Hybrids of Nucleic Acids and Carbon Nanotubes for Nanobiotechnology
Abstract
:1. Introduction
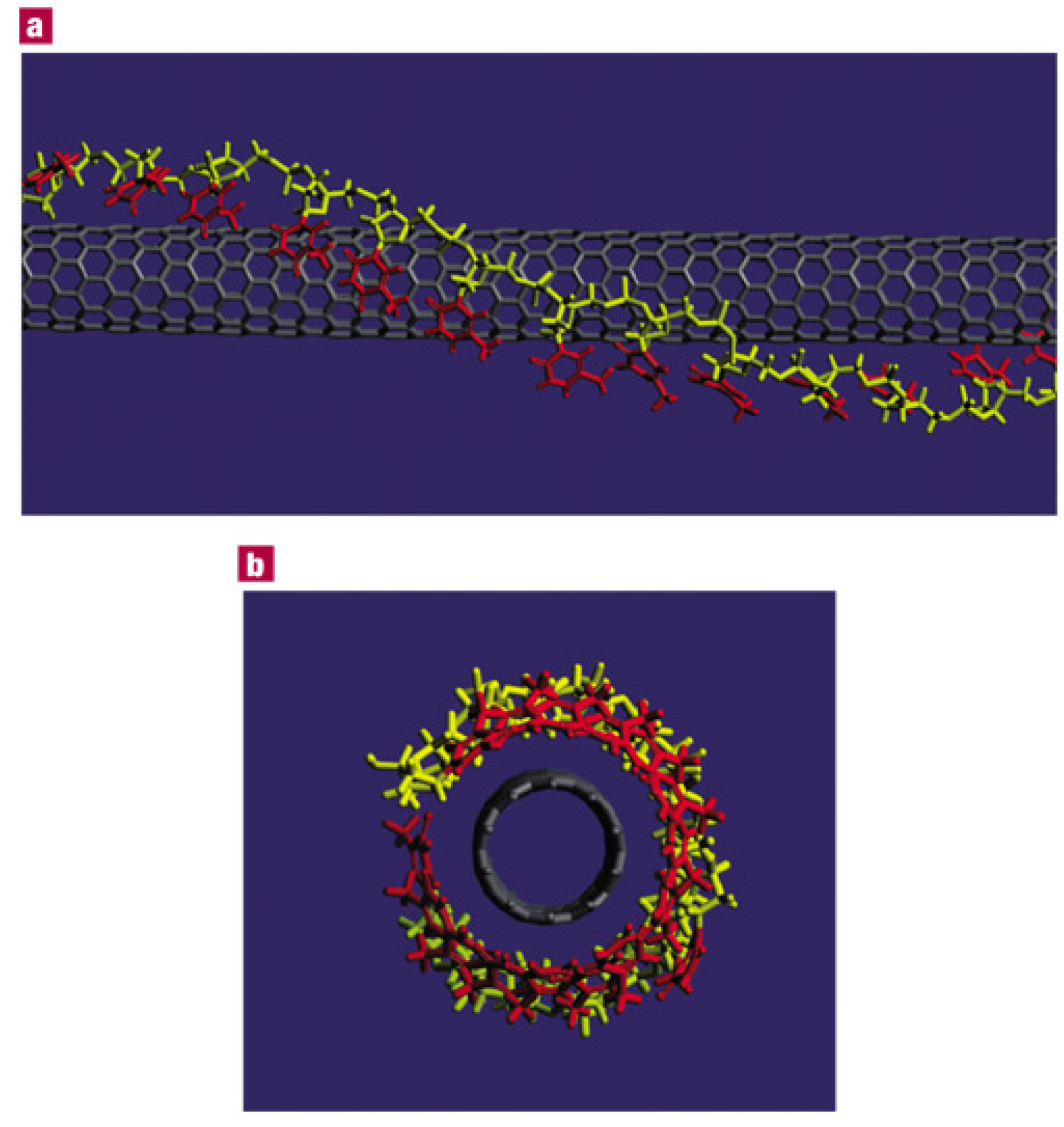
2. Non-Covalent Hybridization of DNA Molecules onto SWNT Surfaces
3. Hybridization with dsDNA
4. Hybridization with ssDNA
5. Comparative Studies Using ssDNA, dsDNA, and Other Nucleic Acids
6. Selective Adsorption and Separation of CNTs
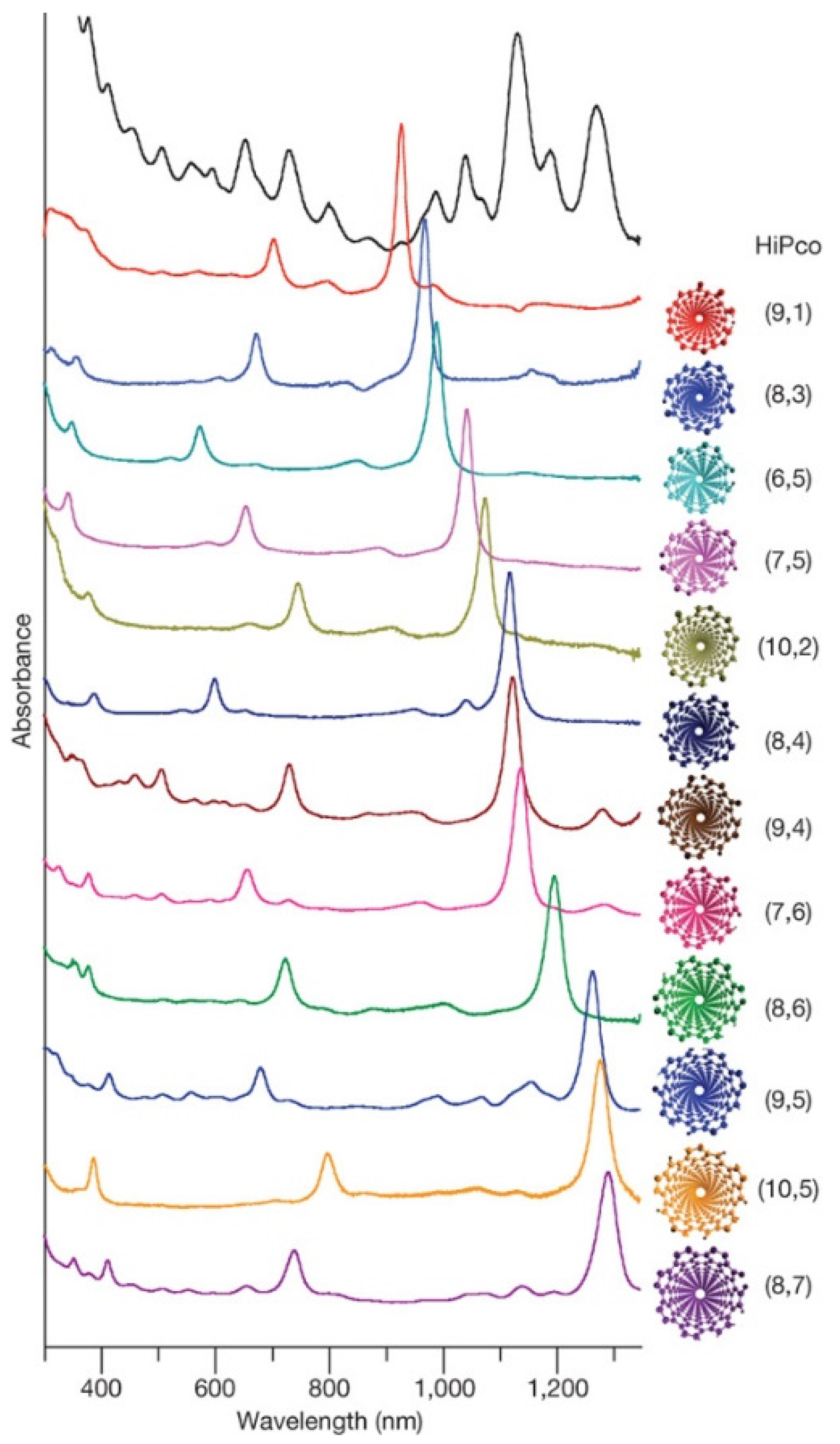
7. Characterization of DNA-CNT Hybrids
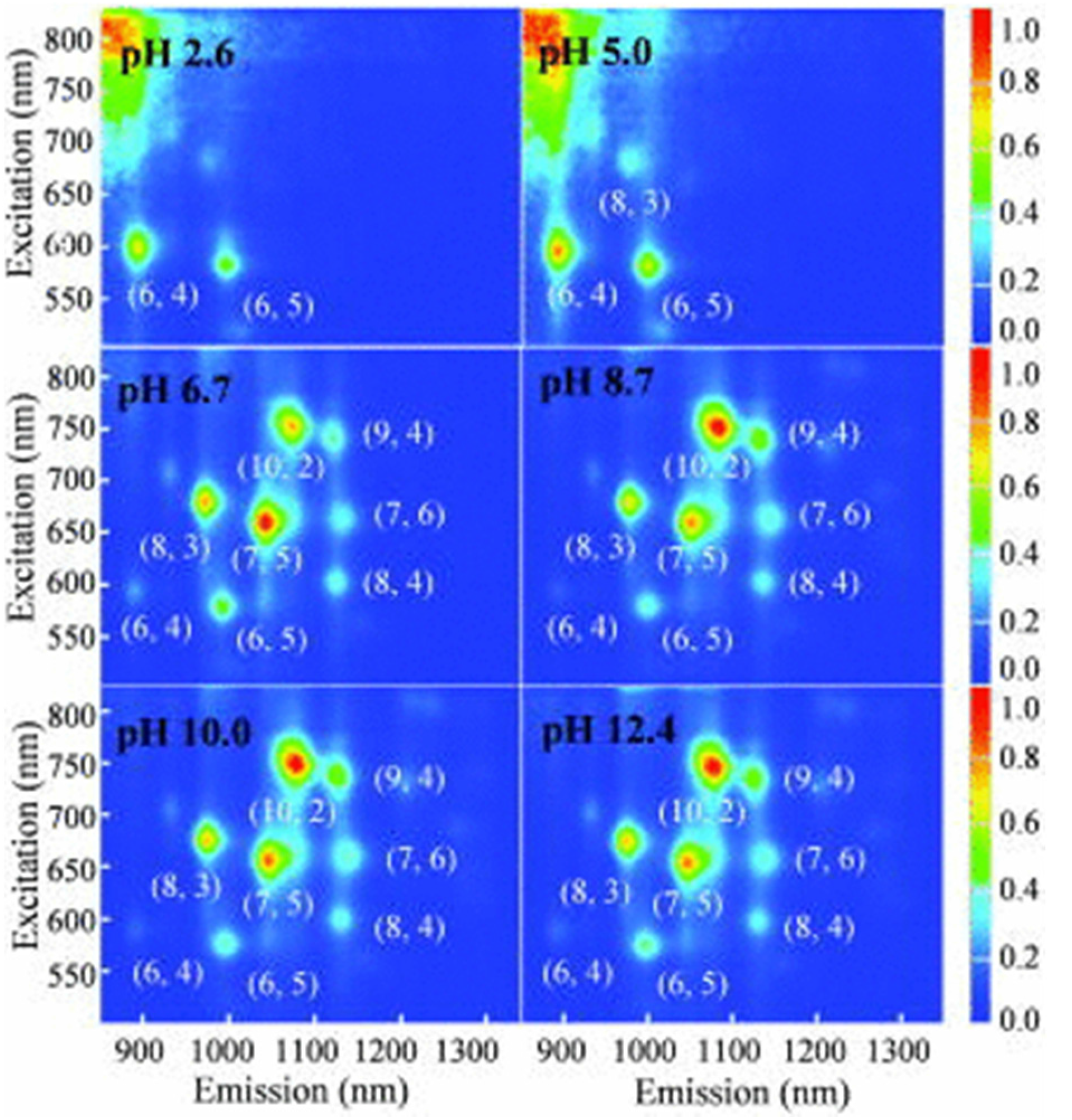
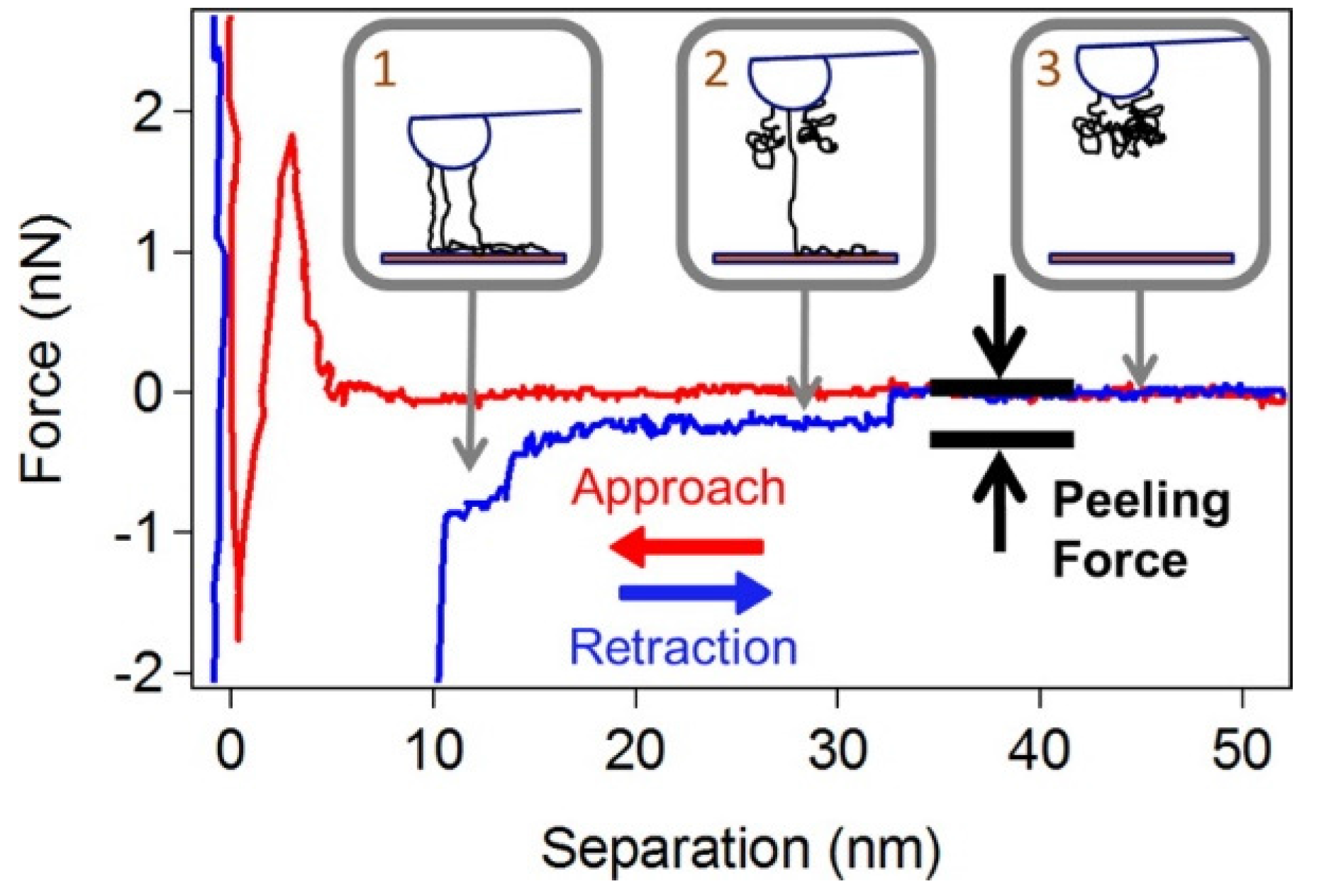

8. Functionalization of CNTs for DNA Attachment
9. Theoretical Studies of Nucleic Acid Adsorption to CNTs
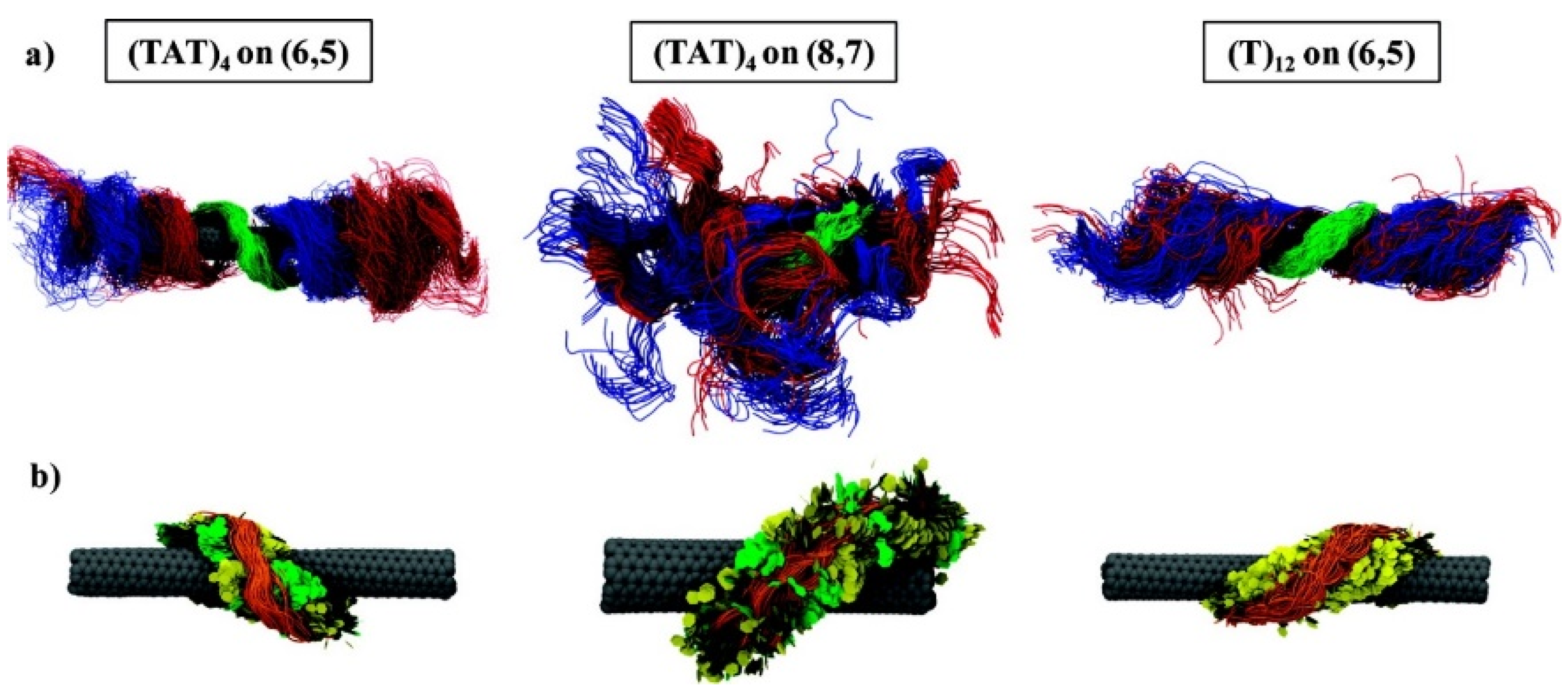
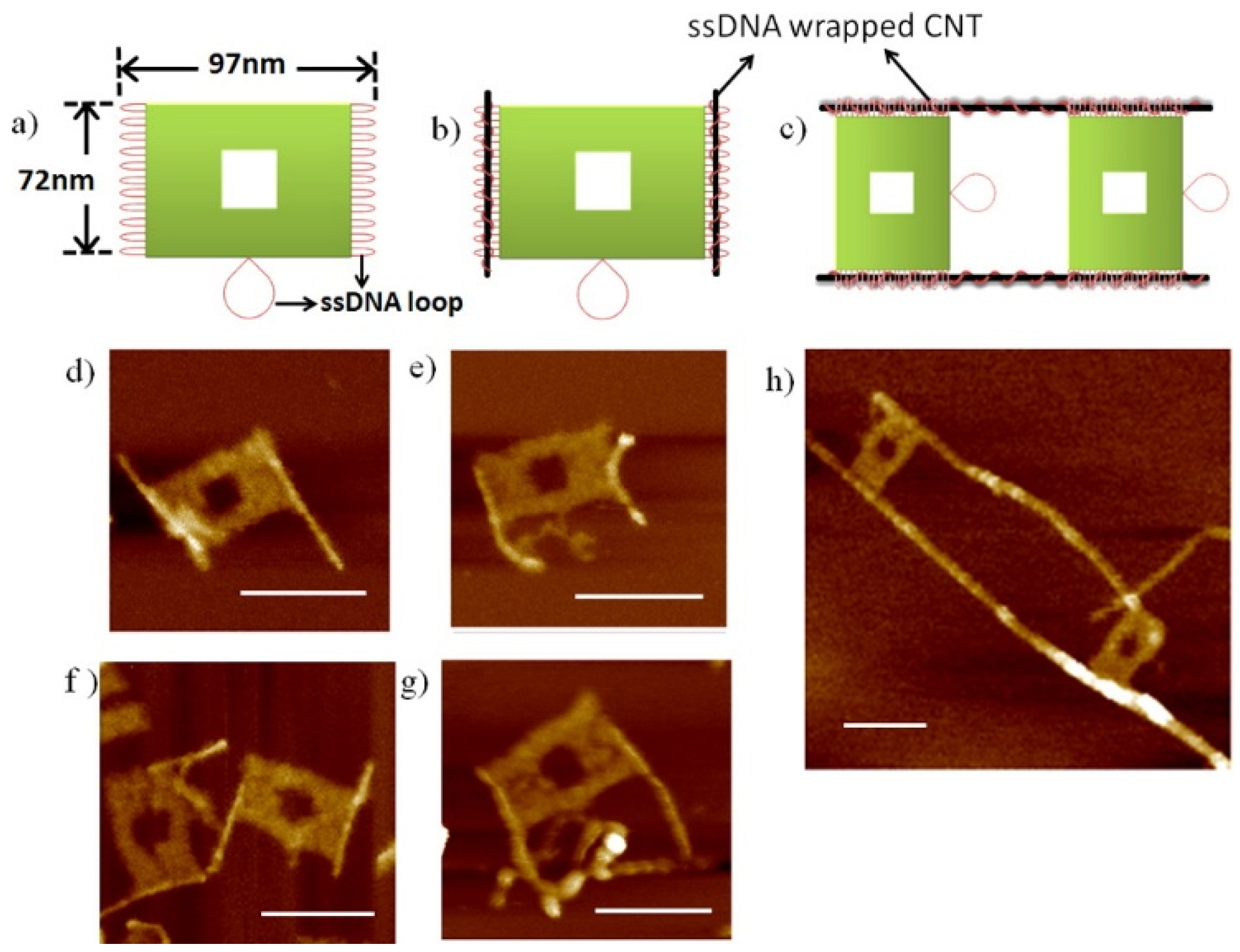
10. Biological Applications
11. Conclusions
Acknowledgments
Conflicts of Interest
References
- Zheng, M.; Jagota, A.; Semke, E.D.; Diner, B.A.; McLean, R.S.; Lustig, S.R.; Richardson, R.E.; Tassi, N.G. DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater. 2003, 2, 338–342. [Google Scholar]
- Nakashima, N.; Okuzono, S.; Murakami, H.; Nakai, T.; Yoshikawa, K. DNA dissolves single-walled carbon nanotubes in water. Chem. Lett. 2003, 32, 456–457. [Google Scholar]
- O’Connell, M.J.; Boul, P.; Ericson, L.M.; Huffman, C.; Wang, Y.H.; Haroz, E.; Kuper, C.; Tour, J.; Ausman, K.D.; Smalley, R.E. Reversible water-solubilization of single-walled carbon nanotubes by polymer wrapping. Chem. Phys. Lett. 2001, 342, 265–271. [Google Scholar]
- Fu, K.F.; Huang, W.J.; Lin, Y.; Zhang, D.H.; Hanks, T.W.; Rao, A.M.; Sun, Y.P. Functionalization of carbon nanotubes with bovine serum albumin in homogeneous aqueous solution. J. Nanosci. Nanotechnol. 2002, 2, 457–461. [Google Scholar]
- Huang, W.J.; Lin, Y.; Taylor, S.; Gaillard, J.; Rao, A.M.; Sun, Y.P. Sonication-assisted functionalization and solubilization of carbon nanotubes. Nano Lett. 2002, 2, 231–234. [Google Scholar]
- Star, A.; Stoddart, J.F. Dispersion and solubilization of single-walled carbon nanotubes with a hyperbranched polymer. Macromolecules 2002, 35, 7516–7520. [Google Scholar]
- Dodziuk, H.; Ejchart, A.; Anczewski, W.; Ueda, H.; Krinichnaya, E.; Dolgonos, G.; Kutner, W. Water solubilization, determination of the number of different types of single-wall carbon nanotubes and their partial separation with respect to diameters by complexation with eta-cyclodextrin. Chem. Commun. 2003, 8, 986–987. [Google Scholar]
- Jiang, L.Q.; Gao, L.; Sun, J. Production of aqueous colloidal dispersions of carbon nanotubes. J. Colloid Interface Sci. 2003, 260, 89–94. [Google Scholar]
- Petrov, P.; Stassin, F.; Pagnoulle, C.; Jerome, R. Noncovalent functionalization of multi-walled carbon nanotubes by pyrene containing polymers. Chem. Commun. 2003, 23, 2904–2905. [Google Scholar]
- Wang, J.; Musameh, M.; Lin, Y.H. Solubilization of carbon nanotubes by nafion toward the preparation of amperometric biosensors. J. Am. Chem. Soc. 2003, 125, 2408–2409. [Google Scholar]
- Hasegawa, T.; Fujisawa, T.; Numata, M.; Umeda, M.; Matsumoto, T.; Kimura, T.; Okumura, S.; Sakurai, K.; Shinkai, S. Single-walled carbon nanotubes acquire a specific lectin-affinity through supramolecular wrapping with lactose-appended schizophyllan. Chem. Commun. 2004, 19, 2150–2151. [Google Scholar]
- Zhang, M.G.; Smith, A.; Gorski, W. Carbon nanotube-chitosan system for electrochemical sensing based on dehydrogenase enzymes. Anal. Chem. 2004, 76, 5045–5050. [Google Scholar]
- Liu, P. Modifications of carbon nanotubes with polymers. Eur. Polym. J. 2005, 41, 2693–2703. [Google Scholar]
- Sinani, V.A.; Gheith, M.K.; Yaroslavov, A.A.; Rakhnyanskaya, A.A.; Sun, K.; Mamedov, A.A.; Wicksted, J.P.; Kotov, N.A. Aqueous dispersions of single-wall and multiwall carbon nanotubes with designed amphiphilic polycations. J. Am. Chem. Soc. 2005, 127, 3463–3472. [Google Scholar]
- Yan, Y.M.; Zhang, M.N.; Gong, K.P.; Su, L.; Guo, Z.X.; Mao, L.Q. Adsorption of methylene blue dye onto carbon nanotubes: A route to an electrochemically functional nanostructure and its layer-by-layer assembled nanocomposite. Chem. Mater. 2005, 17, 3457–3463. [Google Scholar]
- Wang, D.; Chen, L.W. Temperature and ph-responsive single-walled carbon nanotube dispersions. Nano Lett. 2007, 7, 1480–1484. [Google Scholar]
- Wise, A.J.; Smith, J.R.; Bouropoulos, N.; Yannopoulos, S.N.; van der Merwe, S.M.; Fatouros, D.G. Single-wall carbon nanotube dispersions stabilised with N-trimethyl-chitosan. J. Biomed. Nanotechnol. 2008, 4, 67–72. [Google Scholar]
- Plisko, T.V.; Bildyukevich, A.V. Debundling of multiwalled carbon nanotubes in N, N-dimethylacetamide by polymers. Colloid Polym. Sci. 2014, 292, 2571–2580. [Google Scholar]
- Niyogi, S.; Hamon, M.A.; Hu, H.; Zhao, B.; Bhowmik, P.; Sen, R.; Itkis, M.E.; Haddon, R.C. Chemistry of single-walled carbon nanotubes. Accounts Chem. Res. 2002, 35, 1105–1113. [Google Scholar]
- Williams, K.A.; Veenhuizen, P.T.M.; de la Torre, B.G.; Eritja, R.; Dekker, C. Nanotechnology—Carbon nanotubes with DNA recognition. Nature 2002, 420, 761–761. [Google Scholar]
- Fu, K.F.; Sun, Y.P. Dispersion and solubilization of carbon nanotubes. J. Nanosci. Nanotechnol. 2003, 3, 351–364. [Google Scholar]
- Martin, C.R.; Kohli, P. The emerging field of nanotube biotechnology. Nat. Rev. Drug Discov. 2003, 2, 29–37. [Google Scholar]
- Jin, W.J.; Sun, X.F.; Wang, Y. Solubilization and functionalization of carbon nanotubes. New Carbon Mater. 2004, 19, 312–318. [Google Scholar]
- Lin, Y.; Taylor, S.; Li, H.P.; Fernando, K.A.S.; Qu, L.W.; Wang, W.; Gu, L.R.; Zhou, B.; Sun, Y.P. Advances toward bioapplications of carbon nanotubes. J. Mater. Chem. 2004, 14, 527–541. [Google Scholar]
- Pividori, M.I.; Alegret, S. DNA Adsorption on Carbonaceous Materials. In Immobilisation of DNA on Chips I; Wittmann, C., Ed.; Springer-Verlag Berlin: Berlin, Germany, 2005; Volume 260, pp. 1–36. [Google Scholar]
- Nakashima, N. Solubilization of single-walled carbon nanotubes with condensed aromatic compounds. Sci. Technol. Adv. Mater. 2006, 7, 609–616. [Google Scholar]
- Daniel, S.; Rao, T.P.; Rao, K.S.; Rani, S.U.; Naidu, G.R.K.; Lee, H.Y.; Kawai, T. A review of DNA functionalized/grafted carbon nanotubes and their characterization. Sens. Actuator B 2007, 122, 672–682. [Google Scholar]
- Wei, W.; Sethuraman, A.; Jin, C.; Monteiro-Riviere, N.A.; Narayan, R.J. Biological properties of carbon nanotubes. J. Nanosci. Nanotechnol. 2007, 7, 1284–1297. [Google Scholar]
- Tu, X.M.; Zheng, M. A DNA-based approach to the carbon nanotube sorting problem. Nano Res. 2008, 1, 185–194. [Google Scholar]
- Becerril, H.A.; Woolley, A.T. DNA-templated nanofabrication. Chem. Soc. Rev. 2009, 38, 329–337. [Google Scholar]
