation of particulate samples. Deep Sea Res. Part I Oceanogr. Res. Pap. 2015, 96, 69–79. [Google Scholar] [CrossRef]
- Ottesen, E.A.; Marin, R.; Preston, C.M.; Young, C.R.; Ryan, J.P.; Scholin, C.A.; DeLong, E.F. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011, 5, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Arduino, Open Source Hardware and Software. Available online: https://www.arduino.cc/ (accessed on 31 May 2017).
- OpenROV. Available online: https://www.openrov.com/ (accessed on 31 May 2017).
- RAPID Arduino Code. Available online: https://github.com/Teaguey08/RNA-preservation-Device/tree/Files (accessed on 31 May 2017).
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).