- Cheung, W.; Chiu, P.L.; Parajuli, R.R.; Ma, Y.F.; Ali, S.R.; He, H.X. Fabrication of high performance conducting polymer nanocomposites for biosensors and flexible electronics: Summary of the multiple roles of DNA dispersed and functionalized single walled carbon nanotubes. J. Mater. Chem. 2009, 19, 6465–6480. [Google Scholar]
- Sanchez-Pomales, G.; Santiago-Rodriguez, L.; Cabrera, C.R. DNA-functionalized carbon nanotubes for biosensing applications. J. Nanosci. Nanotechnol. 2009, 9, 2175–2188. [Google Scholar]
- Zhao, Y.L.; Stoddart, J.F. Noncovalent functionalization of single-walled carbon nanotubes. Accounts Chem. Res. 2009, 42, 1161–1171. [Google Scholar]
- Fujigaya, T.; Tanaka, Y.; Nakashima, N. Soluble carbon nanotubes and application to electrochemistry. Electrochemistry 2010, 78, 2–15. [Google Scholar]
- Lei, J.P.; Ju, H.X. Nanotubes in biosensing. Nanomed. Nanobiotechnol. 2010, 2, 496–509. [Google Scholar]
- Contreras-Torres, F.F.; Martinez-Loran, E. DNA insertion in and wrapping around carbon nanotubes. Comput. Mol. Sci. 2011, 1, 902–919. [Google Scholar]
- Zhang, W.X.; Zhang, Z.Z.; Zhang, Y.G. The application of carbon nanotubes in target drug delivery systems for cancer therapies. Nanoscale Res. Lett. 2011, 6. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Xu, X.Y.; Lam, R.; Giljohann, D.; Ho, D.; Mirkin, C.A. Strategy for increasing drug solubility and efficacy through covalent attachment to polyvalent DNA—Nanoparticle conjugates. ACS Nano 2011, 5, 6962–6970. [Google Scholar]
- Barisci, J.N.; Tahhan, M.; Wallace, G.G.; Badaire, S.; Vaugien, T.; Maugey, M.; Poulin, P. Properties of carbon nanotube fibers spun from DNA-stabilized dispersions. Adv. Funct. Mater. 2004, 14, 133–138. [Google Scholar]
- Badaire, S.; Zakri, C.; Maugey, M.; Derre, A.; Barisci, J.N.; Wallace, G.; Poulin, P. Liquid crystals of DNA-stabilized carbon nanotubes. Adv. Mater. 2005, 17, 1673–1676. [Google Scholar]
- He, P.G.; Bayachou, M. Layer-by-layer fabrication and characterization of DNA-wrapped single-walled carbon nanotube particles. Langmuir 2005, 21, 6086–6092. [Google Scholar]
- Hobbie, E.K.; Bauer, B.J.; Stephens, J.; Becker, M.L.; McGuiggan, P.; Hudson, S.D.; Wang, H. Colloidal particles coated and stabilized by DNA-wrapped carbon nanotubes. Langmuir 2005, 21, 10284–10287. [Google Scholar]
- Hu, C.G.; Zhang, Y.Y.; Bao, G.; Zhang, Y.L.; Liu, M.L.; Wang, Z.L. DNA functionalized single-walled carbon nanotubes for electrochemical detection. J. Phys. Chem. B 2005, 109, 20072–20076. [Google Scholar]
- Yan, W.; Shen, X.C.; Zhang, Z.L.; Chen, C.; Pang, D.W. Electrochemical behavior of daunorubicin at DNA-MWCNT bioconjugates modified glassy carbon electrodes. Anal. Lett. 2005, 38, 2579–2595. [Google Scholar]
- Chen, R.J.; Zhang, Y.G. Controlled precipitation of solubilized carbon nanotubes by delamination of DNA. J. Phys. Chem. B 2006, 110, 54–57. [Google Scholar]
- Fagan, J.A.; Landi, B.J.; Mandelbaum, I.; Simpson, J.R.; Bajpai, V.; Bauer, B.J.; Migler, K.; Walker, A.R.H.; Raffaelle, R.; Hobbie, E.K. Comparative measures of single-wall carbon nanotube dispersion. J. Phys. Chem. B 2006, 110, 23801–23805. [Google Scholar]
- Gigliotti, B.; Sakizzie, B.; Bethune, D.S.; Shelby, R.M.; Cha, J.N. Sequence-independent helical wrapping of singles-walled carbon nanotubes by long genomic DNA. Nano Lett. 2006, 6, 159–164. [Google Scholar]
- Gladchenko, G.O.; Karachevtsev, M.V.; Leontiev, V.S.; Valeev, V.A.; Glamazda, A.Y.; Plokhotnichenko, A.M.; Stepanian, S.G. Interaction of fragmented double-stranded DNA with carbon nanotubes in aqueous solution. Mol. Phys. 2006, 104, 3193–3201. [Google Scholar]
- Ikeda, A.; Hamano, T.; Hayashi, K.; Kikuchi, J. Water-solubilization of nucleotides-coated single-walled carbon nanotubes using a high-speed vibration milling technique. Org. Lett. 2006, 8, 1153–1156. [Google Scholar]
- Ishibashi, A.; Yamaguchi, Y.; Murakami, H.; Nakashima, N. Layer-by-layer assembly of RNA/single-walled carbon nanotube nanocomposites. Chem. Phys. Lett. 2006, 419, 574–577. [Google Scholar]
- Onoa, B.; Zheng, M.; Dresselhaus, M.S.; Diner, B.A. Carbon nanotubes and nucleic acids: Tools and targets. Phys. Status Solidi A 2006, 203, 1124–1131. [Google Scholar]
- Zhao, W.; Gao, Y.; Brook, M.A.; Li, Y.F. Wrapping single-walled carbon nanotubes with long single-stranded DNA molecules produced by rolling circle amplification. Chem. Commun. 2006, 3582–3584. [Google Scholar]
- Bertoncini, P.; Gresil, M.; Lardoux, J.; Riou, I.; Chauvet, O. Morphology of DNA/single walled nanotubes complexes. Dig. J. Nanomater. Biostruct. 2007, 2, 293–297. [Google Scholar]
- Cathcart, H.; Quinn, S.; Nicolosi, V.; Kelly, J.M.; Blau, W.J.; Coleman, J.N. Spontaneous debundling of single-walled carbon nanotubes in DNA-based dispersions. J. Phys. Chem. C 2007, 111, 66–74. [Google Scholar]
- Han, X.G.; Li, Y.L.; Deng, Z.X. DNA-wrapped single-walled carbon nanotubes as rigid templates for assembling linear gold nanoparticle arrays. Adv. Mater. 2007, 19, 1518–1522. [Google Scholar]
- Hopkins, A.R.; Kruk, N.A.; Lipeles, R.A. Macroscopic alignment of single-walled carbon nanotubes (SWNTs). Surf. Coat. Technol. 2007, 202, 1282–1286. [Google Scholar]
- Jeng, E.S.; Barone, P.W.; Nelson, J.D.; Strano, M.S. Hybridization kinetics and thermodynamics of DNA adsorbed to individually dispersed single-walled carbon nanotubes. Small 2007, 3, 1602–1609. [Google Scholar]
- Jin, H.; Jeng, E.S.; Heller, D.A.; Jena, P.V.; Kirmse, R.; Langowski, J.; Strano, M.S. Divalent ion and thermally induced DNA conformational polymorphism on single-walled carbon nanotubes. Macromolecules 2007, 40, 6731–6739. [Google Scholar]
- Vogel, S.R.; Kappes, M.M.; Hennrich, F.; Richert, C. An unexpected new optimum in the structure space of DNA solubilizing single-walled carbon nanotubes. Chemistry 2007, 13, 1815–1820. [Google Scholar]
- Vogel, S.R.; Muller, K.; Plutowski, U.; Kappes, M.M.; Richert, C. DNA-carbon nanotube interactions and nanostructuring based on DNA. Phys. Status Solidi B 2007, 244, 4026–4029. [Google Scholar]
- Haggenmueller, R.; Rahatekar, S.S.; Fagan, J.A.; Chun, J.H.; Becker, M.L.; Naik, R.R.; Krauss, T.; Carlson, L.; Kadla, J.F.; Trulove, P.C.; et al. Comparison of the quality of aqueous dispersions of single wall carbon nanotubes using surfactants and biomolecules. Langmuir 2008, 24, 5070–5078. [Google Scholar]
- Noguchi, Y.; Fujigaya, T.; Niidome, Y.; Nakashima, N. Single-walled carbon nanotubes/DNA hybrids in water are highly stable. Chem. Phys. Lett. 2008, 455, 249–251. [Google Scholar]
- Cooper, L.; Amano, H.; Hiraide, M.; Houkyou, S.; Jang, I.Y.; Kim, Y.J.; Muramatsu, H.; Kim, J.H.; Hayashi, T.; Kim, Y.A.; et al. Freestanding, bendable thin film for supercapacitors using DNA-dispersed double walled carbon nanotubes. Appl. Phys. Lett. 2009. [Google Scholar] [CrossRef]
- Ghosh, S.; Dutta, S.; Gomes, E.; Carroll, D.; D’Agostino, R.; Olson, J.; Guthold, M.; Gmeiner, W.H. Increased heating efficiency and selective thermal ablation of malignant tissue with DNA-encased multiwalled carbon nanotubes. ACS Nano 2009, 3, 2667–2673. [Google Scholar]
- Jovanovic, S.P.; Markovic, Z.M.; Kleut, D.N.; Romcevic, N.Z.; Trajkovic, V.S.; Dramicanin, M.D.; Markovic, B.M.T. A novel method for the functionalization of gamma-irradiated single wall carbon nanotubes with DNA. Nanotechnology 2009, 20. [Google Scholar] [CrossRef]
- Pease, L.F.; Tsai, D.H.; Fagan, J.A.; Bauer, B.J.; Zangmeister, R.A.; Tarlov, M.J.; Zachariah, M.R. Length distribution of single-walled carbon nanotubes in aqueous suspension measured by electrospray differential mobility analysis. Small 2009, 5, 2894–2901. [Google Scholar]
- Asada, Y.; Miyata, Y.; Ohno, Y.; Kitaura, R.; Sugai, T.; Mizutani, T.; Shinohara, H. High-performance thin-film transistors with DNA-assisted solution processing of isolated single-walled carbon nanotubes. Adv. Mater. 2010, 22, 2698–2701. [Google Scholar]
- Hughes, J.M.; Cathcart, H.; Coleman, J.N. Dispersion and exfoliation of nanotubes with synthetic oligonucleotides: Variation of dispersion efficiency and oligo-nanotube interaction with base type. J. Phys. Chem. C 2010, 114, 11741–11747. [Google Scholar]
- Li, Z.H.; Yang, W. Capture and manipulation of hybrid DNAs by carbon nanotube bundles. Nanotechnology 2010, 21. [Google Scholar] [CrossRef]
- Wang, R.R.; Sun, J.; Gao, L.A.; Zhang, J. Dispersion of single-walled carbon nanotubes by DNA for preparing transparent conductive films. J. Mater. Chem. 2010, 20, 6903–6909. [Google Scholar]
- Yamamoto, Y.; Fujigaya, T.; Niidome, Y.; Nakashima, N. Fundamental properties of oligo double-stranded DNA/single-walled carbon nanotube nanobiohybrids. Nanoscale 2010, 2, 1767–1772. [Google Scholar]
- Ao, G.Y.; Nepal, D.; Aono, M.; Davis, V.A. Cholesteric and nematic liquid crystalline phase behavior of double-stranded DNA stabilized single-walled carbon nanotube dispersions. ACS Nano 2011, 5, 1450–1458. [Google Scholar]
- Sanz, V.; Borowiak, E.; Lukanov, P.; Galibert, A.M.; Flahaut, E.; Coley, H.M.; Silva, S.R.P.; McFadden, J. Optimising DNA binding to carbon nanotubes by non-covalent methods. Carbon 2011, 49, 1775–1781. [Google Scholar] [Green Version]
- Chhikara, B.S.; Misra, S.K.; Bhattacharya, S. Cnt loading into cationic cholesterol suspensions show improved DNA binding and serum stability and ability to internalize into cancer cells. Nanotechnology 2012, 23. [Google Scholar] [CrossRef]
- Wang, Y.W.; Liu, H.; Wang, F.; Gao, Y.M. Electrochemical oxidation behavior of methotrexate at DNA/SWCNT/nafion composite film-modified glassy carbon electrode. J. Solid State Electrochem. 2012, 16, 3227–3235. [Google Scholar]
- Primo, E.N.; Canete-Rosales, P.; Bollo, S.; Rubianes, M.D.; Rivas, G.A. Dispersion of bamboo type multi-wall carbon nanotubes in calf-thymus double stranded DNA. Colloid Surf. B 2013, 108, 329–336. [Google Scholar]
- Qiu, X.Y.; Khripin, C.Y.; Ke, F.Y.; Howell, S.C.; Zheng, M. Electrostatically driven interactions between hybrid DNA-carbon nanotubes. Phys. Rev. Lett. 2013, 111. [Google Scholar] [CrossRef]
- Schmucker, W.; Klumpp, S.; Hennrich, F.; Kappes, M.; Wagenknecht, H.A. A simple pyrene “click”-type modification of DNA affects solubilisation and photoluminescence of single-walled carbon nanotubes. RSC Adv. 2013, 3, 6331–6333. [Google Scholar]
- Tardani, F.; Strobbia, P.; Scipioni, A.; La Mesa, C. Encapsulating carbon nanotubes in aqueous ds-DNA anisotropic phases: Shear orientation and rheological properties. RSC Adv. 2013, 3, 25917–25923. [Google Scholar]
- Albertorio, F.; Hughes, M.E.; Golovchenko, J.A.; Branton, D. Base dependent DNA-carbon nanotube interactions: Activation enthalpies and assembly-disassembly control. Nanotechnology 2009, 20. [Google Scholar] [CrossRef]
- Noguchi, Y.; Fujigaya, T.; Niidome, Y.; Nakashima, N. Regulation of the near-IR spectral properties of individually dissolved single-walled carbon nanotubes in aqueous solutions of dsDNA. Chemistry 2008, 14, 5966–5973. [Google Scholar]
- Ao, G.Y.; Khripin, C.Y.; Zheng, M. DNA-controlled partition of carbon nanotubes in polymer aqueous two-phase systems. J. Am. Chem. Soc. 2014, 136, 10383–10392. [Google Scholar]
- Iliafar, S.; Mittal, J.; Vezenov, D.; Jagota, A. Interaction of single-stranded DNA with curved carbon nanotube is much stronger than with flat graphite. J. Am. Chem. Soc. 2014, 136, 12947–12957. [Google Scholar]
- Vetcher, A.A.; Srinivasan, S.; Vetcher, I.A.; Abramov, S.M.; Kozlov, M.; Baughman, R.H.; Levene, S.D. Fractionation of SWNT/nucleic acid complexes by agarose gel electrophoresis. Nanotechnology 2006, 17, 4263–4269. [Google Scholar]
- Bauer, B.J.; Becker, M.L.; Bajpai, V.; Fagan, J.A.; Hobbie, E.K.; Migler, K.; Guttman, C.M.; Blair, W.R. Measurement of single-wall nanotube dispersion by size exclusion chromatography. J. Phys. Chem. C 2007, 111, 17914–17918. [Google Scholar]
- Sanchez-Pomales, G.; Santiago-Rodriguez, L.; Rivera-Velez, N.E.; Cabrera, C.R. Characterization of the DNA-assisted purification of single-walled carbon nanotubes. Phys. Status Solidi A 2007, 204, 1791–1796. [Google Scholar]
- Bauer, B.J.; Fagan, J.A.; Hobbie, E.K.; Chun, J.; Bajpai, V. Chromatographic fractionation of SWNT/DNA dispersions with on-line multi-angle light scattering. J. Phys. Chem. C 2008, 112, 1842–1850. [Google Scholar]
- Chun, J.; Fagan, J.A.; Hobbie, E.K.; Bauer, B.J. Size separation of single-wall carbon nanotubes by flow-field flow fractionation. Anal. Chem. 2008, 80, 2514–2523. [Google Scholar]
- Kim, J.H.; Kataoka, M.; Kim, Y.A.; Shimamoto, D.; Muramatsu, H.; Hayashi, T.; Endo, M.; Terrones, M.; Dresselhaus, M.S. Diameter-selective separation of double-walled carbon nanotubes. Appl. Phys. Lett. 2008, 93. [Google Scholar] [CrossRef]
- Asada, Y.; Sugai, T.; Kitaura, R.; Shinohara, H. Chromatographic length separation and photoluminescence study on DNA-wrapped single-wall and double-wall carbon nanotubes. J. Nanomater. 2009, 2009. [Google Scholar] [CrossRef]
- Tu, X.M.; Manohar, S.; Jagota, A.; Zheng, M. DNA sequence motifs for structure-specific recognition and separation of carbon nanotubes. Nature 2009, 460, 250–253. [Google Scholar]
- Shakhmaeva, I.I.; Bulatov, E.R.; Bondar, O.V.; Saifullina, D.V.; Culha, M.; Rizvanov, A.A.; Abdullin, T.I. Binding and purification of plasmid DNA using multi-layered carbon nanotubes. J. Biotechnol. 2011, 152, 102–107. [Google Scholar]
- Tardani, F.; la Mesa, C.; Poulin, P.; Maugey, M. Phase behavior of DNA-based dispersions containing carbon nanotubes: Effects of added polymers and ionic strength on excluded volume. J. Phys. Chem. C 2012, 116, 9888–9894. [Google Scholar]
- Glamazda, A.Y.; Dettlaff-Weglikowska, U.; Leontiev, V.S.; Mateichenko, P.V.; Karachevtsev, V.A. Raman spectroscopy and sem study of swnts in aqueous solution and films with surfactant or polymer surroundings. Fuller. Nanotub. Carbon Nanostruct. 2006, 14, 221–225. [Google Scholar]
- Jeng, E.S.; Moll, A.E.; Roy, A.C.; Gastala, J.B.; Strano, M.S. Detection of DNA hybridization using the near-infrared band-gap fluorescence of single-walled carbon nanotubes. Nano Lett. 2006, 6, 371–375. [Google Scholar]
- Karachevtsev, V.A.; Glamazda, A.Y.; Dettlaff-Weglikowska, U.; Leontiev, V.S.; Mateichenko, P.V.; Roth, S.; Rao, A.M. Spectroscopic and SEM studies of SWNTs: Polymer solutions and films. Carbon 2006, 44, 1292–1297. [Google Scholar]
- Lacerda, L.; Pastorin, G.; Wu, W.; Prato, M.; Bianco, A.; Kostarelos, K. Luminescence of functionalized carbon nanotubes as a tool to monitor bundle formation and dissociation in water: The effect of plasmid-DNA complexation. Adv. Funct. Mater. 2006, 16, 1839–1846. [Google Scholar]
- Asada, Y.; Dohi, H.; Kuwahara, S.; Sugai, T.; Kitaura, R.; Shinohara, H. Synthesis and spectroscopic characterization of salmon DNA-wrapped single-wall carbon nanotubes. Nano 2007, 2, 295–299. [Google Scholar]
- Karachevtsev, V.A.; Glamazda, A.Y.; Leontiev, V.S.; Lytvyn, O.S.; Dettlaff-Weglikowska, U. Glucose sensing based on nir fluorescence of DNA-wrapped single-walled carbon nanotubes. Chem. Phys. Lett. 2007, 435, 104–108. [Google Scholar]
- Tu, X.M.; Pehrsson, P.E.; Zhao, W. Redox reaction of DNA-encased HiPCO carbon nanotubes with hydrogen peroxide: A near infrared optical sensitivity and kinetics study. J. Phys. Chem. C 2007, 111, 17227–17231. [Google Scholar]
- Xu, Y.; Pehrsson, P.E.; Chen, L.W.; Zhang, R.; Zhao, W. Double-stranded DNA single-walled carbon nanotube hybrids for optical hydrogen peroxide and glucose sensing. J. Phys. Chem. C 2007, 111, 8638–8643. [Google Scholar]
- Cao, C.F.; Kim, J.H.; Yoon, D.; Hwang, E.S.; Kim, Y.J.; Baik, S. Optical detection of DNA hybridization using absorption spectra of single-walled carbon nanotubes. Mater. Chem. Phys. 2008, 112, 738–741. [Google Scholar]
- Cathcart, H.; Nicolosi, V.; Hughes, J.M.; Blau, W.J.; Kelly, J.M.; Quinn, S.J.; Coleman, J.N. Ordered DNA wrapping switches on luminescence in single-walled nanotube dispersions. J. Am. Chem. Soc. 2008, 130, 12734–12744. [Google Scholar]
- Qian, H.; Araujo, P.T.; Georgi, C.; Gokus, T.; Hartmann, N.; Green, A.A.; Jorio, A.; Hersam, M.C.; Novotny, L.; Hartschuh, A. Visualizing the local optical response of semiconducting carbon nanotubes to DNA-wrapping. Nano Lett. 2008, 8, 2706–2711. [Google Scholar]
- Yoon, D.; Cao, C.; Choi, J.B.; Kim, Y.J.; Baik, S. Optical characterization of DNA-wrapped single walled carbon nanotubes irradiated with ultraviolet light. J. Nanosci. Nanotechnol. 2008, 8, 5135–5138. [Google Scholar]
- Shoda, M.; Bandow, S.; Maruyama, Y.; Iijima, S. Probing interaction between ssDNA and carbon nanotubes by raman scattering and electron microscopy. J. Phys. Chem. C 2009, 113, 6033–6036. [Google Scholar]
- Simpson, J.R.; Fagan, J.A.; Becker, M.L.; Hobbie, E.K.; Walker, A.R.H. The effect of dispersant on defects in length-separated single-wall carbon nanotubes measured by raman spectroscopy. Carbon 2009, 47, 3238–3241. [Google Scholar]
- Chen, Z.; Liu, R.; Wang, Y.L.; Zhu, H.R.; Sun, Z.P.; Zuo, T.S.; Chang, X.L.; Zhao, F.; Xing, G.M.; Yuan, H.; et al. Ag nanoparticles coated swcnt with surface enhanced raman scattering (SERS) signals. J. Nanosci. Nanotechnol. 2010, 10, 8538–8543. [Google Scholar]
- Park, J.J.; Fagan, J.A.; Huh, J.Y.; Migler, K.B.; Karim, A.; Raghavan, D. Spr imaging study of DNA wrapped single wall carbon nanotube (ssDNA-SWCNT) adsorption on a model biological (collagen) substrate. Soft Matter 2010, 6, 5581–5588. [Google Scholar]
- Ignatova, T.; Najafov, H.; Ryasnyanskiy, A.; Biaggio, I.; Zheng, M.; Rotkin, S.V. Significant fret between swnt/DNA and rare earth ions: A signature of their spatial correlations. ACS Nano 2011, 5, 6052–6059. [Google Scholar]
- Shao, J.Y.; Sun, T.; Guo, Q.Y.; Li, H.; Lan, S. In situ electrochemically tuned photoluminescence of [Ru(bpy)2(dppz)]2+ aggregates with single-walled carbon nanotubes and DNA monitored by guanine oxidation. Transit. Met. Chem. 2011, 36, 499–504. [Google Scholar]
- Ensafi, A.A.; Amini, M.; Rezaei, B. Biosensor based on ds-DNA decorated chitosan modified multiwall carbon nanotubes for voltammetric biodetection of herbicide amitrole. Colloid Surf. B 2013, 109, 45–51. [Google Scholar]
- Nii, D.; Hayashida, T.; Umemura, K. Controlling the adsorption and desorption of double-stranded DNA on functionalized carbon nanotube surface. Colloid Surf. B 2013, 106, 234–239. [Google Scholar]
- Ozturk, S.A.; Kelestemur, S.; Culha, M. Influence of oligonucleotide interaction on electronic properties of single walled carbon nanotubes. J. Raman Spectrosc. 2013, 44, 183–189. [Google Scholar]
- Ito, M.; Kobayashi, T.; Ito, Y.; Hayashida, T.; Nii, D.; Umemura, K.; Homma, Y. Intense photoluminescence from dried double-stranded DNA and single-walled carbon nanotube hybrid. Appl. Phys. Lett. 2014, 104. [Google Scholar] [CrossRef]
- Takahashi, H.; Numao, S.; Bandow, S.; Iijima, S. Afm imaging of wrapped multiwall carbon nanotube in DNA. Chem. Phys. Lett. 2006, 418, 535–539. [Google Scholar]
- Campbell, J.F.; Tessmer, I.; Thorp, H.H.; Erie, D.A. Atomic force microscopy studies of DNA-wrapped carbon nanotube structure and binding to quantum dots. J. Am. Chem. Soc. 2008, 130, 10648–10655. [Google Scholar]
- Toita, S.; Kang, D.; Kobayashi, K.; Kawamoto, H.; Kojima, K.; Tachibana, A. Atomic force microscopic study on DNA-wrapping for different diameter single-wall carbon nanotubes. Diam. Relat. Mater. 2008, 17, 1389–1393. [Google Scholar]
- Xu, Y.Q.; Barnard, A.; McEuen, P.L. Bending and twisting of suspended single-walled carbon nanotubes in solution. Nano Lett. 2009, 9, 1609–1614. [Google Scholar]
- Arnett, C.M.; Marsh, C.P.; Welch, C.R.; Strano, M.S.; Han, J.H.; Gray, J.H.; Carlson, T.A. Enzyme-mediated assimilation of DNA-functionalized single-walled carbon nanotubes. Langmuir 2010, 26, 613–617. [Google Scholar]
- Hayashida, T.; Kawashima, T.; Nii, D.; Ozasa, K.; Umemura, K. Kelvin probe force microscopy of single-walled carbon nanotubes modified with DNA or poly(ethylene glycol). Chem. Lett. 2013, 42, 666–668. [Google Scholar]
- Hayashida, T.; Umemura, K. Surface morphology of hybrids of double-stranded DNA and single-walled carbon nanotubes studied by atomic force microscopy. Colloid Surf. B 2013, 101, 49–54. [Google Scholar]
- Nii, D.; Hayashida, T.; Yamaguchi, Y.; Ikawa, S.; Shibata, T.; Umemura, K. Selective binding of single-stranded DNA-binding proteins onto DNA molecules adsorbed on single-walled carbon nanotubes. Colloid Surf. B 2014, 121, 325–330. [Google Scholar]
- Iijima, M.; Watabe, T.; Ishii, S.; Koshio, A.; Yamaguchi, T.; Bandow, S.; Iijima, S.; Suzuki, K.; Maruyama, Y. Fabrication and STM-characterization of novel hybrid materials of DNA/carbon nanotube. Chem. Phys. Lett. 2005, 414, 520–524. [Google Scholar]
- Karachevtsev, M.V.; Lytvyn, O.S.; Stepanian, S.G.; Leontiev, V.S.; Adamowicz, L.; Karachevtsev, V.A. SWNT-DNA and SWNT-polyC hybrids: AFM study and computer modeling. J. Nanosci. Nanotechnol. 2008, 8, 1473–1480. [Google Scholar]
- Lulevich, V.; Kim, S.; Grigoropoulos, C.P.; Noy, A. Frictionless sliding of single-stranded DNA in a carbon nanotube pore observed by single molecule force spectroscopy. Nano Lett. 2011, 11, 1171–1176. [Google Scholar]
- Rao, R.; Lee, J.; Lu, Q.; Keskar, G.; Freedman, K.O.; Floyd, W.C.; Rao, A.M.; Ke, P.C. Single-molecule fluorescence microscopy and raman spectroscopy studies of RNA bound carbon nanotubes. Appl. Phys. Lett. 2004, 85, 4228–4230. [Google Scholar]
- Yang, R.H.; Jin, J.Y.; Chen, Y.; Shao, N.; Kang, H.Z.; Xiao, Z.; Tang, Z.W.; Wu, Y.R.; Zhu, Z.; Tan, W.H. Carbon nanotube-quenched fluorescent oligonucleotides: Probes that fluoresce upon hybridization. J. Am. Chem. Soc. 2008, 130, 8351–8358. [Google Scholar]
- Zhang, L.B.; Tao, L.; Li, B.L.; Jing, L.; Wang, E.K. Carbon nanotube-DNA hybrid fluorescent sensor for sensitive and selective detection of mercury(II) ion. Chem. Commun. 2010, 46, 1476–1478. [Google Scholar]
- Zhao, C.; Qu, K.G.; Song, Y.J.; Xu, C.; Ren, J.S.; Qu, X.G. A reusable DNA single-walled carbon-nanotube-based fluorescent sensor for highly sensitive and selective detection of Ag+ and cysteine in aqueous solutions. Chemistry 2010, 16, 8147–8154. [Google Scholar]
- D’Souza, F.; Das, S.K.; Zandler, M.E.; Sandanayaka, A.S.D.; Ito, O. Bionano donor-acceptor hybrids of porphyrin, ssDNA, and semiconductive single-wall carbon nanotubes for electron transfer via porphyrin excitation. J. Am. Chem. Soc. 2011, 133, 19922–19930. [Google Scholar]
- Qin, W.L.; Yang, K.Q.; Tang, H.; Tan, L.A.; Xie, Q.J.; Ma, M.; Zhang, Y.Y.; Yao, S.Z. Improved GFP gene transfection mediated by polyamidoamine dendrimer-functionalized multi-walled carbon nanotubes with high biocompatibility. Colloid Surf. B 2011, 84, 206–213. [Google Scholar]
- Zhang, J.Q.; Boghossian, A.A.; Barone, P.W.; Rwei, A.; Kim, J.H.; Lin, D.H.; Heller, D.A.; Hilmer, A.J.; Nair, N.; Reuel, N.F.; et al. Single molecule detection of nitric oxide enabled by d(AT)(15) DNA adsorbed to near infrared fluorescent single-walled carbon nanotubes. J. Am. Chem. Soc. 2011, 133, 567–581. [Google Scholar]
- Martinez, M.T.; Tseng, Y.C.; Gonzalez, M.; Bokor, J. Streptavidin as cnts and DNA linker for the specific electronic and optical detection of DNA hybridization. J. Phys. Chem. C 2012, 116, 22579–22586. [Google Scholar]
- Wang, L.Y.; Cheng, Y.Q.; Wang, H.; Li, Z.P. A homogeneous fluorescence sensing platform with water-soluble carbon nanoparticles for detection of microrna and nuclease activity. Analyst 2012, 137, 3667–3672. [Google Scholar]
- Bergler, F.F.; Schoppler, F.; Brunecker, F.K.; Hailman, M.; Hertel, T. Fluorescence spectroscopy of gel-immobilized single-wall carbon nanotubes with microfluidic control of the surfactant environment. J. Phys. Chem. C 2013, 117, 13318–13323. [Google Scholar]
- Judkins, J.; Lee, H.H.; Tung, S.; Kim, J.W. Diffusion of single-walled carbon nanotube under physiological conditions. J. Biomed. Nanotechnol. 2013, 9, 1065–1070. [Google Scholar]
- Hazani, M.; Naaman, R.; Hennrich, F.; Kappes, M.M. Confocal fluorescence imaging of DNA-functionalized carbon nanotubes. Nano Lett. 2003, 3, 153–155. [Google Scholar]
- Stevens, J.L.; Huang, A.Y.; Peng, H.Q.; Chiang, L.W.; Khabashesku, V.N.; Margrave, J.L. Sidewall amino-functionalization of single-walled carbon nanotubes through fluorination and subsequent reactions with terminal diamines. Nano Lett. 2003, 3, 331–336. [Google Scholar]
- Jung, D.H.; Kim, B.H.; Ko, Y.K.; Jung, M.S.; Jung, S.; Lee, S.Y.; Jung, H.T. Covalent attachment and hybridization of DNA oligonucleotides on patterned single-walled carbon nanotube films. Langmuir 2004, 20, 8886–8891. [Google Scholar]
- Singh, K.V.; Pandey, R.R.; Wang, X.; Lake, R.; Ozkan, C.S.; Wang, K.; Ozkan, M. Covalent functionalization of single walled carbon nanotubes with peptide nucleic acid: Nanocomponents for molecular level electronics. Carbon 2006, 44, 1730–1739. [Google Scholar]
- Wang, X.; Liu, F.; Andavan, G.T.S.; Jing, X.Y.; Singh, K.; Yazdanpanah, V.R.; Bruque, N.; Pandey, R.R.; Lake, R.; Ozkan, M.; et al. Carbon nanotube-DNA nanoarchitectures and electronic functionality. Small 2006, 2, 1356–1365. [Google Scholar]
- Yuan, W.K.; Wu, H.; Jiang, Z.Y.; Xu, S.W. Covalent functionalization of carbon nanotubes. Chin. J. Org. Chem. 2006, 26, 1508–1517. [Google Scholar]
- Yang, W.R.; Moghaddam, M.J.; Taylor, S.; Bojarski, B.; Wieczorek, L.; Herrmann, J.; McCall, M.J. Single-walled carbon nanotubes with DNA recognition. Chem. Phys. Lett. 2007, 443, 169–172. [Google Scholar]
- Moghaddam, M.J.; Yang, W.R.; Bojarski, B.; Gengenbach, T.R.; Gao, M.; Zareie, H.; McCall, M.J. Azide photochemistry for facile modification of graphitic surfaces: Preparation of DNA-coated carbon nanotubes for biosensing. Nanotechnology 2012. [Google Scholar] [CrossRef]
- Alidori, S.; Asqiriba, K.; Londero, P.; Bergkvist, M.; Leona, M.; Scheinberg, D.A.; McDevitt, M.R. Deploying rna and DNA with functionalized carbon nanotubes. J. Phys. Chem. C 2013, 117, 5982–5992. [Google Scholar]
- Dolash, B.D.; Lahiji, R.R.; Zemlyanov, D.Y.; Drachev, V.P.; Reifenberger, R.; Bergstrom, D.E. Sonication mediated covalent cross-linking of DNA to single-walled carbon nanotubes. Chem. Phys. 2013, 413, 11–19. [Google Scholar]
- Singh, P.; Menard-Moyon, C.; Battigelli, A.; Toma, F.M.; Raya, J.; Kumar, J.; Nidamanuri, N.; Verma, S.; Bianco, A. Double functionalization of carbon nanotubes with purine and pyrimidine derivatives. Chemistry 2013, 8, 1472–1481. [Google Scholar]
- Canete-Rosales, P.; Gonzalez, M.; Anson, A.; Martinez, M.T.; Yanez, C.; Bollo, S. Electrochemical characterization of oligonucleotide-carbon nanotube functionalized using different strategies. Electrochim. Acta 2014, 140, 489–496. [Google Scholar]
- Kaufmann, A.; Kunhardt, D.; Cirillo, G.; Hampel, S.; Schwenzer, B. Functionalized carbon nanotubes as transporters for antisense oligodeoxynucleotides. J. Mater. Chem. B 2014, 2, 7000–7008. [Google Scholar]
- Gao, H.J.; Kong, Y.; Cui, D.X.; Ozkan, C.S. Spontaneous insertion of DNA oligonucleotides into carbon nanotubes. Nano Lett. 2003, 3, 471–473. [Google Scholar]
- Okada, T.; Kaneko, T.; Hatakeyama, R. Single-stranded DNA insertion into single-walled carbon nanotubes by ion irradiation in an electrolyte plasma. Jpn. J. Appl. Phys. Part 1 Regul. Pap. Brief Commun. Rev. Pap. 2006, 45, 8335–8339. [Google Scholar]
- Okada, T.; Kaneko, T.; Hatakeyama, R.; Tohji, K. Electrically triggered insertion of single-stranded DNA into single-walled carbon nanotubes. Chem. Phys. Lett. 2006, 417, 288–292. [Google Scholar]
- Kaneko, T.; Okada, T.; Hatakeyama, R. DNA encapsulation inside carbon nanotubes using micro electrolyte plasmas. Contrib. Plasma Phys. 2007, 47, 57–63. [Google Scholar]
- Pei, Q.X.; Lim, C.G.; Cheng, Y.; Gao, H.J. Molecular dynamics study on DNA oligonucleotide translocation through carbon nanotubes. J. Chem. Phys. 2008, 129. [Google Scholar] [CrossRef]
- Zimmerli, U.; Koumoutsakos, P. Simulations of electrophoretic rna transport through transmembrane carbon nanotubes. Biophys. J. 2008, 94, 2546–2557. [Google Scholar]
- Kamiya, K.; Okada, S. Energetics and electronic structure of encapsulated single-stranded DNA in carbon nanotubes. Phys. Rev. B 2011, 83. [Google Scholar] [CrossRef]
- Enyashin, A.N.; Gemming, S.; Seifert, G. DNA-wrapped carbon nanotubes. Nanotechnology 2007, 18. [Google Scholar] [CrossRef]
- Frischknecht, A.L.; Martin, M.G. Simulation of the adsorption of nucleotide monophosphates on carbon nanotubes in aqueous solution. J. Phys. Chem. C 2008, 112, 6271–6278. [Google Scholar]
- Zheng, M.; Eom, K.; Ke, C.H. Calculations of the resonant response of carbon nanotubes to binding of DNA. J. Phys. D 2009, 42, 8. [Google Scholar]
- Bobadilla, A.D.; Seminario, J.M. DNA-CNT interactions and gating mechanism using MD and DFT. J. Phys. Chem. C 2011, 115, 3466–3474. [Google Scholar]
- Ding, Z.J.; Jiao, Y.; Meng, S. Quantum simulation of molecular interaction and dynamics at surfaces. Front. Phys. 2011, 6, 294–308. [Google Scholar]
- Makarucha, A.J.; Todorova, N.; Yarovsky, I. Nanomaterials in biological environment: A review of computer modelling studies. Eur. Biophys. J. Biophys. Lett. 2011, 40, 103–115. [Google Scholar]
- Roxbury, D.; Jagota, A.; Mittal, J. Sequence-specific self-stitching motif of short single-stranded DNA on a single-walled carbon nanotube. J. Am. Chem. Soc. 2011, 133, 13545–13550. [Google Scholar]
- Umadevi, D.; Sastry, G.N. Quantum mechanical study of physisorption of nucleobases on carbon materials: Graphene versus carbon nanotubes. J. Phys. Chem. Lett. 2011, 2, 1572–1576. [Google Scholar]
- Xiao, Z.T.; Wang, X.; Xu, X.; Zhang, H.; Li, Y.; Wang, Y.H. Base- and structure-dependent DNA dinucleotide-carbon nanotube interactions: Molecular dynamics simulations and thermodynamic analysis. J. Phys. Chem. C 2011, 115, 21546–21558. [Google Scholar]
- Akdim, B.; Pachter, R.; Day, P.N.; Kim, S.S.; Naik, R.R. On modeling biomolecular-surface nonbonded interactions: Application to nucleobase adsorption on single-wall carbon nanotube surfaces. Nanotechnology 2012, 23. [Google Scholar] [CrossRef]
- Alegret, N.; Santos, E.; Rodriguez-Fortea, A.; Rius, F.X.; Poblet, J.M. Disruption of small double stranded DNA molecules on carbon nanotubes: A molecular dynamics study. Chem. Phys. Lett. 2012, 525–526, 120–124. [Google Scholar]
- Roxbury, D.; Mittal, J.; Jagota, A. Molecular-basis of single-walled carbon nanotube recognition by single-stranded DNA. Nano Lett. 2012, 12, 1464–1469. [Google Scholar]
- Santosh, M.; Panigrahi, S.; Bhattacharyya, D.; Sood, A.K.; Maiti, P.K. Unzipping and binding of small interfering RNA with single walled carbon nanotube: A platform for small interfering RNA delivery. J. Chem. Phys. 2012, 136. [Google Scholar] [CrossRef]
- Yamazaki, T.; Fenniri, H. Imaging carbon nanotube interaction with nucleobases in water using the statistical mechanical theory of molecular liquids. J. Phys. Chem. C 2012, 116, 15087–15092. [Google Scholar]
- Amirani, M.C.; Tang, T.; Cuervo, J. Quantum mechanical treatment of binding energy between DNA nucleobases and carbon nanotube: A DFT analysis. Physica E 2013, 54, 65–71. [Google Scholar]
- Clavier, A.; Kraszewski, S.; Ramseyer, C.; Picaud, F. Insertion kinetics of small nucleotides through single walled carbon nanotube. J. Biotechnol. 2013, 164, 13–18. [Google Scholar]
- Wu, N.; Wang, Q.; Pang, S.S. Dispersion of a bundle of carbon nanotubes by mechanical torsional energy. Carbon 2013, 59, 229–236. [Google Scholar]
- Ostojic, G.N.; Ireland, J.R.; Hersam, M.C. Noncovalent functionalization of DNA-wrapped single-walled carbon nanotubes with platinum-based DNA cross-linkers. Langmuir 2008, 24, 9784–9789. [Google Scholar]
- Zhang, L.; Huang, C.Z.; Li, Y.F.; Xiao, S.J.; Xie, J.P. Label-free detection of sequence-specific DNA with multiwalled carbon nanotubes and their light scattering signals. J. Phys. Chem. B 2008, 112, 7120–7122. [Google Scholar]
- Zhou, Z.P.; Kang, H.; Clarke, M.L.; Lacerda, S.H.D.; Zhao, M.H.; Fagan, J.A.; Shapiro, A.; Nguyen, T.; Hwang, J. Water-soluble DNA-wrapped single-walled carbon-nanotuble/quantum-dot complexes. Small 2009, 5, 2149–2155. [Google Scholar]
- Campbell, J.F.; Napier, M.E.; Feldberg, S.W.; Thorp, H.H. Metal-mediated electrochemical oxidation of DNA-wrapped carbon nanotubes. J. Phys. Chem. B 2010, 114, 8861–8870. [Google Scholar]
- Weizmann, Y.; Chenoweth, D.M.; Swager, T.M. Addressable terminally linked DNA-CNT nanowires. J. Am. Chem. Soc. 2010, 132, 14009–14011. [Google Scholar]
- Aravind, S.S.J.; Ramaprabhu, S. Noble metal dispersed multiwalled carbon nanotubes immobilized ss-DNA for selective detection of dopamine. Sens. Actuator B 2011, 155, 679–686. [Google Scholar]
- Asada, Y.; Miyata, Y.; Shiozawa, K.; Ohno, Y.; Kitaura, R.; Mizutani, T.; Shinohara, H. Thin-film transistors with length-sorted DNA-wrapped single-wall carbon nanotubes. J. Phys. Chem. C 2011, 115, 270–273. [Google Scholar]
- Cao, S.R.; Wang, J.F.; Yuan, R.; Chai, Y.Q. An amperometric immunoassay for human chorionic gonadotrophin based on DNA-multiwalled carbon nanotubes and methylene blue as sensing interface. Sens. Lett. 2011, 9, 1636–1642. [Google Scholar]
- Fujigaya, T.; Yamamoto, Y.; Kano, A.; Maruyama, A.; Nakashima, N. Enhanced cell uptake via non-covalent decollation of a single-walled carbon nanotube-DNA hybrid with polyethylene glycol-grafted poly(l-lysine) labeled with an alexa-dye and its efficient uptake in a cancer cell. Nanoscale 2011, 3, 4352–4358. [Google Scholar]
- Guo, Q.Y.; Shao, J.Y.; Sun, T.; Li, H.; Lan, S.; Xu, Z.H. Electrochemical fabrication and potential-enhanced luminescence of [Ru(bpy)2tatp]2+ incorporating DNA-stabilized single-wall carbon nanotubes on an indium tin oxide electrode. Electrochim. Acta 2011, 56, 1432–1438. [Google Scholar]
- Shi, J.; Cha, T.G.; Claussen, J.C.; Diggs, A.R.; Choi, J.H.; Porterfield, D.M. Microbiosensors based on DNA modified single-walled carbon nanotube and Pt black nanocomposites. Analyst 2011, 136, 4916–4924. [Google Scholar]
- Han, S.P.; Maune, H.T.; Barish, R.D.; Bockrath, M.; Goddard, W.A. DNA-linker-induced surface assembly of ultra dense parallel single walled carbon nanotube arrays. Nano Lett. 2012, 12, 1129–1135. [Google Scholar]
- Kawaguchi, M.; Yamazaki, J.; Ohno, J.; Fukushima, T. Preparation and binding study of a complex made of DNA-treated single-walled carbon nanotubes and antibody for specific delivery of a “molecular heater” platform. Int. J. Nanomed. 2012, 7, 4363–4371. [Google Scholar]
- Shin, S.R.; Lee, C.K.; Eom, T.W.; Lee, S.H.; Kwon, C.H.; So, I.; Kim, S.J. DNA-coated MWNT microfibers for electrochemical actuator. Sens. Actuator B 2012, 162, 173–177. [Google Scholar]
- Thuy, N.T.; Tam, P.D.; Tuan, M.A.; Le, A.T.; Tam, L.T.; Thu, V.V.; Hieu, N.V.; Chien, N.D. Detection of pathogenic microorganisms using biosensor based on multi-walled carbon nanotubes dispersed in DNA solution. Curr. Appl. Phys. 2012, 12, 1553–1560. [Google Scholar]
- Yu, T.; Gong, Y.X.; Lu, T.T.; Wei, L.; Li, Y.Q.; Mu, Y.G.; Chen, Y.; Liao, K. Recognition of carbon nanotube chirality by phage display. RSC Adv. 2012, 2, 1466–1476. [Google Scholar]
- Zhang, L.Y.; Guo, C.X.; Cui, Z.M.; Guo, J.; Dong, Z.L.; Li, C.M. DNA-directed growth of Pd nanocrystals on carbon nanotubes towards efficient oxygen reduction reactions. Chemistry 2012, 18, 15693–15698. [Google Scholar]
- Gong, J.L.; Sarkar, T.; Badhulika, S.; Mulchandani, A. Label-free chemiresistive biosensor for mercury (II) based on single-walled carbon nanotubes and structure-switching DNA. Appl. Phys. Lett. 2013, 102. [Google Scholar] [CrossRef]
- Mangalum, A.; Rahman, M.; Norton, M.L. Site-specific immobilization of single-walled carbon nanotubes onto single and one-dimensional DNA origami. J. Am. Chem. Soc. 2013, 135, 2451–2454. [Google Scholar]
- Sha, J.J.; Hasan, T.; Milana, S.; Bertulli, C.; Bell, N.A.W.; Privitera, G.; Ni, Z.H.; Chen, Y.F.; Bonaccorso, F.; Ferrari, A.C.; et al. Nanotubes complexed with DNA and proteins for resistive-pulse sensing. ACS Nano 2013, 7, 8857–8869. [Google Scholar]
- Su, H.C.; Zhang, M.L.; Bosze, W.; Lim, J.H.; Myung, N.V. Metal nanoparticles and DNA co-functionalized single-walled carbon nanotube gas sensors. Nanotechnology 2013, 24. [Google Scholar] [CrossRef]
- Gong, X.; Sharma, A.K.; Strano, M.S.; Mukhopadhyay, D. Selective assembly of DNA-conjugated single-walled carbon nanotubes from the vascular secretome. ACS Nano 2014, 8, 9126–9136. [Google Scholar]
- Guo, C.X.; Chitre, A.A.; Lu, X.M. DNA-assisted assembly of carbon nanotubes and MnO2 nanospheres as electrodes for high-performance asymmetric supercapacitors. Phys. Chem. Chem. Phys. 2014, 16, 4672–4678. [Google Scholar]
- Tardani, F.; Pucci, C.; La Mesa, C. Confining ss-DNA/carbon nanotube complexes in ordered droplets. Soft Matter 2014, 10, 1024–1031. [Google Scholar]
- Tardani, F.; Sennato, S. Phase behavior of DNA-stabilized carbon nanotubes dispersions: Association with oppositely-charged additives. J. Phys. Chem. C 2014, 118, 9268–9274. [Google Scholar]
- Williams, R.M.; Nayeem, S.; Dolash, B.D.; Sooter, L.J. The effect of DNA-dispersed single-walled carbon nanotubes on the polymerase chain reaction. PLoS One 2014, 9. [Google Scholar] [CrossRef]
- Gowtham, S.; Scheicher, R.H.; Ahuja, R.; Pandey, R.; Karna, S.P. Physisorption of nucleobases on graphene: Density-functional calculations. Phys. Rev. B 2007, 76. [Google Scholar] [CrossRef]
- Varghese, N.; Mogera, U.; Govindaraj, A.; Das, A.; Maiti, P.K.; Sood, A.K.; Rao, C.N.R. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem 2009, 10, 206–210. [Google Scholar]
- He, S.J.; Song, B.; Li, D.; Zhu, C.F.; Qi, W.P.; Wen, Y.Q.; Wang, L.H.; Song, S.P.; Fang, H.P.; Fan, C.H. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar]
- Liu, F.; Choi, J.Y.; Seo, T.S. DNA mediated water-dispersible graphene fabrication and gold nanoparticle-graphene hybrid. Chem. Commun. 2010, 46, 2844–2846. [Google Scholar]
- Liu, J.B.; Li, Y.L.; Li, Y.M.; Li, J.H.; Deng, Z.X. Noncovalent DNA decorations of graphene oxide and reduced graphene oxide toward water-soluble metal-carbon hybrid nanostructures via self-assembly. J. Mater. Chem. 2010, 20, 900–906. [Google Scholar]
- Lv, W.; Guo, M.; Liang, M.H.; Jin, F.M.; Cui, L.; Zhi, L.J.; Yang, Q.H. Graphene-DNA hybrids: Self-assembly and electrochemical detection performance. J. Mater. Chem. 2010, 20, 6668–6673. [Google Scholar]
- Zhang, Q.A.; Qiao, Y.; Hao, F.; Zhang, L.; Wu, S.Y.; Li, Y.; Li, J.H.; Song, X.M. Fabrication of a biocompatible and conductive platform based on a single-stranded DNA/graphene nanocomposite for direct electrochemistry and electrocatalysis. Chemistry 2010, 16, 8133–8139. [Google Scholar]
- Tang, L.H.; Wang, Y.; Liu, Y.; Li, J.H. DNA-directed self-assembly of graphene oxide with applications to ultrasensitive oligonucleotide assay. ACS Nano 2011, 5, 3817–3822. [Google Scholar]
- Wei, L.L.; Borowiec, J.; Zhu, L.H.; Zhang, J.D. Electrochemical sensor for monitoring the photodegradation of catechol based on DNA-modified graphene oxide. Microchim. Acta 2011, 173, 439–443. [Google Scholar]
- Wu, M.; Kempaiah, R.; Huang, P.J.J.; Maheshwari, V.; Liu, J.W. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir 2011, 27, 2731–2738. [Google Scholar]
- Zhao, X.C. Self-assembly of DNA segments on graphene and carbon nanotube arrays in aqueous solution: A molecular simulation study. J. Phys. Chem. C 2011, 115, 6181–6189. [Google Scholar]
- Kabelac, M.; Kroutil, O.; Predota, M.; Lankas, F.; Sip, M. Influence of a charged graphene surface on the orientation and conformation of covalently attached oligonucleotides: A molecular dynamics study. Phys. Chem. Chem. Phys. 2012, 14, 4217–4229. [Google Scholar]
- Le, D.; Kara, A.; Schroder, E.; Hyldgaard, P.; Rahman, T.S. Physisorption of nucleobases on graphene: A comparative van der waals study. J. Phys. Condes. Matter 2012, 24. [Google Scholar] [CrossRef]
- Premkumar, T.; Geckeler, K.E. Graphene-DNA hybrid materials: Assembly, applications, and prospects. Prog. Polym. Sci. 2012, 37, 515–529. [Google Scholar]
- Qu, K.G.; Wu, L.; Ren, J.S.; Qu, X.G. Natural DNA-modified graphene/pd nanoparticles as highly active catalyst for formic acid electro-oxidation and for the Suzuki reaction. ACS Appl. Mater. Interfaces 2012, 4, 5001–5009. [Google Scholar]
- Manna, A.K.; Pati, S.K. Theoretical understanding of single-stranded DNA assisted dispersion of graphene. J. Mat. Chem. B 2013, 1, 91–100. [Google Scholar]
- Zhang, X.; Gao, F.; Cai, X.L.; Zheng, M.X.; Gao, F.; Jiang, S.L.; Wang, Q.X. Application of graphene-pyrenebutyric acid nanocomposite as probe oligonucleotide immobilization platform in a DNA biosensor. Mater. Sci. Eng. C 2013, 33, 3851–3857. [Google Scholar]
- Joseph, D.; Seo, S.; Williams, D.R.; Geckeer, K.E. Double-stranded DNA-graphene hybrid: Preparation and anti-proliferative activity. ACS Appl. Mater. Interfaces 2014, 6, 3347–3356. [Google Scholar]
- Shekar, S.C.; Swathi, R.S. Stability of nucleobases and base pairs adsorbed on graphyne and graphdiyne. J. Phys. Chem. C 2014, 118, 4516–4528. [Google Scholar]
- Belyanskaya, L.; Weigel, S.; Hirsch, C.; Tobler, U.; Krug, H.F.; Wick, P. Effects of carbon nanotubes on primary neurons and glial cells. Neurotoxicology 2009, 30, 702–711. [Google Scholar]
- Deng, X.Y.; Wu, F.; Liu, Z.; Luo, M.; Li, L.; Ni, Q.S.; Jiao, Z.; Wu, M.H.; Liu, Y.F. The splenic toxicity of water soluble multi-walled carbon nanotubes in mice. Carbon 2009, 47, 1421–1428. [Google Scholar]
- Folkmann, J.K.; Risom, L.; Jacobsen, N.R.; Wallin, H.; Loft, S.; Moller, P. Oxidatively damaged DNA in rats exposed by oral gavage to C60 fullerenes and single-walled carbon nanotubes. Environ. Health Perspect. 2009, 117, 703–708. [Google Scholar]
- Antonelli, A.; Serafini, S.; Menotta, M.; Sfara, C.; Pierige, F.; Giorgi, L.; Ambrosi, G.; Rossi, L.; Magnani, M. Improved cellular uptake of functionalized single-walled carbon nanotubes. Nanotechnology 2010, 21. [Google Scholar] [CrossRef]
- Asakura, M.; Sasaki, T.; Sugiyama, T.; Takaya, M.; Koda, S.; Nagano, K.; Arito, H.; Fukushima, S. Genotoxicity and cytotoxicity of multi-wall carbon nanotubes in cultured chinese hamster lung cells in comparison with chrysotile a fibers. J. Occup. Health 2010, 52, 155–166. [Google Scholar]
- Cveticanin, J.; Joksic, G.; Leskovac, A.; Petrovic, S.; Sobot, A.V.; Neskovic, O. Using carbon nanotubes to induce micronuclei and double strand breaks of the DNA in human cells. Nanotechnology 2010, 21. [Google Scholar] [CrossRef]
- Lima, A.; Melo, A.M.D.; Pires, E.V.; Ferreira, R.C.D.; Sant’Ana, A.E.G.; Goulart, M.O.F.; de Abreu, F.C. Electroanalytical studies of sulfentrazone in protic medium, its degradation by the electro-fenton process, and toxicity assessment using ss-DNA. Chemosphere 2010, 81, 884–889. [Google Scholar]
- Luszczyn, J.; Plonska-Brzezinska, M.E.; Palkar, A.; Dubis, A.T.; Simionescu, A.; Simionescu, D.T.; Kalska-Szostko, B.; Winkler, K.; Echegoyen, L. Small noncytotoxic carbon nano-onions: First covalent functionalization with biomolecules. Chemistry 2010, 16, 4870–4880. [Google Scholar]
- Raffa, V.; Ciofani, G.; Vittorio, O.; Riggio, C.; Cuschieri, A. Physicochemical properties affecting cellular uptake of carbon nanotubes. Nanomedicine 2010, 5, 89–97. [Google Scholar]
- Sabuncu, A.C.; Kalluri, B.S.; Qian, S.Z.; Stacey, M.W.; Beskok, A. Dispersion state and toxicity of mwCNTs in cell culture medium with different T80 concentrations. Colloid Surf. B 2010, 78, 36–43. [Google Scholar]
- Shen, C.X.; Zhang, Q.F.; Li, J.A.; Bi, F.C.; Yao, N. Induction of programmed cell death in arabidopsis and rice by single-wall carbon nanotubes. Am. J. Bot. 2010, 97, 1602–1609. [Google Scholar]
- Nam, C.W.; Kang, S.J.; Kang, Y.K.; Kwak, M.K. Cell growth inhibition and apoptosis by sds-solubilized single-walled carbon nanotubes in normal rat kidney epithelial cells. Arch. Pharm. Res. 2011, 34, 661–669. [Google Scholar]
- Dong, P.X.; Wan, B.; Guo, L.H. In vitro toxicity of acid-functionalized single-walled carbon nanotubes: Effects on murine macrophages and gene expression profiling. Nanotoxicology 2012, 6, 288–303. [Google Scholar]
- Handy, R.D.; van den Brink, N.; Chappell, M.; Muhling, M.; Behra, R.; Dusinska, M.; Simpson, P.; Ahtiainen, J.; Jha, A.N.; Seiter, J.; et al. Practical considerations for conducting ecotoxicity test methods with manufactured nanomaterials: What have we learnt so far? Ecotoxicology 2012, 21, 933–972. [Google Scholar]
- Kim, J.S.; Sung, J.H.; Song, K.S.; Lee, J.H.; Kim, S.M.; Lee, G.H.; Ahn, K.H.; Lee, J.S.; Shin, J.H.; Park, J.D.; et al. Persistent DNA damage measured by comet assay of sprague dawley rat lung cells after five days of inhalation exposure and 1 month post-exposure to dispersed multi-wall carbon nanotubes (MWCNTs) generated by new MWCNT aerosol generation system. Toxicol. Sci. 2012, 128, 439–448. [Google Scholar]
- Mogurampelly, S.; Panigrahi, S.; Bhattacharyya, D.; Sood, A.K.; Maiti, P.K. Unraveling siRNA unzipping kinetics with graphene. J. Chem. Phys. 2012, 137. [Google Scholar] [CrossRef]
- Van Berlo, D.; Clift, M.; Albrecht, C.; Schins, R. Carbon nanotubes: An insight into the mechanisms of their potential genotoxicity. Swiss Med. Wkly. 2012, 142. [Google Scholar] [CrossRef]
- Awasthi, K.K.; John, P.J.; Awasthi, A.; Awasthi, K. Multi walled carbon nano tubes induced hepatotoxicity in Swiss albino mice. Micron 2013, 44, 359–364. [Google Scholar]
- Mao, H.L.; Kawazoe, N.; Chen, G.P. Uptake and intracellular distribution of collagen-functionalized single-walled carbon nanotubes. Biomaterials 2013, 34, 2472–2479. [Google Scholar]
- Zhang, Y.L.; Deng, J.J.; Zhang, Y.X.; Guo, F.; Li, C.G.; Zou, Z.; Xi, W.; Tang, J.; Sun, Y.; Yang, P.; et al. Functionalized single-walled carbon nanotubes cause reversible acute lung injury and induce fibrosis in mice. J. Mol. Med. 2013, 91, 117–128. [Google Scholar]
- Chen, W.; Xiong, Q.; Ren, Q.X.; Guo, Y.K.; Li, G. Can amino-functionalized carbon nanotubes carry functional nerve growth factor? Neural Regen. Res. 2014, 9, 285–292. [Google Scholar]
- Clichici, S.; Biris, A.R.; Catoi, C.; Filip, A.; Tabaran, F. Short-term splenic impact of single-strand DNA functionalized multi-walled carbon nanotubes intraperitoneally injected in rats. J. Appl. Toxicol. 2014, 34, 332–344. [Google Scholar]
- Hu, C.W.; Zhang, L.J.; Wang, W.L.; Cui, Y.B.; Li, M. Evaluation of the combined toxicity of multi-walled carbon nanotubes and sodium pentachlorophenate on the earthworm Eisenia fetida using avoidance bioassay and comet assay. Soil Biol. Biochem. 2014, 70, 123–130. [Google Scholar]
- Liu, Z.B.; Liu, Y.F.; Peng, D.M. Hydroxylation of multi-walled carbon nanotubes reduces their cytotoxicity by limiting the activation of mitochondrial mediated apoptotic pathway. J. Mater. Sci. Mater. Med. 2014, 25, 1033–1044. [Google Scholar]
- McShan, D.; Yu, H.T. DNA damage in human skin keratinocytes caused by multiwalled carbon nanotubes with carboxylate functionalization. Toxicol. Ind. Health 2014, 30, 489–498. [Google Scholar]
- Tavares, A.M.; Louro, H.; Antunes, S.; Quarre, S.; Simar, S.; de Temmerman, P.J.; Verleysen, E.; Mast, J.; Jensen, K.A.; Norppa, H.; et al. Genotoxicity evaluation of nanosized titanium dioxide, synthetic amorphous silica and multi-walled carbon nanotubes in human lymphocytes. Toxicol. Vitro 2014, 28, 60–69. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umemura, K. Hybrids of Nucleic Acids and Carbon Nanotubes for Nanobiotechnology. Nanomaterials 2015, 5, 321-350. https://doi.org/10.3390/nano5010321
Umemura K. Hybrids of Nucleic Acids and Carbon Nanotubes for Nanobiotechnology. Nanomaterials. 2015; 5(1):321-350. https://doi.org/10.3390/nano5010321
Chicago/Turabian StyleUmemura, Kazuo. 2015. "Hybrids of Nucleic Acids and Carbon Nanotubes for Nanobiotechnology" Nanomaterials 5, no. 1: 321-350. https://doi.org/10.3390/nano5010321




